Back to Journals » International Journal of Nanomedicine » Volume 11
Nucleic acid binding and other biomedical properties of artificial oligolysines
Authors Roviello GN, Vicidomini C, Costanzo V, Roviello V
Received 1 September 2016
Accepted for publication 8 October 2016
Published 10 November 2016 Volume 2016:11 Pages 5897—5904
DOI https://doi.org/10.2147/IJN.S121247
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Giovanni N Roviello,1 Caterina Vicidomini,1 Vincenzo Costanzo,1 Valentina Roviello2
1CNR Istituto di Biostrutture e Bioimmagini, Via Mezzocannone site and Headquarters, 2Centro Regionale di Competenza (CRdC) Tecnologie, Via Nuova Agnano, Napoli, Italy
Abstract: In the present study, we report the interaction of an artificial oligolysine (referred to as AOL) realized in our laboratory with targets of biomedical importance. These included polyinosinic acid (poly rI) and its complex with polycytidylic acid (poly I:C), RNAs with well-known interferon-inducing ability, and double-stranded (ds) DNA. The ability of the peptide to bind both single-stranded poly rI and ds poly I:C RNAs emerged from our circular dichroism (CD) and ultraviolet (UV) studies. In addition, we found that AOL forms complexes with dsDNA, as shown by spectroscopic binding assays and UV thermal denaturation experiments. These findings are encouraging for the possible use of AOL in biomedicine for nucleic acid targeting and oligonucleotide condensation, with the latter being a key step preceding their clinical application. Moreover, we tested the ability of AOL to bind to proteins, using serum albumin as a model protein. We demonstrated the oligolysine–protein binding by CD experiments which suggested that AOL, positively charged under physiological conditions, binds to the protein regions rich in anionic residues. Finally, the morphology characterization of the solid oligolysine, performed by scanning electron microscopy, showed different crystal forms including cubic-shaped crystals confirming the high purity of AOL.
Keywords: nucleic acid binding, polyinosinic acid, double-stranded nucleic acids, oligolysine, circular dichroism
Introduction
The importance of nucleic acids in biology relies on their ability to form complexes driven by complementary base pairing or to form other unique architectures.1 Taken together, these features make them important targets for biomedical strategies aimed at modulating the nucleic-acid-driven processes on the basis of pathological dysfunctions. Different approaches to interfere with the nucleic acid cellular machinery make use of different classes of oligonucleotide analogs, such as nucleobase-containing peptides.2–11 Apart from these latter, whose nucleic acid binding ability is due to their nucleobase-decorated nature, other compounds can serve as efficient nucleic acid binders. These include alkaloids,12 benzodifurans,13,14 or pure peptides, which are more often used for modulating protein-based processes.15 In particular, in case of basic peptides the nucleic acid binding is due to their ionic interaction with anionic phosphate groups in nucleic acids.16
Polylysines (PLLs) are important cationic peptides with several biomedical applications. In fact, they suppress prion accumulation in spleen in neurodegenerative diseases17 and are part of contrast agent probes for magnetic resonance imaging18 and gene/drug delivery tools.19 The PLLs, produced chemically or microbiologically for various biotechnological purposes,20–22 are oligomers/polymers formed by amidation of ε-amino groups of L-lysines. Moreover, intensive efforts led to the synthesis of dendrimeric α,ε-poly-L-lysines, tested as oligonucleotide delivery agents,23 and linear α,ε-oligo-L-lysines (AOL),24 object of our preliminary biological studies.24,25
The encouraging properties such as water solubility and serum stability, previously found for the artificial AOL, prompted us to deepen its evaluation as a nucleic acid condensation agent. In addition, we explored the interaction of AOL with proteins, using bovine serum albumin (BSA) as a protein model.
