Back to Journals » Cancer Management and Research » Volume 12
NUDT21 Suppresses Breast Cancer Tumorigenesis Through Regulating CPSF6 Expression
Authors Wang BJ, Liu DC, Guo QY, Han XW, Bi XM, Wang H, Wu ZS, Wu WY
Received 22 November 2019
Accepted for publication 9 April 2020
Published 1 May 2020 Volume 2020:12 Pages 3069—3078
DOI https://doi.org/10.2147/CMAR.S239664
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
Bi-Jun Wang,1,* Da-Chao Liu,1,* Qian-Ying Guo,2,* Xiao-Wen Han,2 Xiao-Min Bi,2 Hao Wang,2 Zheng-Sheng Wu,2 Wen-Yong Wu1
1Department of General Surgery, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, People’s Republic of China; 2Department of Pathology, Anhui Medical University, Hefei, Anhui, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zheng-Sheng Wu
Department of Pathology, Anhui Medical University, Hefei, Anhui, People’s Republic of China
Email [email protected]
Wen-Yong Wu
Department of General Surgery, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, People’s Republic of China
Email [email protected]
Background: NUDT21, an RNA binding protein, has been reported to play an important role in the regulation of multiple biological responses. Detection of NUDT21 expression may lead to the identification of a novel marker for breast cancer.
Purpose: The aim of this study was to investigate the clinical significance and functional role of NUDT21 in breast cancer.
Methods: The protein expression of NUDT21 was examined by immunohistochemistry (IHC) in 100 paraffin-embedded, archived breast cancer samples and 100 benign breast tissues. Then, the correlations between the NUDT21 expression and clinicopathologic characteristics and prognoses of the breast cancer patients were analyzed. In addition, the function of NUDT21 in breast cancer cell lines was detected by the methyl thiazolyl tetrazolium, colony formation and transwell assays. Finally, mass spectrometry analysis and Western blotting were used to identify the proteins that interact directly with NUDT21.
Results: IHC analysis revealed that the expression of NUDT21 was significantly lower in breast cancer tissues compared with benign breast disease tissues. The correlation analysis revealed that low expression of NUDT21 was positively correlated with tumor size, lymph node metastasis, and TNM stage. Also, Kaplan–Meier survival curves showed that patients with lower NUDT21 expression had shorter overall survival and relapse-free survival compared with higher NUDT21 expression. In addition, the knockdown of NUDT21 enhanced cell proliferation, migration, invasion and epithelial-mesenchymal transition (EMT). Consistently, the overexpression of NUDT21 inhibited cell proliferation, migration, invasion, and EMT. In addition, NUDT21 directly interacted with CPSF6 and negatively regulated its expression. Moreover, the knockdown of CPSF6 reversed NUDT21 expression-induced cancer cell migration and invasion.
Conclusion: NUDT21 might play a tumor-suppressive role by inhibiting cell proliferation and invasion via the NUDT21/CPSF6 signaling pathway in breast cancer cells.
Keywords: NUDT21, EMT, CPSF6, breast cancer
Introduction
Breast cancer is one of the leading causes of death in females around the world, and despite great advances in basic research, breast cancer is a major health concern and represents a top biomedical research priority.1 The incidence of this aggressive disease remains alarmingly high with approximately 1.700.000 new cases each year, these rates suggest that advances in prevention have been limited.1,2 A complex biological process of breast cancer that involves not only the interaction of numerous genes and the activation of multiple tumor-associated pathways but also the activation of complex molecular mechanisms. A variety of factors, for example, heredity, estrogen levels, growth factors and environmental factors, which are carcinogenic contributors to breast cancer.3,4 The accumulation of genomic and epigenetic changes is also associated with the development and progression of breast cancer.4,5 At present, the specific pathogenesis of breast cancer remains to be elucidated. With intensive study in the field of genomics, the effects of altered epigenetics on tumorigenesis has gradually attracted the attention of researchers.
