Back to Journals » Neuropsychiatric Disease and Treatment » Volume 12
Olig1 expression pattern in neural cells during rat spinal cord development
Authors Qi Q, Zhang Y, Shen L, Wang R, Zhou J, Lü H, Hu J
Received 27 October 2015
Accepted for publication 16 February 2016
Published 18 April 2016 Volume 2016:12 Pages 909—916
DOI https://doi.org/10.2147/NDT.S99257
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Wai Kwong Tang
Qi Qi,1,2 Yuxin Zhang,1 Lin Shen,1 Rui Wang,1 Jiansheng Zhou,1 Hezuo Lü,1,3 Jianguo Hu1,3
1Anhui Key Laboratory of Tissue Transplantation, The First Affiliated Hospital of Bengbu Medical College, Bengbu, Anhui, People’s Republic of China; 2Department of Histology and Embryology, Bengbu Medical College, Bengbu, Anhui, People’s Republic of China; 3Department of Clinical Laboratory, The First Affiliated Hospital of Bengbu Medical College, Bengbu, Anhui, People’s Republic of China
Purpose: Our purpose was to systematically investigate the expression pattern and role of Olig1 in neural cells during rat spinal cord development.
Animals and methods: Spinal cord tissues were dissected from Sprague–Dawley rats at embryonic day 14.5 (E14.5) and E18.5, postnatal day 0 (P0), P3, P7, postnatal 2 weeks (P2W), P4W, and adults (more than 2 months after birth), respectively. The expression of Olig1 was determined by Western blot and immunostaining. To observe expression of Olig1 in different neural cell types, a double immunohistochemical staining was performed using antibodies against Olig1 with O4, ß-tubulin, glial fibrillary acidic protein (GFAP), and myelin basic protein, respectively.
Results: The expression of Olig1 protein shows a significant level change in rat spinal cord at different developmental time points. Starting with E14.5, the expression gradually increased and peaked at E18.5. Olig1 decreased gradually from P3 and reached its lowest level on P7. However, interestingly, the Olig1 expression increased again from P2W, until adulthood. Olig1 was coexpressed with O4-positive oligodendrocyte progenitor cells (OPCs) and β-tubulin-positive neurons at all time points during development. Olig1 was also coexpressed transiently with GFAP-positive astrocytes at only E14.5. Olig1 was localized in the cytoplasm of O4- and β-tubulin-positive cells during the period from E14.5 to adult.
Conclusion: The expression of Olig1 in OPCs and neurons at all time points during development and in astrocytes at E14.5 suggests that Olig1 may play an important role in the generation and maturation of specific neural cells during development of spinal cord. Our results contribute to understanding the mechanism underlying developmental regulation of neural cells by Olig1.
Keywords: Olig1, spinal cord, development, neural cells
Introduction
Neurons, oligodendrocytes (OLs), and astrocytes are unique cell types in the central nervous system (CNS) of vertebrates.1 Neurons process and transmit information electrochemically.2 OLs generate myelin sheaths around most axons of the vertebrate CNS, enabling a faster conduction of the nerve impulses. Astrocytes provide support and nutrients for the nerve tissue.3
The different cell types in the nervous system are regulated by the precise spatiotemporal expression of specific genes.4,5 Transcriptional regulation plays an important role in the determination of neural cell differentiation.6,7 Olig genes belong to the basic helix–loop–helix transcription factor family, which encode OL lineage transcription factors 1, 2, and 3 (Olig1, Olig2, and Olig3). With the exception of Olig3, Olig genes are specifically expressed in the CNS, and play a critical role in CNS development by controlling differentiation and maturation of OLs, motor neurons (MNs), and astrocytes.8,9 Olig2 null mice die at birth from a lack of MNs.10 Both gain- and loss-of-function studies were performed in an Olig1 null mouse with normal myelin during development, but which were unable to remyelinate on experimental challenge.11 A second Olig1 null mouse with less compensatory effect by Olig2 had a more severe phenotype and died around postnatal day 14 from a complete lack of myelin. This mutant had mature OLs, but failed to wrap myelin or even deposit lipid around axons.12 Knocking out Olig1 and Olig2 individually or together affected differentiation and maturation of OLs, suggesting functional overlap in the CNS.10,13,14 Until now, the role of Olig2 during development of spinal cord attracted more attention. However, studies investigating the expression and function of Olig1 in development and disease are limited.