Materials and methods
Materials
Polyinosinic acid (poly rI), polyribocytidylic acid (poly rC), and BSA were purchased from Sigma-Aldrich Co., while dA12 and dT12 (DNA) were obtained from Biomers. To obtain dA12/dT12 and poly rI:poly rC double-stranded (ds) systems, single-stranded (ss) nucleic acids were mixed in a 1:1 ratio and annealed by heating the sample to 95°C (over 5 minutes) and slowly cooling it to 4°C over 12 hours. Synthesis of (alpha-epsilon-lys)8–NH2 (AOL) was conducted according to a literature procedure.24 Circular dichroism (CD) spectra were recorded at 15°C on a Jasco J-715 spectropolarimeter, while ultraviolet (UV) spectra were recorded on an UV-vis Jasco model V-550 spectrophotometer equipped with a Peltier ETC-505T temperature controller, using a Hellma quartz cell with a light path of 1 cm and a Hellma Tandem quartz cell 2×0.4375 cm. CD and UV spectra were recorded in the 200–260 or 200–320 nm wavelength range.
CD studies
For ssRNA, the CD spectra were recorded in the 200–320 nm wavelength range, before and after AOL/poly rI complexation by a tandem cell. Titration experiments with different AOL/poly rI ratios and fixed RNA concentration were conducted. Regarding ds nucleic acids, sum and complex CD spectra for AOL/dsDNA and AOL/dsRNA were obtained. We recorded them at different time points, to allow the system to reach equilibrium, in the same wavelength range as indicated earlier. Further, titration experiments with different AOL/(poly rI:poly rC) and AOL/(dA12:dT12) ratios were conducted by varying the concentrations of the peptide added to the complex solutions. CD spectra for BSA and AOL/BSA complex were obtained in a tandem cell by using a 4 mg/L concentration of protein.
UV melting studies
We recorded the UV melting curves for the following systems: dA12/dT12, AOL/dA12/dT12, poly rI/poly rC, AOL/poly rI/poly rC, and AOL (pure peptide solution:control). The denaturation profiles were obtained by monitoring the absorbance change at 260 nm with temperature (1.0°C/min). Melting temperature (Tm) values were determined from the first derivatives of the UV melting curves.
SEM analysis
A few drops of a 0.5 mM aqueous solution of AOL were deposited on an aluminum stub and allowed to evaporate in air. The sample metallized with an Au–Pd alloy was observed under a field-emission scanning electron microscope (SEM, Nova NanoSEM 450; FEI) at 3.00 kV using an Everhart-Thornley detector and through the lens detector systems.
Results and discussion
AOL binds to poly rI and poly I:C
Poly rI is a synthetic RNA homopolymer of inosine endowed with interesting properties. In fact, it triggers leukocyte production of important components of inflammatory homeostasis26 and induces innate responses by activating the immune cells such as B lymphocytes, dendritic cells, and macrophages.27 Its complex with poly rC, poly I:C, potentiates nonspecific immunotherapy in bladder cancer treatment28 and provokes prostate cancer cell apoptosis with an enhanced effect when treated with interferon-γ.29 In addition, poly I:C is a potential therapeutic agent useful for attenuating neuronal damage and promoting recovery after brain ischemia.30 However, stability issues limit the clinical application of poly rI and poly I:C that undergo facile enzymatic degradation. Hence is the need for technological approaches aimed to improve the delivery and consequent efficacy of these RNAs.31
Our previous works24,25 showed the ability of AOL to bind ss- and dsRNA. However, we had no information on the interaction of the same oligolysine with RNAs of therapeutic importance. For this purpose, we explored the potential of AOL as delivery agent of particular RNAs: poly rI and poly I:C. First, we studied the binding between AOL and the poly rI by CD spectroscopy by using a two-chamber quartz cell. Upon mixing the poly rI and AOL solutions in a 1:5.4 (rI/–NH3+) ratio, we recorded a CD spectrum different from that preceding the complexation process (Figure 1). In particular, we observed significant changes in intensity and position of the CD bands (bathochromic shift), which suggested, therefore, an AOL-induced conformational change of the poly rI.
After this binding assay, we conducted a CD titration to deduce quantitative information on the complexation process. We added increasing amounts of AOL to the poly rI solution and recorded the consequent CD spectra. From this study, we found a slight variation in the RNA CD spectrum caused by AOL. We observed no further change in the spectroscopic signal at AOL concentrations higher than 3.33 μM (1:6.7=rI/–NH3+ ratio).
