Back to Journals » OncoTargets and Therapy » Volume 7
Oncogene mutational profile in nasopharyngeal carcinoma
Authors Zhang Z, Fu S, Wang F, Wang H, Zeng Y, Shao J
Received 6 December 2013
Accepted for publication 25 January 2014
Published 20 March 2014 Volume 2014:7 Pages 457—467
DOI https://doi.org/10.2147/OTT.S58791
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Zi-Chen Zhang,1,* Sha Fu,1,* Fang Wang,1 Hai-Yun Wang,1 Yi-Xin Zeng,2 Jian-Yong Shao1
1Department of Molecular Diagnostics, 2Department of Experimental Research, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center of Cancer Medicine, Guangzhou, People's Republic of China
*These authors contributed equally to this work
Abstract: Nasopharyngeal carcinoma (NPC) is a common tumor in Southern China, but the oncogene mutational status of NPC patients has not been clarified. Using time-of-flight mass spectrometry, 238 mutation hotspots in 19 oncogenes were examined in 123 NPC patients. The relationships between mutational status and clinical data were assessed with a χ2 or Fisher's exact test. Survival analysis was performed using the Kaplan–Meier method with the log-rank test. In 123 patients, 21 (17.1%) NPC tumors were positive for mutations in eight oncogenes: six patients had PIK3CA mutations (4.9%), five NRAS mutations (4.1%), four KIT mutations (3.3%), two PDGFRA mutations (1.6%), two ABL mutations (1.6%), and one with simultaneous mutations in HRAS, EGFR, and BRAF (1%). Patients with mutations were more likely to relapse or develop metastasis than those with wild-type alleles (P=0.019). No differences or correlations were found in other clinical characteristics or in patient survival. No mutations were detected in oncogenes AKT1, AKT2, CDK, ERBB2, FGFR1, FGFR3, FLT3, JAK2, KRAS, MET, and RET. These results demonstrate an association between NPC and mutations in NRAS, KIT, PIK3CA, PDGFRA, and ABL, which are associated with patient relapse and metastasis.
Keywords: NPC, oncogene, mutation
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor that originates in the upper lining epithelium of the human retronasal cavity.1 It is generally rare, but is more common in certain geographic regions, such as Southeast Asia, North Africa, and especially Southern China.2 The annual incidence of NPC is up to 30 per 100,000 in Guangdong (a province in Southern China), which is 50-fold higher than that in the Western world.3 There is a clear relationship between NPC and this specific region, with convincing evidence of a correlation with Epstein–Barr virus (EBV) infection.4 Although this tumor is sensitive to radiotherapy, therapy can fail in patients with advanced stage disease, as the disease is highly invasive and metastatic in nature.5,6
Because of the significant correlation between NPC and EBV, most studies of NPC have focused on EBV-related proteins and genes, such as EBER (EBV-encoded RNA) and LMP (EBV-associated membrane antigen).7,8 Gene linkage studies have also been conducted.9 Moreover, several chromosome regions, such as 3p21.3-1-21.2, and the human leukocyte antigen (HLA) haplotypes have been linked to the development of NPC.10–12 Previous studies have also investigated the expression of other genes, such as TP53 in NPC.13–16 C-KIT and PIK3CA mutations have been detected in NPC cell lines and NPC specimens.17,18 However, few studies have examined the genomic mutations of NPC.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) can detect multiple gene mutations with high sensitivity and accuracy. Using this technology, Patrick et al showed that RAS mutations are more frequent in cutaneous squamous cell tumor patients treated with RAF inhibitors than in those not so treated.19 Kang et al also detected EGFR T790M mutations in patients with non-small-cell lung cancer using MALDI-TOF MS, which detected and quantified the mutations highly sensitively.20 In this study, a panel of 19 oncogenes including EGFR, RAS family genes, KIT, and PIK3CA, were analyzed for 238 possible mutations in tumor tissues from 123 NPC patients. The purpose of this study was to analyze the mutational status of multiple genes in NPC samples and clarify the possible relationships between these mutations and the characteristics of NPC patients.