Although it is widely known that Olig1 promotes the differentiation and maturation of OLs, it is unclear how these occur during development. Exploring temporal and spatial expression and distribution of Olig1 will contribute to our understanding of the role of Olig1 in specialization of neural cells during development. Therefore, in this study, we determined the expression pattern of Olig1 in neural cells during rat spinal cord development.
Animals and methods
Animals and tissue preparation
Sprague–Dawley rats were obtained from the Laboratory Animal Center, Bengbu Medical College (Bengbu, People’s Republic of China). All experimental protocols involving animals and their care were approved by the Ethics Committee of Laboratory Animal Services Center of Bengbu Medical College.
To produce embryonic and newborn rats, one female was cohabited with two males, and gestational age (embryo, E) was designated as day 0.5 (when vaginal plugs in female rat were observed). Pregnant rats were bred in separate cages. Eighty rats were randomized to eight groups and subgroups: embryonic day 14.5 (E14.5) (n=10), E18.5 (n=10), postnatal day 0 (P0) (n=10), P3 (n=10), P7 (n=10), postnatal 2 weeks (P2W) (n=10), P4W (n=10), and adults (n=10). Each group was randomized into two subgroups equally. In the first subgroup (n=5), the spinal cords were immunohistochemically stained, and in the second subgroup (n=5), the spinal cords were subjected to Western blot. Spinal cords from embryos (E14.5 and E18.5) were dissected following cervical dislocation of the pregnant rats. Spinal cords were dissected from postnatal rats (P0, P3, P7, P2W, P4W, and adults) and perfused intracardially with phosphate-buffered saline (PBS), followed by 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). For immunohistochemistry, spinal cord tissues were postfixed for 2 hours and immersed into a graded series of sucrose solution (Sigma, St Louis, MO, USA) at 4°C for another 5–7 days. Using a cryostat, transverse 15 μm thick sections were obtained, mounted onto gelatin-coated slides (Fisher Scientific, Fairlawn, NJ, USA), and stored at −80°C until use. For Western blot, spinal cord tissues were stored immediately at −80°C until use.
Immunohistochemistry
For immunohistochemistry staining, the frozen slides were air-dried at room temperature (RT) for 10 minutes and rinsed with PBS for 10 minutes. They were blocked with 10% donkey serum in Tris-buffered saline (TBS) containing 0.3% Triton X-100 (TBST) for 1 hour at RT. The primary antibody was used in the same blocking solution overnight at 4°C. The following primary antibodies were used: monoclonal mouse anti-Olig1 (IgG, 1:100, Millipore, Billerica, MA, USA), monoclonal rabbit anti-myelin basic protein (anti-MBP; 1:300, Millipore), rabbit anti-glial fibrillary acidic protein (anti-GFAP; 1:100, Millipore), monoclonal mouse anti-O4 (IgM, 1:100, Millipore), and rabbit anti-β-tubulin (1:100, Millipore). The next day, the slides were washed in PBS three times and incubated with rhodamine-conjugated goat anti-mouse IgM, IgG or goat anti-rabbit IgM, IgG (all 1:200; Jackson ImmunoResearch Lab, West Grove, PA, USA) for 1 hour at 37°C. The slides were washed three times with PBS and coverslipped with gel aqueous mounting media (Biomeda, Foster City, CA, USA) containing Hoechst 33342 (Sigma-Aldrich) to counterstain the nuclei. Images were captured with an Olympus IX71 microscope. At least five randomly selected fields with a total of more than 500 cells were counted. In all experiments, primary antibody omission controls were used to confirm the specificity of immunofluorescence labeling.