Similarly, we investigated the ability of AOL to bind a preformed poly I:C complex by CD and UV spectroscopies. To this scope, we realized a complex by mixing equimolar amounts of the two RNAs and heating at 95°C, with subsequent slow cooling (annealing, 12 hours). At this point, we performed a CD binding study in a tandem quartz cell containing AOL in a chamber and dsRNA in the other one. AOL provoked drastic changes in the poly I:C CD spectrum, corresponding to a conformational alteration in the secondary structure of the dsRNA, condensed by AOL (Figure 2A). In addition, we studied the kinetic behavior of the AOL/poly I:C complex by recording the CD spectra at different time points after the first addition of AOL. We found that the complex was completely formed only after 30 minutes from the solution mixing. Indeed, CD spectra varied continuously (Figure 2A and B) before that time, whereas that obtained after 37 minutes did not differ significantly from the previous one (Figure 2A and B).
Moreover, we investigated the stoichiometry of the AOL/poly I:C complex by performing another CD experiment, ie, by adding increasing amounts of oligolysine to the poly I:C solution and recording the associated spectra. The complex stabilized after the addition of 85 nmol (in –NH3+) of AOL, which corresponds to a ~11 –NH3+/rI/rC ratio (Figure 3). We evaluated the influence of the interaction with AOL on the thermal stability of poly I:C by UV thermal melting experiments. In particular, we recorded the UV denaturation curves for the following: 1) poly I:C, 2) AOL, and 3) AOL/poly I:C in 1× phosphate-buffered saline (PBS) buffer (Figure 4). The thermal denaturation curve for the oligolysine/dsRNA complex (3, Figure 4) is not the superimposition of the other two melting curves (1 and 2, Figure 4). This means that the AOL/poly I:C thermal behavior is peculiar and is certainly influenced by peptide/RNA interactive forces, confirming, thus, the above-reported AOL/poly I:C binding hypothesis.
AOL-dsDNA interaction: kinetics, quantitative aspects, and thermal stabilization
Previous studies24,25 showed that AOL was ineffective in binding ssDNA. However, the importance of dsDNA in biomedicine led us to study the ability of our oligolysine to bind dsDNA for possible DNA-delivery applications of this oligocation. For this study, we used dA12/dT12 as a dsDNA model, following an approach analogous to that used for the AOL/RNA binding studies (Figures 5 and 6).
Furthermore, the dsDNA CD spectrum changed immediately upon mixing the ligand solutions due to the effect of the interaction with AOL. Interestingly, CD bands of the AOL/dA12/dT12 complex showed a progressive temporal variation with certain stabilization observable only after 24 minutes from the addition (Figures 5 and 6).
After this kinetic study, we performed a titration experiment on the AOL/dA12/dT12 complex solution and recorded the CD spectra consequent to the addition of different aliquots of oligolysine.
The CD signal underwent stabilization after the addition of 85 nmol (in –NH3+) of AOL or, in other terms, at a ~11 –NH3+/dA/dT ratio (Figure 7A and B).
In addition, we examined the thermal stability of the dsDNA in the presence of AOL. In particular, the dA12/dT12 complex in 1× PBS buffer (pH 7.5) at a concentration of 5 μM showed a Tm of ~31°C. On the other hand, under the same experimental conditions, but in the presence of AOL (5 μM), we found a Tm increase of ~4°C (Figure 8). This finding sustains the hypothesized AOL/dsDNA binding, essential for dsDNA condensation, and suggests that AOL could reinforce the interaction occurring between the DNA strands, thus stabilizing the dsDNA.
  | Figure 8 UV melting curves for dA12/dT12 ( |
BSA binding study
Protein–protein interactions or, more in general, protein-associated processes are subjects of several studies aimed at modulating these biomolecular events by organic molecules or peptides.32,33
Thus, in order to investigate the interaction of AOL with biomolecules different from nucleic acids, we took into consideration the family of proteins. More in detail, we carried out CD binding experiments on the protein BSA as a model protein using a quartz tandem cell. We placed BSA (8 mg/L, 1× PBS, pH 7.5) in one cell chamber, while the other hosted AOL (1.67 μM, 1× PBS, pH 7.5). Therefore, we recorded a sum CD spectrum (blue line, Figure 9A), corresponding, ie, to the two separate ligands, under the above-mentioned experimental conditions. After mixing, we obtained a CD spectrum (green line), which was different from the sum spectrum (Figure 9A). The observed CD spectrum was not the superimposition of the “mix” CD spectra of the two controls (Figure 9B). These latter were: 1) pure buffer in one chamber and BSA solution in the other one and 2) AOL solution in one chamber and buffer in the other one.