Materials and methods
Clinical samples
Formalin-fixed paraffin-embedded samples were obtained from 123 patients with pathologically diagnosed NPC between October 1991 and July 2002 at Sun Yat-sen University Cancer Center (SYSUCC) (Guangzhou, People’s Republic of China). Informed consent and clinicopathological information were obtained from all patients. Disease stage was classified or reclassified according to the People’s Republic of China 1992 NPC TNM staging system.21 The clinicopathological characteristics of the 123 NPC patients are summarized in Table 1. Institute Research Medical Ethics Committee of SYSUCC granted approval for this study.
DNA extraction
We chose paraffin blocks containing more than 60% tumor cells from hematoxylin and eosin stained sections of each tumor. Sections (4–6 μm) were cut and transferred to 1.5 mL Eppendorf tubes for DNA extraction. DNA was extracted using the QIAamp DNA Formalin-fixed Paraffin-embedded Tissue Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. The quantity and quality of the isolated DNA were tested using a Nanodrop ND-2000 Spectrophotometer (Thermo Scientific, Niederelbert, Germany). The final DNA samples were diluted to 10 ng/μL for analysis.
OncoCarta assay
A total of 238 possible mutations in 19 oncogenes were investigated in 123 NPC samples using the OncoCarta Panel (v 1.0; Sequenom Inc., San Diego, CA, USA). This panel is a set of predesigned and prevalidated assays for sensitive and efficient mutation screening by the parallel analysis of 238 possible mutations across the following 19 common oncogenes: ABL1, AKT1, AKT2, BRAF, CDK, EGFR, ERBB2, FGFR1, FGFR3, FLT3, HRAS, JAK2, KIT, KRAS, MET, NRAS, PDGFRA, PIK3CA, and RET. The mutation types of each gene are list in Table S1.
In brief, 20 ng of DNA was amplified using 24 sets of OncoCarta PCR primers. An extension reaction based on the OncoCarta extension primers was then performed. After a cation exchange resin was used to remove salts, the products were spotted onto a 384-well SpectroChipII using the MassARRAY Nanodispenser RS1000 (Sequenom Inc.) and analyzed on a MALDI-TOF mass spectrometer (Sequenom Inc.). We chose high performance liquid chromatography purified water as the blank control and normal human somatic cells as the negative control in each experiment.
Data analysis
Mutation data was analyzed using the software MassARRAY Typer (v4.0; Sequenom Inc.), using a cutoff mutation frequency of 1%. A successful experiment should show that the sample figure was typical and the blank control had no peak (Figure 1).
Statistical analysis
The statistical analysis was performed using SPSS software (v 16.0; SPSS Inc., Chicago, IL, USA). The relationships between the patients’ mutational status and clinical data were assessed with a χ2 or Fisher’s exact test. Kaplan–Meier analysis was used to compare differences in the survival rate of the groups. A P-value of less than 0.05 was considered statistically significant. A multivariate analysis was used to check the possible relationship between factors. Overall survival (OS) and progression-free survival (PFS) were calculated for the 123 NPC patients from the date of surgery until death or the last follow-up. The clinical data follow-up date was updated to October 2012.
Results
Mutation profiles of NPC tumors
Mutations were detected in 17.1% (21/123) of NPC tumors, with one NPC patient having two simultaneous mutations. Therefore, a total of 21 patients presented with 22 mutations. The mutations affected eight oncogenes, as follows: six in PIK3CA (three E542K, two H1047Y, and one R38H), five in NRAS (one G12D, two G13D, and two Q61K), four in KIT (two V559I, one V559A, and one D52N), two in PDGFRA (two T647I), and two in ABL (two E255K); HRAS (G13S) and EGFR (E709A) mutations were found in one NPC tumor each; one NPC tumor had a BRAF mutation (G464E) together with a PIK3CA mutation (E545K). No mutation was detected in the remaining 102 (82.9%) NPC tissues (with no mutations in AKT1, AKT2, CDK, ERBB2, FGFR1, FGFR3, FLT3, JAK2, KRAS, MET, or RET). The mutational profiles and distributions of the NPC tumors are shown in Table 2.