NUDT21 is an RNA binding protein complex. Cleavage factor I mammalian (CFIm) is composed of a large subunit CFIm68 designated as CPSF6 and a small subunit CFIm25 designated as NUDT21 (CPSF5).6 Previous studies have shown that the production of a eukaryotic protein-coding messenger RNA (mRNA) requires the recognition of a specific poly(A) site sequence at the end of the gene. The deletion of CFIm25 or CFIm68 promotes the use of a proximal poly(A) site, thereby affecting the function of the 3ʹ untranslated region (3ʹUTR) of many mRNAs.7,8 In addition, Elkon et al suggested that when located in the last or 3ʹ-most exons, alternative poly(A) sites could lead to the production of mRNAs with variable 3ʹ-untranslated regions, resulting in protein products that vary at the C-terminus.9 This suggests that CFIm generally promotes the recognition of distal poly(A) sites. Related studies have shown that the misregulation of CFIm is associated with the tumorigenicity of glioblastoma and some neuropsychiatric diseases by altering the length of the 3ʹUTR of mRNA.10,11 Therefore, we decided to explore the role of NUDT21 in breast cancer by regulating its expressions.
In the current study, we first determined the protein expression of NUDT21 in different breast cancer cells. Then, we explored NUDT21 role in breast cancer cell proliferation, migration, and invasion as well as its necessity for targeting genes. Finally, our findings may offer novel therapeutic targets for the treatment of breast cancer patients.
Materials and Methods
Cell Lines and Culture
All human breast cancer cell lines (MDA-MB-231, MCF-7, BT549, SKBR3, and T47D) and human breast epithelial cell line (MCF-10A) were purchased from the American Type Culture Collection (ATCC, Manassas, VA). MDA-MB-231 and MDA-MB-468 cells were cultured in Leibovitz’s medium with 10% fetal bovine serum (FBS, Gibco) streptomycin (50 mg/mL). MCF-7, BT549, T47D and SKBR3 cells were cultured in RPMI-1640 medium with 10% fetal bovine serum (FBS, Gibco) streptomycin (50 mg/mL). MCF-10A cells were cultured in Ham F-12 medium with 5% fetal bovine serum (FBS, Gibco), 5 µg/mL insulin and 1 µg/mL hydrocortisone.
Patients and Breast Cancer Specimens
In this study, human tissue specimens were collected from the First Affiliated Hospital of Anhui Medical University (Hefei, People’s Republic of China). We selected 100 breast cancer samples and 100 benign breast disease samples were enrolled in this work. All breast cancer patients were female and treated with surgery. The use of the group was approved by the Biomedical Ethics Committee of Anhui Medical University with informed consent was signed by all relevant patients (institutional review board-approved protocol number: 20200091).
Immunohistochemistry
Immunohistochemistry (IHC) was used to analyze NUDT21 protein expression in paraffin sections of tissues from breast cancer patients. Formalin-fixed and paraffin-embedded tissue samples were obtained from 100 breast cancer samples and 100 benign breast disease samples. Tissue samples were cut into sections approximately 4 μm thick, dewaxed using xylene and dehydrated in a fractionated series. The antigen was then buffered with 0.01M citrate for 3 minutes to boil. Peroxidase was blocked with hydrogen peroxide, and the slides were incubated with normal goat serum. Light microscopy (Olympus) scores were recorded by two pathologists who were blinded to the clinical and histopathological features of each slide, and both pathologists had similar accuracy. The patients were divided into two groups according to the extent of NUDT21-specific nuclear immunostaining: one group was negative, and one group was positive. The expression levels of NUDT21 in patient tissues were evaluated using the 0, 1, and 2 scoring system; sections with 0% stained cells were designated as no expression of NUDT21 (score of 0); sections with 1–50% stained cells were designated as low expression of NUDT21 (score of 1); sections with 51–100% stained cells were designated as high expression of NUDT21 (score of 2).12 In the Kaplan–Meier analysis, patients with scores of 0 and 1 of NUDT21 expression were designated negative for NUDT21, and patients with scores of 2 were designated positive for NUDT21.
Western Blotting
Transfected cells were lysed with lysis buffer, centrifuged and extracted, and proteins were fractionated by 10% SDS-PAGE and transferred onto a PVDF membrane (Merck Millipore, Darmstadt, Germany). Then, blocking for 40 minutes at room temperature in 5% skim milk, the membrane was incubated with a primary anti-NUDT21 (Proteintech) for 2 hours at room temperature, then washed three times with phosphate buffer saline with Tween-20 (PBST) and incubated with the matched secondary antibody for 1 hour. After washing three times with PBST again, immunoreactive protein signals were detected using the Pierce ECL Substrate (Advansta) and ChemiDoc Mp System (BioRad). Densitometric analysis of the bands was performed using ImageJ software (National Institute of Health, USA). The antibodies used for immunoblotting are listed as follows: antibodies against NUDT21 (Proteintech), CPSF6 (Proteintech), β-actin (BD Bioscience), E-cadherin (Proteintech), Occludin (Proteintech), ZEB-1 (Santa CRUZ) or Vimentin (BD Bioscience) were used.