Western blot
Western blot was used to detect the expression of Olig1 in rat spinal cord tissues at different developmental stages. Briefly, tissue selected from the same segment of spinal cord was immediately transferred to a homogenizer containing 1 mL lysis buffer and 10 μL phenylmethanesulfonyl fluoride (PMSF) (10 mg/mL) and thoroughly homogenized for 5 minutes on ice, and then centrifuged at 12,000× g for 30 minutes at 4°C to collect the supernatant. The protein concentrations of the supernatant were determined using a BCA Protein Assay kit (Pierce, Rockford, IL, USA). The protein concentration was determined using the Bio-Rad DC protein assay (every 10 μL sample contain 120 μg protein). Protein samples containing an equal amount of protein (20 μg) were electrophoresed on Sodium dodecyl sulfate (SDS)-polyacrylamide gels, and transferred to polyvinylidene difluoride filters (Millipore). The filters were blocked with 5% nonfat dry milk in TBS for 1 hour at RT and then incubated overnight at 4°C with primary antibodies (in TBST-5% bovine serum albumin [BSA]) including Olig1 (1:200, Millipore) and β-actin (1:400, Sigma) as markers of differentiated neural cells. After rinsing with TBST, the membranes were incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (all from KPL, Gaithersburg, MD, USA) for 1 hour at RT. To visualize the immunoreactive proteins, the enhanced chemiluminescence (ECL) kit (Pierce, Rockford, IL, USA) was used, following the manufacturer’s instructions. Films were digitized, and densitometry was performed using Gel-Pro analyzer (Media Cybernetics, Silver Spring, MD, USA). The intensity of immunoreactive bands for Olig1 protein was normalized by the intensity of β-actin.
Statistical analyses
The data were analyzed using SPSS16.0 (IBM, Armonk, NY, USA) software. Data are presented as mean ± standard deviation of the mean. The data with two groups were analyzed by Student’s t-test. The data with three or more groups were analyzed by one-way analysis of variance (ANOVA) followed by post hoc Tukey’s t-test to determine whether there were significant differences between individual groups. A P-value of <0.05 was considered statistically significant.
Results
Olig1 expression in rat spinal cord development
Western blot was used to determine Olig1 expression in the rat spinal cord from E14.5 to adulthood. Our results showed a significant change in the expression of Olig1 protein in rat spinal cord at different developmental time points. The Olig1 expression started at E14.5, peaking at E18.5. However, its expression decreased gradually from P3 to its lowest level by P7. However, interestingly, the expression of Olig1 increased again from P2W to a high level until adulthood (Figure 1).
Olig1 expression and localization in neural cells
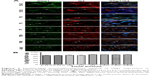
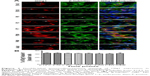
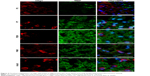
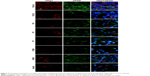
To determine the expression and localization of Olig1 in different types of neural cells, we performed double immunostaining using the antibody against Olig1 and another antibody against O4, MBP, β-tubulin, and GFAP, respectively. Our results showed that Olig1 was expressed in both O4-positive oligodendrocyte progenitor cells (OPCs; Figure 2) and β-tubulin-positive neurons (Figure 3) at all time points. Furthermore, we failed to observe expression of MBP in the spinal cord during the embryonic period (data not shown). MBP expression was not observed prior to P3, and no cellular coexpression of MBP and Olig1 was observed (Figure 4). Finally, as shown in Figure 5, GFAP expression was observed during development. The coexpression of GFAP and Olig1 was observed at E14.5 but absent from day E18.5 to adulthood. We also analyzed subcellular localization of Olig1 in neural cells and found that Olig1 was localized in the cytoplasm of coexpressed cells at all time points during rat spinal cord development.
Discussion
The CNS originates in the neuroepithelial stem cells (NSCs), which ultimately generate the three major cell types: neurons, OLs, and astrocytes.1 In this process, transcriptional regulation plays an important role.6,7 In mice, genetic studies revealed that Olig transcription factors play key roles in the genesis of MNs and OLs. Gain- and loss-of-function analysis confirmed the fundamental role of Olig1 in the survival and maturation of OPCs.12,15–17 Regulation of Olig1 gene expression in spinal cord of demyelinating disease promotes myelin repair after spinal cord injury. However, the distribution and expression of Olig1 in neural cells during development is unclear. In this study, we systematically investigated the expression pattern of Olig1 in neural cells during rat spinal cord development.