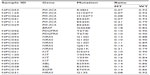  | Table 2 Mutation status of the 21 positive cases |
Correlations between oncogene mutations and patient clinicopathological characteristics
We divided the patients into groups according to their clinical characteristics, and calculated the mutation rate in each group. We then assessed the relationships between the mutational status and clinical data using the χ2 or Fisher’s exact test. The results are presented in Table 3. There was an association between oncogene mutations and relapse/metastasis of NPC (P=0.019, Table 3). We also tested the intersubject effects with a univariate analysis and found no effect between all factors (Table S2). No significant correlation between the presence of an oncogenic mutation and other clinicopathological parameters, such as age, sex, clinical stage, or WHO histological grade was found (P>0.05, Table 3).
Correlation between oncogene mutation and patient survival
We estimated the survival of NPC patients by comparing the OS and PFS of the mutation and wild-type subgroups (Figure 2). The mean survival time (MST) in the mutation subgroup did not differ significantly from that of the wild-type subgroup (OS: 86.5 vs 90.0, P>0.05, Figure 2A; PFS: 83.6 vs 88.6, P>0.05, Figure 2B).
Because the PIK3CA, KIT, and NRAS oncogenes had higher mutation frequencies of the oncogenes analyzed, we also assessed the correlation between the clinicopathological characteristics of the patients and the presence of these mutations, but found no significant correlation (Table S3).
Discussion
Various aspects of NPC have been widely investigated because it is an important cancer of the head and neck. However, few studies have examined the role of mutations in NPC, and the results of such studies are controversial. In this study, our data show that PIK3CA, KIT, and RAS are the oncogenes most susceptible to mutations in NPC, whereas mutations of BRAF, PDGFRA, ABL1, and EGFR occur less frequently. Many of the mutations described here have never been previously reported in NPC samples. A summary of oncogenes’ mutations in NPC in our study and other literature is shown in Table 4.17,18,22–33
In a comparison of the mutation frequencies in the subgroups of patients with and without relapse or metastasis, we found that NPC patients who relapsed or developed metastases had higher mutation frequencies (28.9% vs 11.8%, respectively, P=0.019).
The PIK3CA gene encodes the p110α catalytic subunit of PI3K and plays an important role in many tumors. Mutations of this gene are reportedly located in exons 9 and 20, with hotspots at E542K, E545K, and H1047Y.34 In our study, 83.3% (5/6) of all the PIK3CA mutations identified occurred at these hotspots. We found that NPC patient survival did not correlate significantly with the presence of PIK3CA mutations, which is consistent with a previous study (Figure S1).33 In NPC cell lines, the inhibitor NVP-BEZ235 was found to selectively inhibit the proliferation of NPC cells carrying PIK3CA mutations.35 Currently, mTOR inhibitors are used as therapies for cancers in which the PI3K/AKT/mTOR pathway is activated. Although mutation rate is not so high, PIK3CA is also worthy as a research object of targeted therapy in NPC.
It is well established that the RAS/RAF/ERK pathway plays an important role in tumor development. KRAS, HRAS, and NRAS mutations occur in at least one-third of all human cancers, with KRAS mutations being the most common.28,36,37 In the present study, we detected mutations of NRAS and HRAS, but not in KRAS. KRAS mutation rate in all tumors is estimated to be 25%–30%.38 But here, in NPC, KRAS mutation is particularly scarce. We detected NRAS mutations at Q61K, G13D, and G12D and a HRAS mutation at G13S, all of which are acknowledged hotspots. Consistent with our results, previous studies have detected no mutations in codons 12, 13, or 61 of KRAS in NPC specimens or NPC cell lines.28,33 These data suggest that RAS mutations exist in NPC, but that KRAS mutations are rare.
KIT is a type III receptor tyrosine kinase that initiates multiple downstream signaling pathways, such as the PI3K/AKT and JAK/STAT pathways. KIT gene mutations are mainly found in melanomas, and imatinib is an effective inhibitor of this oncogene. Here, we observed mutations V559I and V559A, which are the most common KIT mutation types. V559I is considered to confer resistance to imatinib, whereas V559A reportedly confers sensitivity to imatinib.39,40 PDGFRA belongs to the type III tyrosine kinase family. In fact, there appears to be a close relationship between KIT and PDGFRA, and the correlation between them has been widely investigated.41,42 PDGFRA mutation T674I confers imatinib resistance. In the present study, both mutations of KIT (3.3%) and PDGFR (1.6%) were detected in NPC tissues; this result is consistent with other reports.43,44 Further clinical trials are required to evaluate the correlation between NPC patients with KIT and PDGFR mutations and their response to the drug imatinib.