RNA Isolation and RT-qPCR
Total cellular RNA was extracted with TRIzol reagent (Life Technologies) according to the manufacturer’s instructions, and reverse transcription was performed using the RevertAidTM First Strand cDNA Synthesis Kit (Life Technologies). Then, qRT-PCR was performed to detect the expression of NUDT21 and GAPDH. The primer sequences are listed in Table 1.
 |
Table 1 The Primer Sequences of NUDT21 and GAPDH |
MTT Assays
Cell viability was measured using the methyl thiazolyl tetrazolium (MTT) assay (Sigma). The cells were harvested, counted (1x103 cells per well) and inoculated into 96-well plates with 6 parallel subwells per condition. For 5 days, 100 μL of MTT solution (0.5 mg/mL) was added to each well. After 1–2 hours of incubation, the medium was removed, and the cells were washed with PBS (Phosphate buffer solution).13 Incubation was carried out by adding 100 μL of DMSO for 20 minutes. Absorbance was detected at 570 nm using a microplate reader (BioTek, Vermont, USA).
Colony Formation Assays
Cells were harvested, counted and plated in 6-well plates at approximately 1000 cells per well, and three parallel subwells were established. The medium was changed for three days. After two weeks of observation, cell colonies were formed. They were first washed three times with PBS solution, then fixed with ethanol for 30 minutes, and finally stained with crystal violet solution for 5 minutes. Colonies were counted, and the data were combined to calculate the average ± standard deviation.
Migration and Invasion Assays
First, cells were trypsinized, resuspended in FBS-free medium and counted. Second, for determine the migration, add 1x105 cells to the upper chamber without Matrigel, for determine the invasion, 1x105 cells were added to the Matrigel-coated upper chamber and the lower chamber was filled with a medium containing 10% serum, after 24–48 hours of incubation, the cells were washed with PBS and fixed with alcohol (90%) and then stained with 0.1% crystal violet solution for 5 minutes.13 Finally, noninvasive cells were removed from the inner surface with a cotton swab. Images were taken using an Olympus IX-70 microscope.
Plasmid Construction
In our experiment, we used the expression vector pSin-flag (Invitrogen) to construct NUDT21-overexpressing plasmids. The NUDT21 coding sequence transcript was cloned into pSin-flag and designated as NUDT21-pSin-flag, sh-NUDT21, si-CPSF6; these were synthesized by Genepharma (Shanghai, China). All plasmids were transfected using Lipofectamine 3000 (Qiagen). The specific siRNA sequences used are listed in Table 2.
 |
Table 2 Sequence of the Oligonucleotides Used for Experiments |
Immunoprecipitation (IP)
For IP assays, MCF-7 cells were lysed in IP lysis buffer. For endogenous proteins, 0.1 μg of anti-NUDT21 (Proteintech) antibody and anti-IgG antibody (Sigma) were bound to 20 μL mixed protein A/G beads and incubated with 0.6 mL of cell lysate for 6 h at 4 °C. The beads were washed three times with IP buffer.14 The level of IP-enriched NUDT21 was measured by Western blotting.
Statistical Analyses
The chi-squared test was used to analyze the NUDT21 expression of the clinicopathological parameters. The statistical differences of the experimental data were evaluated by one-way analysis of variance (ANOVA) using the GraphPad Prism 7 software package. Overall survival(OS) and relapse-free survival (RFS) rate in patients were analyzed through Kaplan–Meier curves, and the significance of the differences was analyzed using the Log rank test. P<0.05 indicated statistically significant differences.
