Back to Journals » OncoTargets and Therapy » Volume 10
Nuclear expression of XBP1s is correlated with breast cancer survival: a retrospective analysis based on tissue microarray
Authors Wang M, Ruan S, Ming J, Dong F
Received 24 July 2017
Accepted for publication 28 September 2017
Published 13 December 2017 Volume 2017:10 Pages 5927—5934
DOI https://doi.org/10.2147/OTT.S147102
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Mengyi Wang, Shengnan Ruan, Jie Ming, Fang Dong
Department of Breast and Thyroid Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Abstract: An alternatively spliced transcription factor that participates in the unfolded protein response, XBP1 is a novel protein involved in cancer progression and outcome. This study aimed to investigate the relationship of spliced XBP1 (XBP1s) with the clinicopathological characteristics and prognosis of breast cancer by using tissue-microarray analysis. A consecutive series of 170 patients with breast cancer diagnosed between 2001 and 2004 in hospitals in eastern and southern China were included. Immunohistochemical staining for XBP1s was performed, and the expression of XBP1s was separately examined in nuclei and cytoplasm. We found that a higher expression of XBP1s in nuclei strongly correlated with poorer survival (46.7% versus 75%, P=0.018); however, the expression of XBP1s in the cytoplasm had no relationship with survival. Multivariate Cox regression analysis indicated that the expression of XBP1s was not an independent prognostic factor (RR 2.074, 95% CI 0.909–4.736; P=0.083). None of the other clinicopathological characteristics – age, pathology grade, T stage, N stage, TNM stage, estrogen receptor, progesterone receptor, or HER2 status – was found to be correlated with XBP1s expression in the nuclei. In conclusion, independently of other clinicopathological factors, the nuclear expression of XBP1s is correlated with shorter breast cancer survival; however, whether nuclear XBP1s is an independent prognostic biomarker needs to be confirmed by further studies with larger samples and detailed sample stratification.
Keywords: breast cancer, tissue microarray, XBP1s, endoplasmic reticulum stress, biomarker
Introduction
Breast cancer (BC) is a heterogeneous disease sustained by complex growth pathways. Chemotherapy, hormone therapy, and molecular target therapy have had a significant impact on the survival of BC patients;1,2 however, adjuvant treatments do not always guarantee optimal results.3–5 In the past few decades, a better understanding of the molecular pathways involved in BC growth and progression allowed the identification of molecular targets that could be selectively inhibited by targeting agents.6,7 Nowadays, we are going through a bottleneck period of identifying new targets, allowing the possibility to develop novel approaches to further increasing the survival of BC patients.
XBP1, a key molecule in the unfolded protein response (UPR), was discovered to have a close relationship with cancer progression.8–10 The alternatively spliced form of XBP1 (XBP1s) is a major active form that as a transcriptional factor translocates into the nucleus and regulates gene expression, which helps resolve ER stress and support cell survival during the UPR.11–14 Evidence has emerged that XBP also plays a role in BC development.15–19 Serial analysis of gene expression showed XBP1 to be highly expressed in cancerous mammary epithelial cells.16 Immunohistochemistry analysis demonstrated expression of XBP1 in 90% of breast tumors, and in vitro UPR activation induced resistance to some chemotherapy drugs.17 An analysis of independent cohorts of patients with triple-negative BC revealed a specific XBP1 gene-expression signature that was strongly associated with poor prognosis.18 Recent work has revealed that increasing expression of XBP1 is associated with BC progression and that the XBP1 protein is significantly overexpressed in metastatic tumors.19
In our previous study,20 we demonstrated that XBP1s may play a role in estrogen-therapy resistance. Increased expression of both XBP1s mRNA and protein was discovered in tamoxifen-resistant MCF7 cells compared with normal MCF7 cells. Moreover, the sensitivity of XBP1s MCF7-TAMR cells to tamoxifen can be reestablished in vitro by a novel XBP1s inhibitor called STF083010. Furthermore, in an analysis of 170 BC patients’ samples, XBP1s expression was discovered to be highly correlated with overall survival of ER+ BC patients, strongly suggesting potential therapeutic value of XBP1 inhibitors in BC treatment.