EGFR is a cell-surface protein that binds to EGF, and mutations in EGFR are associated with a wide variety of tumors. EGFR mutations are very frequent in non-small-cell lung cancer, with exons 18, 19, 20, and 21 being the predominantly mutated regions.45 In our study, only one NPC patient was positive for an EGFR mutation, resulting in a mutation rate of less than 1%. This mutation was E709A, which is encoded in exon 20 and usually reported in lung cancer.46,47 This result may suggest that the treatment of NPC patients with tyrosine kinase inhibitors may not be an effective strategy.
Detection of multiple mutations status in NPC was also one of our aims. It can provide more information about treatment and prognosis than single mutation detection. Herein, one NPC sample was detected to have two simultaneous mutations (PIK3CA and BRAF). Studies have suggested that the concurrent presence of PIK3CA and BRAF mutations predict resistance to everolimus.48,49
We should not ignore the negative results of this study. This report describes the analysis of 238 potential mutations in 19 oncogenes in 123 NPC samples to gain a preliminary understanding of mutational status of these 19 oncogenes. The negative results of this study indicate that mutations are rare in NPC, AKT1, AKT2, CDK, ERBB2, FGFR1, FGFR3, FLT3, JAK2, KRAS, MET, and RET, suggesting that drugs targeting these genes may be ineffective.
This study had several limitations. Compared with deep sequencing, MALDI-TOF MS offers high-throughput and is less expensive. However, the comprehensiveness of detection is inadequate. Many deserted or concealed mutations cannot be detected using this method, so we may miss some important genes in NPC.
Conclusion
In summary, a small number of mutations in NRAS, KIT, PIK3CA, PDGFRA, and ABL are present in NPC, whereas mutations in other genes, including AKT1, AKT2, CDK, ERBB2, FGFR1, FGFR3, FLT3, JAK2, KRAS, MET, and RET, are extremely rare. The presence of oncogene mutations in NPC patients is associated with relapse and metastasis.
Acknowledgments
This work was supported in part by grants from the National High Technology Research and Development Program of China (863 Program) (No 2012AA02A501); the Chinese State Key Basic Research Project (No 2011CB504805); the National Natural Science Fund (No 81272952); and a State Key Laboratory Grant to the Sun Yat-sen University Cancer Center.
Author contributions
JYS and YXZ conceived and designed the experiments; ZCZ and SF performed the experiments; ZCZ and FW analyzed the data; JYS contributed the reagents/materials; ZCZ and JYS wrote the paper; HYW acquired clinical samples and follow-up clinical information. All authors read and approved the final manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005; 365(9476):2041–2054. | |
McDermott AL, Dutt SN, Watkinson JC. The aetiology of nasopharyngeal carcinoma. Clin Otolaryngol Allied Sci. 2001;26(2):82–92. | |
Jia WH, Huang QH, Liao J, et al. Trends in incidence and mortality of nasopharyngeal carcinoma over a 20–25 year period (1978/1983–2002) in Sihui and Cangwu counties in southern China. BMC cancer. 2006;6:178. | |
Liu P, Fang X, Feng Z, et al. Direct sequencing and characterization of a clinical isolate of Epstein-Barr virus from nasopharyngeal carcinoma tissue by using next-generation sequencing technology. J Virol. 2011;85(21):11291–11299. | |
Chua DT, Ma J, Sham JS, et al. Long-term survival after cisplatin-based induction chemotherapy and radiotherapy for nasopharyngeal carcinoma: a pooled data analysis of two phase III trials. J Clin Oncol. 2005;23(6):1118–1124. | |
Ma BB, Chan AT. Recent perspectives in the role of chemotherapy in the management of advanced nasopharyngeal carcinoma. Cancer. 2005;103(1):22–31. | |