Results
Expression of NUDT21 in Breast Cancer Cell Lines and Tissue Samples
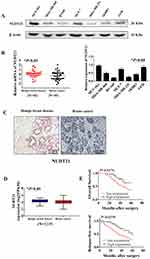
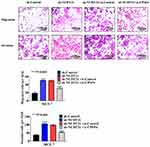
Firstly, to explore the differential expression of NUDT21 in breast cancer cells for migration and invasion, we performed Western blot analysis of the breast common epithelioid cell lines MCF-10A, MDA-MB-468, BT-549, MCF-7, MDA-MB-231, SKBR3 and T47D (Figure 1A). Next, we detected the expression in human breast tissue specimens to further verify the cell lines data. We performed RT-qPCR analysis of NUDT21 expression in tissues from 40 breast cancer patients and 40 patients with benign breast disease. We found that the levels of NUDT21 expression in breast cancer were significantly lower than the levels in benign disease (Figure 1B). Consistently, lower expression of NUDT21 was observed in breast cancer tissues compared with benign breast tissues, as revealed by IHC assay (Figure 1C). NUDT21 protein expression was observed in 74 (74%) of the 100 benign breast disease tissues that was significantly higher than breast cancer specimens (Table 3). In addition, we also analyzed the clinicopathological features of breast cancer patients. We found that lower expression of NUDT21 was positively associated with tumor size, tumor stage and lymph node metastasis in breast cancer patients (Table 4). Furthermore, from data in the Cancer Genome Atlas (TCGA), NUDT21 was also expressed at lower levels in breast cancer tissues than in benign breast tissues (Figure 1D). Moreover, patients with higher expression of NUDT21 mostly had longer overall survival (OS) and relapse-free survival (RFS) than patients with lower expression this protein (Figure 1E).
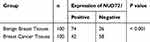 |
Table 3 Expression of NUDT21 in Breast Cancer Tissues and Benign Breast Tissues [n(%)] |
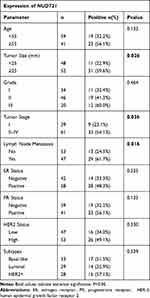 |
Table 4 Association of NUDT21 Expression with Clinicopathological Parameters from Breast Cancer Patients |
Effects of NUDT21 on Breast Cancer Cell Proliferation, Migration, and Invasion
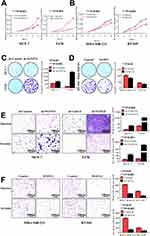
To investigate the functional role of NUDT21 in breast cancer cell lines, we first performed cell assays to observe its ability to regulate tumor cell viability in MCF-7, T47D, MDA-MB-231, and BT-549 cells. We found that knockdown of NUDT21 expression in MCF-7 and T47D cells significantly increased cell viability (Figure 2A and C). In contrast, ectopic NUDT21 expression decreased cell proliferation in MDA-MB-231 and BT-549 cells (Figure 2B and D). We next examined the role of NUDT21 in the capacity of migration and invasion of breast cancer cells. We found that knocking down NUDT21 expression increased the migration and invasion potential of breast cancer cells (Figure 2E), and this was reversed by ectopic NUDT21 expression (Figure 2F). These results indicated the NUDT21 is involved in breast cancer growth and progression in vitro.
Effects of NUDT21 on Breast Cancer Cell EMT
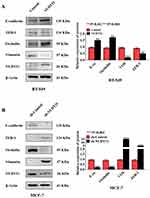
Metastasis is responsible for most cancer-related deaths, and yet the cellular and molecular mechanisms underlying tumor cell spread remain obscure.15 EMT involves the exchange of epithelial features (eg, nonmotility) with mesenchymal traits (eg, motility).16 To explore the role of NUDT21 in EMT of breast cancer, we selected epithelial markers (E-cadherin and Occludin) and mesenchymal markers (ZEB-1 and Vimentin) by using Western blotting confirmed that in NUDT21-overexpressing BT-549 cells, we found that E-cadherin and Occludin expression were upregulated, but Vimentin and ZEB-1 expression were downregulated (Figure 3A). In contrast, in the NUDT21 knockdown MCF-7 cells, we found that E-cadherin and Occludin expression were decreased. However, Vimentin and ZEB-1 expression were increased (Figure 3B).