This finding suggests an interaction between the serum protein and the basic AOL. This binding provokes a change in the CD profile, especially in the spectral region at ~210 nm. The interaction involves, in other words, a variation of the secondary structure of BSA, probably due to ionic interactions involving AOL and negatively charged regions of the protein.
Microscopy analysis
After the above-reported experiments, we conducted a microscopy analysis of a solid sample of AOL in analogy to our previous work8 to obtain information on its morphology. In particular, the substance, examined under a polarizing optical microscope equipped with a micro-furnace, appeared in the form of tiny crystals as can be seen from Figure 10A (30,000×). In the image, tiny aggregates of crystals of hundreds of nanometers are visible at high magnifications. The sample obtained from a solution diluted with two volumes of distilled water appeared instead with a more regular morphology (shown in Figure 10B on a larger scale, 1,000×). Full-grown crystals of a few micrometers are observed in this case, as shown in Figure 10C in more detail (12,000×), that invariably showed a cubic shape. Finally, the absence of non-crystallized material revealed the high purity of the substance.
Conclusion
In this work, we demonstrated that AOL is an interesting oligocation able to condense RNA and dsDNA, provoking in this latter case a stabilization of the DNA complex. In particular, AOL binds to poly rI and poly I:C, potent interferon inducers endowed with several other biomedical characteristics, and could act, thus, as an effective delivery agent for these RNAs. The dsRNA-binding ability of AOL suggests its importance for antiviral strategies through the targeting of viral dsRNA, as well as in the compaction and cell delivery of therapeutic dsRNAs. Our experiments suggest that AOL participates in complexes formed by multiple peptide units and the target, as testified by the high AOL/nucleic acid stoichiometries found in the CD titrations. Moreover, the ability of AOL to interact with proteins, eliciting the perturbation of their secondary structure, has emerged from our CD binding experiments conducted on the BSA protein.
Overall, these findings, together with the serum stability properties that emerged from our previous work,25 highlight the importance of AOL as a potential molecular tool useful in biomedical strategies. In fact, it could play a key role in targeting RNA and dsDNA or, more in general, as a compacting agent useful for protection and cell delivery of therapeutic nucleic acids.
Acknowledgments
We thank Prof Antonio Roviello for his precious suggestions and Regione Campania for the grant received for the research project “Nuovi sistemi nucleopeptidici per applicazioni diagnostiche” under the research program Legge Regionale n. 5 (annualità 2007).
Disclosure
The authors report no conflicts of interest in this work.
References
Musumeci D, Amato J, Randazzo A, et al. G-quadruplex on oligo affinity support (G4-OAS): an easy affinity chromatography-based assay for the screening of G-quadruplex ligands. Anal Chem. 2014;86(9): 4126–4130. | ||
Amato F, Tomaiuolo R, Borbone N, et al. Design, synthesis and biochemical investigation, by in vitro luciferase reporter system, of peptide nucleic acids as new inhibitors of miR-509-3p involved in the regulation of cystic fibrosis disease-gene expression. Med Chem Commun. 2014; 5(1):68–71. | ||
Amato F, Tomaiuolo R, Nici F, et al. Exploitation of a very small peptide nucleic acid as a new inhibitor of miR-509-3p involved in the regulation of cystic fibrosis disease-gene expression. Biomed Res Int. 2014;2014: 610718. | ||