Yoshizaki T. A novel immune evasion mechanism of LMP-1, an EBV-primary oncogene, in nasopharyngeal carcinoma. Adv Otorhinolaryngol. 2011;72:157–159. | |
Takada K. Role of EBER and BARF1 in nasopharyngeal carcinoma (NPC) tumorigenesis. Semin Cancer Biol. 2012;22(2):162–165. | |
Zhou X, Cui J, Macias V, et al. The progress on genetic analysis of nasopharyngeal carcinoma. Comp Funct Genomics. 2007:57513. | |
Lu SJ, Day NE, Degos L, et al. Linkage of a nasopharyngeal carcinoma susceptibility locus to the HLA region. Nature. 1990;346(6283):470–471. | |
Xiong W, Zeng ZY, Xia JH, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64(6):1972–1974. | |
Bei JX, Li Y, Jia WH, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42(7):599–603. | |
Rubio L, Burgos JS, Lopez-Guerrero JA, Morera C, Vera-Sempere FJ. Expression of p53 protein and tumor angiogenesis as prognostic factors in nasopharyngeal carcinoma patients. Pathol Res Pract. 2002;198(2):97–102. | |
Ma BB, Poon TC, To KF, et al. Prognostic significance of tumor angiogenesis, Ki 67, p53 oncoprotein, epidermal growth factor receptor and HER2 receptor protein expression in undifferentiated nasopharyngeal carcinoma – a prospective study. Head Neck. 2003;25(10):864–872. | |
Chen MK, Lee HS, Chang JH, Chang CC. Expression of p53 protein and primary tumour volume in patients with nasopharyngeal carcinoma. J Otolaryngol. 2004;33(5):304–307. | |
Sun Y, Yi H, Yang Y, et al. Functional characterization of p53 in nasopharyngeal carcinoma by stable shRNA expression. Int J Oncol. 2009;34(4):1017–1027. | |
Liu P, Li DJ, Qin HD, Zhang RH, Chen LZ, Zeng YX. [Screening for mutations in the hotspot mutation regions of PIK3CA gene in nasopharyngeal carcinoma]. Ai Zheng. 2007;26(1):15–20. Chinese. | |
Huang PY, Hong MH, Zhang X, Mai HQ, Luo DH, Zhang L. C-KIT overexpression and mutation in nasopharyngeal carcinoma cell lines and reactivity of Imatinib on these cell lines. Chin J Cancer. 2010;29(2):131–135. | |
Oberholzer PA, Kee D, Dziunycz P, et al. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J Clin Oncol. 2012;30(3):316–321. | |
Su KY, Chen HY, Li KC, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30(4):433–440. | |
Wang C, Tran-Thanh D, Moreno JC, et al. Expression of Abl interactor 1 and its prognostic significance in breast cancer: a tissue-array-based investigation. Breast Cancer Res Treat. 2011;129(2):373–386. | |
Jiang Q, Mai C, Yang H, et al. Nuclear expression of CDK4 correlates with disease progression and poor prognosis in human nasopharyngeal carcinoma. Histopathology. Epub October 29, 2013. | |
Naji F, Attaleb M, Laantri N, et al. Identification of G2607A mutation in EGFR gene with a significative rate in Moroccan patients with nasopharyngeal carcinoma. Cell Mol Biol. 2010;56 Suppl:OL1442–OL1446. | |
Lee SC, Lim SG, Soo R, et al. Lack of somatic mutations in EGFR tyrosine kinase domain in hepatocellular and nasopharyngeal carcinoma. Pharmacogenet Genomics. 2006;16(1):73–74. | |
Zhang Y, Liang G, Sun G, Pan Z, Wu G, Liu Z. [Expression of C-erbB-2 and EGFR expression and its relationship with cell proliferation in nasopharyngeal carcinoma]. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2011;25(2):67–70. Chinese. | |
Wang SS, Guan ZZ, Xiang YQ, et al. [Significance of EGFR and p-ERK expression in nasopharyngeal carcinoma]. Zhonghua Zhong Liu Za Zhi. 2006;28(1):28–31. Chinese. | |
Yazici H, Altun M, Alatli C, Dogan O, Dalay N. c-erbB-2 gene amplification in nasopharyngeal carcinoma. Cancer Invest. 2000;18(1):6–10. | |
Ma BB, Lui VW, Poon FF, et al. Preclinical activity of gefitinib in non-keratinizing nasopharyngeal carcinoma cell lines and biomarkers of response. Invest New Drugs. 2010;28(3):326–333. | |