Mass Spectrometry Revealed the Interaction Between NUDT21 and CPSF6
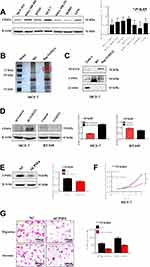
We performed Western blot analysis was carried out in non-transformed cell line (MCF-10A) and breast cancer cell lines (MDA-MB-468, BT-549, MCF-7, MDA-MB-231, SKBR3, and T47D) to explore the differential expression of CPSF6. The results indicated that CPSF6 was highly expressed in MCF-7 cells while its expression was low in T47D cells (Figure 4A). To identify potential mechanisms for how NUDT21 influenced breast cancer cell growth and progression, we employed mass spectrometry (Figure 4B). After that, we focused on CPSF6, a newly identified tumor suppressor as the top candidate for potential interaction with NUDT21 (Table 5). The IP assay confirmed that NUDT21 directly interacted with CPSF6 and that interfering with NUDT21 negatively regulated CPSF6 expression in MCF-7 cells (Figure 4C). Then, we chose T47D cells and MCF-7 cells to perform Western blotting. The results showed that forced expression of NUDT21 noticeably decreased the protein expression of CPSF6. In contrast, knockdown of NUDT21 protein expression in T47D cells reversed this phenomenon (Figure 4D). Afterward, cell function experiments were performed in MCF-7 cells from the depletion by si-CPSF6 (designated as si-CPSF6)/si-NC. The protein levels of CPSF6 decreased significantly after transfection with si-CPSF6 rather than the levels after transfection with the control (Figure 4E). Notably, cell proliferation, migration, and invasion all decreased with the decreased expression of CPSF6 (Figure 4F and G).
 |
Table 5 The Result of Mass Spectrometry |
NUDT21 Functioned as a Tumor Suppressor in Breast Cancer via Down-Regulating CPSF6
Cell function experiments were performed in MCF-7 cells with NUDT21 knockdown and CPSF6 depletion by si-CPSF6 (designated as si-CPSF6)/si-NC. The transwell assay showed that MCF-7 cell migration and invasion rates increased in CPSF6 knockdown cells compared with those in the control cells (Figure 5). This is suggested that NUDT21 can inhibit the migration and invasion of breast cancer cells by regulating the expression of CPSF6.
Discussion
The properties of NUDT21 in diverse human malignancies have been increasingly recognized and documented.8,9 Over the past two decades, the prognosis of breast cancer patients has been improved due to lifestyle modifications, prophylactic mastectomy, and advanced treatment.17 However, the efficacy in using traditional chemotherapy drugs is not satisfactory, and some patients with advanced breast cancer have developed resistance to existing chemotherapy drugs.10 The causal relationship between the expression of NUDT21 in cancer cell proliferation has been confirmed in glioblastoma, hepatocellular carcinoma and bladder cancer.13,18,19 Thus, we examined the functional role of NUDT21 in human breast cancer.
In this study, we report the expression of NUDT21 in breast cancer tissues and its relationship with clinical outcomes of breast cancer patients. After overexpression and knockdown of the expression of NUDT21 in breast cancer cells, we found that the fate of cancer cells was significantly influenced. In vivo, the NUDT21 was lower expressed in breast cancer tissues than in benign breast tissues. The expression level was associated with an increase in tumor size, clinical stage, and metastasis to lymph nodes. Meanwhile, the high expression of NUDT21 in tumors was associated with better OS and RFS outcomes than those with lower expression of NUDT21. Taken together, our results suggest that NUDT21 might play an important role in breast cancer. Although the expression of NUDT21 and CPSF6 was not significantly associated with survival outcomes of breast cancer patients in TCGA cohort, the discrepancy results might be due to the sampling or ethnicity difference. Nevertheless, we have noticed that the decreased expression of NUDT21 was associated with unfavorable prognoses in multiple cancers, which was further concordant with the oncogenic role of NUDT21 in different types of cancers.13,19,20
EMT is an important cellular mechanism during tumor invasion that is closely related to tumor invasion and migration.21 The EMT process is accompanied by the decreased expression of epithelial markers such as cytokeratin or increased expression of Vimentin and N-cadherin, while the most studied change in EMT is the loss of E-cadherin.22,23 However, previous studies have not determined whether the expression of NUDT21 could influence the activity of EMT pathway in breast cancer cells. In this study, the knockdown of NUDT21 was observed to inhibit the activity of EMT by down-regulating the epithelial markers E-cadherin and Occludin, while up-regulating the mesenchymal-associated marker Vimentin and ZEB-1. Western blotting showed that knockdown of NUDT21 enhanced the activity of EMT pathway and that overexpression of NUDT21 suppressed EMT activity in breast cancer cells. To our knowledge, this is first report suggesting that NUDT21 regulates the fate of breast cancer cells via EMT pathway.