First, using the Western blot, we demonstrated the differential expression of Olig1 in rat spinal cord development. The expression of Olig1 protein was observed at E14.5. Olig1 expression increased and peaked at E18.5, which may contribute to the differentiation of a large number of neural cells into OPCs and MN precursors at this stage. However, its expression decreased gradually from P3 to its lowest level on P7. Recently, it has been reported that Olig1 protein expression was significantly decreased during mouse OL progenitor differentiation.18 However, the expression of Olig1 increased again from P2W, which was consistent with a recent report that Olig1 was expressed in human OLs during process outgrowth and maintained at a high level prior to myelination.19
Next, we identified the expression of Olig1 in neural cells at several time points during spinal cord development. We confirmed that Olig1 was expressed not only in OPCs but also in neurons from E14.5 to adulthood. CNS development starts from NSCs, which generate the three major cell types (neurons, OLs, and astrocytes) of the CNS.20,21 However, the intermediate steps in the differentiation of NSCs in vivo have not been clarified. In the embryonic ventral spinal cord, MNs and OLs are derived from a common progenitor pool, suggesting that these lineages share intrinsic and extrinsic regulatory mechanisms.10,13,21,22 OL progenitors were more closely aligned with neuronal subtype progenitors than with astrocytes, at least in the developing CNS.23 A study demonstrated that Olig2 was more likely an early-stage factor for OL precursor cells, while Olig1 played a critical role in late-stage OL maturation and myelin formation.19 However, Jakovcevski and Zecevic24 found Olig1 protein expressed in primitive neuroepithelia that can give rise to interneurons in the human fetal brain. A recent study suggests that Olig1 regulates neuron-glial fate choice in the embryonic telencephalon.25 In conclusion, Olig family members do not act independently but interact with other transcription factors to tightly control CNS development.17,26 Our findings raise the possibility that Olig1 has some function in the formation of MNs during CNS development.
MBP is a major myelin protein, which is expressed in mature OLs.27,28 MBP is the major protein component of myelin at 2 weeks after birth in the rat brain, expressed a week later than in spinal cord.28 In this study, we observed that MBP expression commenced from postnatal period, and Olig1 was not coexpressed with MBP during rat spinal cord development.
OPCs undergo two types of differentiation, resulting in OLs in the absence of serum or Type II astrocytes in the presence of serum in vitro.29–31 Fate-mapping studies have shown that OPCs generate a subset of astrocytes during perinatal development.32 Most neurogenic bHLH transcription factors, for example, mammalian achaete-scute homologue 1, mammalian atonal homologue, and neurogenins 1 and 2, are only expressed transiently in progenitor cells as needed.23 In this study, we observed the transient expression of Olig1 in GFAP-positive cells at E14.5, suggesting that OLs and astrocytes may derive from the same progenitor cell.
Both Olig1 and Olig2 occur in the nucleus of OLs and their progenitors occur on P1 in mouse brain.33 Olig2 continues to be nuclear at all developmental stages, whereas Olig1 is almost completely cytoplasmic in the adult mouse brain. Changes in nuclear or cytoplasmic Olig1 location correspond with altered morphology of OPCs.11 Recently, Othman et al19 reported that O1-positive multipolar OPCs with nuclear expression of Olig1 exhibited fewer processes, while OPCs with cytoplasmic Olig1 manifested more extensive membrane expansion. Nuclear Olig1 facilitated MBP expression, but with greatly diminished membrane expansion.19 However, these experiments were performed in mouse and human. Our results demonstrated that Olig1 was expressed in O4-positive OPCs and β-tubulin-positive neurons, but was localized in the cytoplasm during the period from E14.5 to adulthood. We speculate that the cause for this inconsistency may be attributed to differences between animal species or other unknown factors.
Conclusion
In conclusion, we systematically explored the expression pattern of Olig1 in neural cells during rat spinal cord development. Our results showed that Olig1 was coexpressed with O4-positive OPCs and β-tubulin-positive neurons at all time points during development. Olig1 was also coexpressed transiently with GFAP-positive astrocytes at E14.5. Olig1 was localized in the cytoplasm of O4- or β-tubulin-positive cells during the transition from E14.5 to adult. Our results contribute to understanding the mechanism of developmental regulation of neural cells by Olig1.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Number 81471277; 81271363) and the Project of Anhui Education Department (Number KJ2015B049by; KJ2016A869).