Anghel I, Anghel AG, Dumitru M, Soreanu CC. Nasopharyngeal carcinoma – analysis of risk factors and immunological markers. Chirurgia. 2012;107(5):640–645. | |
Li Z, Lin SX, Liang HZ, He JH. [Expression of hepatocyte growth factor/c-Met system in nasopharyngeal carcinoma and its biological significance]. Zhonghua Bing Li Xue Za Zhi. 2005;34(2):75–79. Chinese. | |
Fendri A, Khabir A, Mnejja W, et al. PIK3CA amplification is predictive of poor prognosis in Tunisian patients with nasopharyngeal carcinoma. Cancer Sci. 2009;100(11):2034–2039. | |
Ching-Shian Leong V, Jabal MF, Leong PP, Abdullah MA, Gul YA, Seow HF. PIK3CA gene mutations in breast carcinoma in Malaysian patients. Cancer Genet Cytogenet. 2008;187(2):74–79. | |
Chou CC, Chou MJ, Tzen CY. PIK3CA mutation occurs in nasopharyngeal carcinoma but does not significantly influence the disease-specific survival. Med Oncol. 2009;26(3):322–326. | |
Gustin JP, Cosgrove DP, Park BH. The PIK3CA gene as a mutated target for cancer therapy. Curr Cancer Drug Targets. 2008;8(8):733–740. | |
Yang F, Qian XJ, Qin W, et al. Dual phosphoinositide 3-kinase/mammalian target of rapamycin inhibitor NVP-BEZ235 has a therapeutic potential and sensitizes cisplatin in nasopharyngeal carcinoma. PLoS One. 2013;8(3):e59879. | |
Hui AB, Lo KW, Teo PM, To KF, Huang DP. Genome wide detection of oncogene amplifications in nasopharyngeal carcinoma by array based comparative genomic hybridization. Int J Oncol. 2002;20(3):467–473. | |
Kratz CP, Schubbert S, Bollag G, Niemeyer CM, Shannon KM, Zenker M. Germline mutations in components of the Ras signaling pathway in Noonan syndrome and related disorders. Cell Cycle. 2006;5(15):1607–1611. | |
Fernandez-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2(3):344–358. | |
Nakagomi N, Hirota S. Juxtamembrane-type c-kit gene mutation found in aggressive systemic mastocytosis induces imatinib-resistant constitutive KIT activation. Lab Invest. 2007;87(4):365–371. | |
Terheyden P, Houben R, Pajouh P, Thorns C, Zillikens D, Becker JC. Response to imatinib mesylate depends on the presence of the V559A-mutated KIT oncogene. J Invest Dermatol. 2010;130(1):314–316. | |
Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342–4349. | |
Heinrich MC, Maki RG, Corless CL, et al. Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J Clin Oncol. 2008;26(33):5352–5359. | |
Heinrich MC, Corless CL, Duensing A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299(5607):708–710. | |
Hirota S, Ohashi A, Nishida T, et al. Gain-of-function mutations of platelet-derived growth factor receptor alpha gene in gastrointestinal stromal tumors. Gastroenterology. 2003;125(3):660–667. | |
Sequist LV, Lynch TJ. EGFR tyrosine kinase inhibitors in lung cancer: an evolving story. Ann Rev Med. 2008;59:429–442. | |
Arcila ME, Nafa K, Chaft JE, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12(2):220–229. | |
Penzel R, Sers C, Chen Y, et al. EGFR mutation detection in NSCLC – assessment of diagnostic application and recommendations of the German Panel for Mutation Testing in NSCLC. Virchows Archiv. 2011;458(1):95–98. | |
Jaiswal BS, Janakiraman V, Kljavin NM, et al. Combined targeting of BRAF and CRAF or BRAF and PI3K effector pathways is required for efficacy in NRAS mutant tumors. PLoS One. 2009;4(5):e5717. | |
Mohseni M, Park BH. PIK3CA and KRAS mutations predict for response to everolimus therapy: now that’s RAD001. J Clin Invest. 2010;120(8):2655–2658. |
Supplementary materials
  | Table S1 Mutation subtypes detected by the OncoCarta Panel (v 1.0; Sequenom Inc., San Diego, CA, USA) MassARRAY |
  | Table S2 Tests of between-subjects effects |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