Hence, we explored the role of NUDT21 in breast cancer cell lines, MCF-7, T47D, MDA-MB-231, and BT-549, which have different invasive and metastatic potentials in vitro. The differential expression of NUDT21 in these cell lines suggests that NUDT21 may play a role in the regulation of breast cancer cell proliferation, migration and invasion. We performed Western blot analysis of the breast common epithelioid cell lines. Interestingly, we found the high expression of NUDT21 in T47D and MCF-7 cells, which are both ER and PR positive cell lines. While MDA-MB-231 and BT549 cells with low expression of NUDT21 are triple-negative breast cancer cell lines. This result indicates that breast cancer is a complex biological mechanism regulated by multiple genes and factors, and brought some new clues for further exploration of the expressing and regulating mechanism of NUDT21.
To further explore how NUDT21 exerts its inhibitory function in breast cancer, we found by mass spectrometry that NUDT21 interacts with CPSF6 to form a complex that mediates breast cancer. Existing research has indicated that CPSF6 is implicated in various human ailments, for example, myeloproliferative neoplasms, breast cancer, and acute myeloid leukemia are closely related to the expression of CPSF6.14,25–29 The current study showed that CPSF6 promoted breast cancer cell growth and metastasis.14 The MCF-7 cells were capable of migration and invasion weaker than T47D cells and the T47D cells have a longer migration and invasion time. We selected MCF-7 cells for further experiments rather than T47D cells which have poor capacity of invasion to better demonstrate the role of NUDT21 in breast cancer cells and improve effectively the experimental efficiency. Our current study also revealed that NUDT21 can inhibit the expression of CPSF6. However, we have not fully characterized the potential mechanisms of how NUDT21 and CPSF6 interact each other. Further functional and mechanistic studies are warranted to prove our findings.
Conclusion
To our knowledge, this is the first report indicating that NUDT21 regulates breast cancer cell fate through the EMT pathway. Our findings suggest a role of NUDT21 in the molecular etiology of breast cancer and provide a possible target for therapeutic strategies.
Ethics and Consent Statements
This study has been approved by the ethics committee of the First Affiliated Hospital of Anhui Medical University in China (institutional review board-approved protocol number: 20200091).The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Project 81572305, Project 81472493 and Project 81972472), Specialized Research Fund for the Doctoral Program of Higher Education (Project 20133420120006). Key Program of Outstanding Young Talents in Higher Education Institutions of Anhui (Project gxyqZD2016046). Anhui provincial academic and technical leader reserve candidate (Project 2016H074).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019; 69(6):438–451.
2. DeSantis C, Howlader N, Cronin KA, Jemal A. Breast cancer incidence rates in U.S. women are no longer declining. Cancer Epidemiol Biomarkers Prev. 2011;20(5):733–739. doi:10.1158/1055-9965.EPI-11-0061
3. Mannello F, Tonti GA, Simone P, Ligi D, Medda V. Iron-binding proteins and C-reactive protein in nipple aspirate fluids: role of Iron-driven inflammation in breast cancer microenvironment? Am J Transl Res. 2010;3(1):100–113.