Roviello GN, Musumeci D, De Cristofaro A, et al. Alternate dab-aegPNAs: synthesis, nucleic acid binding studies and biological activity. Mol Biosyst. 2010;6(1):199–205. | ||
Roviello GN, Ricci A, Bucci EM, Pedone C. Synthesis, biological evaluation and supramolecular assembly of novel analogues of peptidyl nucleosides. Mol Biosyst. 2011;7(5):1773–1778. | ||
Roviello GN, Moccia M, Sapio R, et al. Synthesis, characterization and hybridization studies of new nucleo-gamma-peptides based on diaminobutyric acid. J Pept Sci. 2006;12(12):829–835. | ||
Roviello GN, Roviello G, Musumeci D, Bucci EM, Pedone C. Dakin-West reaction on 1-thyminyl acetic acid for the synthesis of 1,3-bis(1-thyminyl)-2-propanone, a heteroaromatic compound with nucleopeptide-binding properties. Amino Acids. 2012;43(4):1615–1623. | ||
Roviello GN, Roviello V, Autiero I, Saviano M. Solid phase synthesis of TyrT, a thymine–tyrosine conjugate with poly(A) RNA-binding ability. RSC Adv. 2016;6(33):27607–27613. | ||
Roviello GN, Vicidomini C, Di Gaetano S, Capasso D, Musumeci D, Roviello V. Solid phase synthesis and RNA-binding activity of an arginine-containing nucleopeptide. RSC Adv. 2016;6(17):14140–14148. | ||
Roviello GN, Musumeci D. Synthetic approaches to nucleopeptides containing all four nucleobases, and nucleic acid-binding studies on a mixed-sequence nucleo-oligolysine. RSC Adv. 2016;6(68): 63578–63585. | ||
Milano G, Musumeci D, Gaglione M, Messere A. An alternative strategy to synthesize PNA and DNA magnetic conjugates forming nanoparticle assembly based on PNA/DNA duplexes. Mol Biosyst. 2010;6(3):553–561. | ||
Kumar GS, Basu A. The use of calorimetry in the biophysical characterization of small molecule alkaloids binding to RNA structures. Biochim Biophys Acta. 2016;1860(5):930–944. | ||
Caruso U, Panunzi B, Roviello GN, Roviello G, Tingoli M, Tuzi A. Synthesis, structure and reactivity of amino-benzodifurane derivatives. Comptes Rendus Chimie. 2009;12(5):622–634. | ||
Roviello GN, Roviello V, Musumeci D, Pedone C. Synthesis of a novel benzodifuran derivative and its molecular recognition of poly rA RNA. Biol Chem. 2013;394(9):1235–1239. | ||
Russo A, Aiello C, Grieco P, Marasco D. Targeting “Undruggable” proteins: design of synthetic cyclopeptides. Curr Med Chem. 2016;23(8): 748–762. | ||
Roviello GN, Di Gaetano S, Capasso D, et al. RNA-binding and viral reverse transcriptase inhibitory activity of a novel cationic diamino acid-based peptide. J Med Chem. 2011;54(7):2095–2101. | ||
Titlow WB, Waqas M, Lee J, et al. Effect of polylysine on scrapie prion protein propagation in spleen during asymptomatic stage of experimental prion disease in mice. J Microbiol Biotechnol. 2016;26(9): 1657–1660. | ||
Zu G, Liu M, Zhang K, et al. Functional hyperbranched polylysine as potential contrast agent probes for magnetic resonance imaging. Biomacromolecules. 2016;17(6):2302–2308. | ||
Du J, Tian C, Ling J, Wang Y. R8-modified polysarcosine-b-polylysine polypeptide to enhance circulation stability and gene delivery efficiency. J Control Release. 2015;213:e50–e51. | ||
Hirohara H, Takehara M, Saimura M, Ikezaki A, Miyamoto M. Biosynthesis of poly(epsilon-L-lysine)s in two newly isolated strains of Streptomyces sp. Appl Microbiol Biotechnol. 2006;73(2):321–331. | ||
Hirohara H, Saimura M, Takehara M, Miyamoto M, Ikezaki A. Substantially monodispersed poly(epsilon-L-lysine)s frequently occurred in newly isolated strains of Streptomyces sp. Appl Microbiol Biotechnol. 2007;76(5):1009–1016. | ||