Authors’ contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Altman J, Bayer SA. The development of the rat spinal cord. Adv Anat Embryol Cell Biol. 1984;85:1–164. | ||
Bargmann CI, Marder E. From the connectome to brain function. Nat Methods. 2013;10:483–490. | ||
Morrison BM, Lee Y, Rothstein JD. Oligodendroglia: metabolic supporters of axons. Trends Cell Biol. 2013;23:644–651. | ||
Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. | ||
Miller RH. Regulation of oligodendrocyte development in the vertebrate CNS. Prog Neurobiol. 2002;67:451–467. | ||
Nicolay DJ, Doucette JR, Nazarali AJ. Transcriptional control of oligodendrogenesis. Glia. 2007;55:1287–1299. | ||
Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010;330:779–782. | ||
Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. | ||
Zhao JW, Raha-Chowdhury R, Fawcett JW, Watts C. Astrocytes and oligodendrocytes can be generated from NG2+ progenitors after acute brain injury: intracellular localization of oligodendrocyte transcription factor 2 is associated with their fate choice. Eur J Neurosci. 2009;29:1853–1869. | ||
Lu QR, Sun T, Zhu Z, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. | ||
Arnett HA, Fancy SP, Alberta JA, et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. | ||
Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. | ||
Zhou Q, Anderson DJ. The bHLH transcription factors Olig2 and Olig1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. | ||
Tan B, Yu J, Yin Y, Jia G, Jiang W, Yu L. The Olig family affects central nervous system development and disease. Neural Regen Res. 2014;9:329–336. | ||
Lu QR, Cai L, Rowitch D, Cepko CL, Stiles CD. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001;4:973–974. | ||
Balasubramaniyan V, Timmer N, Kust B, Boddeke E, Copray S. Transient expression of Olig1 initiates the differentiation of neural stem cells into oligodendrocyte progenitor cells. Stem Cells. 2004;22:878–882. | ||
Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. | ||
Gong X, Lin T, Sun Z, Fu M, Zuo H, Xie Z. Olig1 is downregulated in oligodendrocyte progenitor cell differentiation. Neuroreport. 2008;19:1203–1207. | ||
Othman A, Frim DM, Polak P, Vujicic S, Arnason BG, Boullerne AI. Olig1 is expressed in human oligodendrocytes during maturation and regeneration. Glia. 2011;59:914–926. | ||
Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. | ||
Wu S, Wu Y, Capecchi MR. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133:581–590. | ||
Rowitch DH, Lu QR, Kessaris N, Richardson WD. An “oligarchy” rules neural development. Trends Neurosci. 2002;25:417–422. | ||
Meijer DH, Kane MF, Mehta S, et al. Separated at birth? The functional and molecular divergence of Olig1 and Olig2. Nat Rev Neurosci. 2012;13:819–831. | ||
Jakovcevski I, Zecevic N. Olig transcription factors are expressed in oligodendrocyte and neuronal cells in human fetal CNS. J Neurosci. 2005;25:10064–10073. | ||
Silbereis JC, Nobuta H, Tsai HH, et al. Olig1 function is required to repress dlx1/2 and interneuron production in mammalian brain. Neuron. 2014;81:574–587. | ||
Liu Z, Hu X, Cai J, et al. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302:683–693. | ||
Dubois-Dalcq M, Behar T, Hudson L, Lazzarini RA. Emergence of three myelin proteins in oligodendrocytes cultured without neurons. J Cell Biol. 1986;102:384–392. | ||
Akiyama K, Ichinose S, Omori A, Sakurai Y, Asou H. Study of expression of myelin basic proteins (MBPs) in developing rat brain using a novel antibody reacting with four major isoforms of MBP. J Neurosci Res. 2002;68:19–28. | ||
Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303:390–396. | ||
Louis JC, Magal E, Muir D, Manthorpe M, Varon S. CG-4, a new bipotential glial cell line from rat brain, is capable of differentiating in vitro into either mature oligodendrocytes or type-2 astrocytes. J Neurosci Res. 1992;31:193–204. | ||
Hu JG, Lu HZ, Wang YX, Bao MS, Zhao BM, Zhou JS. BMP signaling mediates astrocyte differentiation of oligodendrocyte progenitor cells. Tohoku J Exp Med. 2010;222:195–200. | ||
Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135:145–157. | ||
Niu J, Mei F, Wang L, et al. Phosphorylated Olig1 localizes to the cytosol of oligodendrocytes and promotes membrane expansion and maturation. Glia. 2012;60:1427–1436. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.