4. Karsli-Ceppioglu S, Dagdemir A, Judes G, et al. Epigenetic mechanisms of breast cancer: an update of the current knowledge. Epigenomics-Uk. 2014;6(6):651–664. doi:10.2217/epi.14.59
5. Fackler MJ, Umbricht CB, Williams D, et al. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res. 2011;71(19):6195–6207. doi:10.1158/0008-5472.CAN-11-1630
6. Yang Q, Coseno M, Gilmartin GM, Doublie S. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure. 2011;19(3):368–377. doi:10.1016/j.str.2010.12.021
7. Zhu Y, Wang X, Forouzmand E, et al. Molecular mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol Cell. 2018;69(1):62–74. doi:10.1016/j.molcel.2017.11.031
8. Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3ʹ-UTRs. Nucleic Acids Res. 2006;34(21):6264–6271. doi:10.1093/nar/gkl794
9. Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. doi:10.1038/nrg3482
10. Masamha CP, Xia Z, Yang J, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510(7505):412–416. doi:10.1038/nature13261
11. Gennarino VA, Alcott CE, Chen CA, et al. NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. Elife. 2015;4. doi:10.7554/eLife.10782
12. Ding K, Tan S, Huang X, et al. GSE1 predicts poor survival outcome in gastric cancer patients by SLC7A5 enhancement of tumor growth and metastasis. J Biol Chem. 2018;293(11):3949–3964. doi:10.1074/jbc.RA117.001103
13. Tan S, Li H, Zhang W, et al. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene. 2018;37(35):4887–4900. doi:10.1038/s41388-018-0280-6
14. Binothman N, Hachim IY, Lebrun JJ, Ali S. CPSF6 is a clinically relevant breast cancer vulnerability target: role of CPSF6 in breast cancer. Ebiomedicine. 2017;21:65–78. doi:10.1016/j.ebiom.2017.06.023
15. Aiello NM, Maddipati R, Norgard RJ, et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev Cell. 2018;45(6):681–695. doi:10.1016/j.devcel.2018.05.027
16. Chang L, Weiner LS, Hartman SJ, et al. Breast cancer treatment and its effects on aging. J Geriatr Oncol. 2019;10(2):346–355. doi:10.1016/j.jgo.2018.07.010
17. Liu X, Duan B, Dong Y, et al. MicroRNA-139-3p indicates a poor prognosis of colon cancer. Int J Clin Exp Pathol. 2014;7(11):8046–8052.
18. Nathanson KL, Domchek SM. Therapeutic approaches for women predisposed to breast cancer. Annu Rev Med. 2011;62:295–306. doi:10.1146/annurev-med-010910-110221
19. Xiong M, Chen L, Zhou L, et al. NUDT21 inhibits bladder cancer progression through ANXA2 and LIMK2 by alternative polyadenylation. Theranostics. 2019;9(24):7156–7167. doi:10.7150/thno.36030
20. Weng T, Huang J, Wagner EJ, et al. Downregulation of CFIm25 amplifies dermal fibrosis through alternative polyadenylation. J Exp Med. 2020;217:2. doi:10.1084/jem.20181384
21. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi:10.1016/j.cell.2009.11.007
22. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–428. doi:10.1038/nrc2131
23. Gjerdrum C, Tiron C, Hoiby T, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci U S A. 2010;107(3):1124–1129. doi:10.1073/pnas.0909333107
24. Chu Y, Elrod N, Wang C, et al. Nudt21 regulates the alternative polyadenylation of Pak1 and is predictive in the prognosis of glioblastoma patients. Oncogene. 2019;38(21):4154–4168. doi:10.1038/s41388-019-0714-9
25. Hidalgo-Curtis C, Chase A, Drachenberg M, et al. The t(1;9)(p34;q34) and t(8;12)(p11;q15) fuse pre-mRNA processing proteins SFPQ (PSF) and CPSF6 to ABL and FGFR1. Genes Chromosomes Cancer. 2008;47(5):379–385. doi:10.1002/gcc.20541
26. Naumann N, Schwaab J, Metzgeroth G, et al. Fusion of PDGFRB to MPRIP, CPSF6, and GOLGB1 in three patients with eosinophilia-associated myeloproliferative neoplasms. Genes Chromosomes Cancer. 2015;54(12):762–770. doi:10.1002/gcc.22287
27. Liu T, Wen L, Yuan H, et al. Identification of novel recurrent CPSF6-RARG fusions in acute myeloid leukemia resembling acute promyelocytic leukemia. Blood. 2018;131(16):1870–1873. doi:10.1182/blood-2017-11-818716
28. Qin YZ, Huang XJ, Zhu HH. Identification of a novel CPSF6-RARG fusion transcript in acute myeloid leukemia resembling acute promyelocytic leukemia. Leukemia. 2018;32(10):2285–2287. doi:10.1038/s41375-018-0095-z
29. Henning MS, Dubose BN, Burse MJ, Aiken C, Yamashita M. In vivo functions of CPSF6 for HIV-1 as revealed by HIV-1 capsid evolution in HLA-B27-positive subjects. PLoS Pathog. 2014;10(1):e1003868. doi:10.1371/journal.ppat.1003868
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.