Takehara M, Saimura M, Inaba H, Hirohara H. Poly(gamma-L-diaminobutanoic acid), a novel poly(amino acid), coproduced with poly(epsilon-L-lysine) by two strains of Streptomyces celluloflavus. FEMS Microbiol Lett. 2008;286(1):110–117. | ||
Eom KD, Park SM, Tran HD, Kim MS, Yu RN, Yoo H. Dendritic alpha,epsilon-poly(L-lysine)s as delivery agents for antisense oligonucleotides. Pharm Res. 2007;24(8):1581–1589. | ||
Moccia M, Roviello GN, Bucci EM, Pedone C, Saviano M. Synthesis of a l-lysine-based alternate alpha,epsilon-peptide: a novel linear polycation with nucleic acids-binding ability. Int J Pharm. 2010;397(1–2): 179–183. | ||
Roviello GN, Musumeci D, Roviello V. Cationic peptides as RNA compaction agents: a study on the polyA compaction activity of a linear alpha, epsilon-oligo-L-lysine. Int J Pharm. 2015;485(1–2):244–248. | ||
Zare F, Magnusson M, Möllers LN, Jin T, Tarkowski A, Bokarewa M. Single-stranded polyinosinic acid oligonucleotides trigger leukocyte production of proteins belonging to fibrinolytic and coagulation cascades. J Leukoc Biol. 2008;84(3):741–747. | ||
Marshall-Clarke S, Downes JE, Haga IR, et al. Polyinosinic acid is a ligand for toll-like receptor 3. J Biol Chem. 2007;282(34):24759–24766. | ||
Ayari C, Besançon M, Bergeron A, LaRue H, Bussières V, Fradet Y. Poly(I:C) potentiates Bacillus Calmette-Guérin immunotherapy for bladder cancer. Cancer Immunol Immunother. 2016;65(2):223–234. | ||
Tan H, Zeng C, Xie J, et al. Effects of interferons and double-stranded RNA on human prostate cancer cell apoptosis. Oncotarget. 2015;6(36): 39184–39195. | ||
Li Y, Xu XL, Zhao D, et al. TLR3 ligand poly IC attenuates reactive astrogliosis and improves recovery of rats after focal cerebral ischemia. CNS Neurosci Ther. 2015;21(11):905–913. | ||
Hafner AM, Corthésy B, Merkle HP. Particulate formulations for the delivery of poly(I:C) as vaccine adjuvant. Adv Drug Deliv Rev. 2013; 65(10):1386–1399. | ||
Poletto M, Malfatti MC, Dorjsuren D, et al. Inhibitors of the apurinic/apyrimidinic endonuclease 1 (APE1)/nucleophosmin (NPM1) interaction that display anti-tumor properties. Mol Carcinog. 2015;55(5): 688–704. | ||
Wójcik P, Berlicki Ł. Peptide-based inhibitors of protein-protein interactions. Bioorg Med Chem Lett. 2016;26(3):707–713. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


 ) and complex (
) and complex ( ) CD spectra, recorded at 15°C in a tandem cell, relative to poly rI (8 nmol in I) and AOL (43 nmol in –NH3+) dissolved each in 0.8 mL of 1× PBS buffer (pH 7.5). Blue and green lines correspond to further additions of AOL (concentrations 3.3 [
) CD spectra, recorded at 15°C in a tandem cell, relative to poly rI (8 nmol in I) and AOL (43 nmol in –NH3+) dissolved each in 0.8 mL of 1× PBS buffer (pH 7.5). Blue and green lines correspond to further additions of AOL (concentrations 3.3 [ ]and 5 μM [
]and 5 μM [ ]respectively). (B) Variation of the CD values recorded at 270 nm relative to the solution of poly rI as function of the AOL concentration.
]respectively). (B) Variation of the CD values recorded at 270 nm relative to the solution of poly rI as function of the AOL concentration.
 , and 37
, and 37 minutes) after mixing. (B) Variation of the CD values recorded at 236 nm for the AOL/poly I:C complex in 1× PBS buffer (pH 7.5) as function of time.
minutes) after mixing. (B) Variation of the CD values recorded at 236 nm for the AOL/poly I:C complex in 1× PBS buffer (pH 7.5) as function of time.


 minutes) in 1× PBS buffer (pH 7.5).
minutes) in 1× PBS buffer (pH 7.5).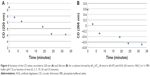


 ); and AOL/buffer (sum
); and AOL/buffer (sum ; mix
; mix