The purpose of this study was to investigate the effect of XBP1s expression and subcellular localization on survival of BC patients through immunohistochemistry of a tissue microarray containing 170 patients with invasive BC. Kaplan–Meier survival analysis indicated a significant correlation between nuclear XBP1s (XBP1s-N) expression and overall survival. In addition, correlations among XBP1s expression and clinicopathological variables was explored to determine its potential value in BC classification and prognosis.
Materials and methods
Ethics statement
Ethical approval was given by the medical ethics committee of Tongji Medical College, Huazhong Science and Technology University (S025), and was approved for a project of the National Natural Science Foundation of China (81202094). This study was a part of this project. Patients provided written informed consent for the use of their tissue for future research prior to treatment.
Patients and tissue
A consecutive series of 170 patients diagnosed with invasive BC who underwent surgery between 2001 and 2004 in hospitals in Jiangsu, Zhejiang, and Shanghai were selected. Their information was obtained from the tissue bank of the Shanghai Biochip Center. Tissues were collected immediately after surgical resection and snap-frozen in liquid nitrogen, then stored in the tissue bank until later use. After exclusion of patients lost to follow-up or lacking full data, 160 patients were included. Detailed information on histopathological variables, clinical data, and long-term follow-up was available for these patients and registered in a database. The median follow-up of these patients was 9–12.5 years.
Tissue-microarray preparation
Formalin-fixed paraffin-embedded tumor samples of patients were obtained from the Shanghai Biochip Center. Histopathological characteristics of these samples are shown in Table 1. Three morphologically representative tumor regions were chosen from each of the 160 samples. Three corresponding normal adjacent tissue samples were obtained simultaneously. Cylindrical core-tissue specimens (5×15×15 mm) were acquired from each tumor sample and then arrayed into a newly designed recipient paraffin block using a custom-built precision instrument (Outdo Biotech, Shanghai, China). After being heated at 52°C, the core tissues were melted and closely fitted into the paraffin block.
  | Table 1 Patient characteristics |
Immunohistochemistry
All tissue-microarray samples were cut with a microtome into 4 μm sections that were mounted on poly-L-lysine-coated glass slides. Immunohistochemical staining for XBP1s was recorded separately for the cytoplasm and nuclei of the 160 BC samples and ten normal adjacent tissue samples. ER, PR, and HER2 staining had been performed previously, and results were recorded after the initial surgery was done. After a second incubation with biotinylated antigoat antibodies, slides were incubated with peroxidase-labeled streptavidin. Reaction products were visualized by immersing the slides in diaminobenzidine tetrachloride and counterstaining with Harris hematoxylin. Staining for XBP1s was considered positive only if a minimum of 10% definite tumor cells showed a positive reaction.
Statistical analysis
All analyses were completed using SPSS 16.0 software. Survival curves were calculated using the Kaplan–Meier method, with significance evaluated using the Mantel–Cox log-rank test. The prognostic significance of parameters was assessed using the Cox proportional-hazard model with overall survival as an end point. A multivariate analysis was performed using a Cox model; previously identified prognostic factors for BC were included in the model. Relationships between XBP1s expression and clinicopathological parameters were calculated using nonparametric Kruskal–Wallis and Mann–Whitney methods using Spearman’s correlation analysis. P<0.05 was considered statistically significant. Correlations with numerical variables were analyzed by the Mann–Whitney U test.
Results
Patient and tumor characteristics
Data of the patients and tumor characteristics are outlined in Table 1. The median age of the study group was 51 years (range 29–83 years). The majority of tumors (95.1%) were categorized as invasive ductal carcinoma; other presentations included infiltrative lobular carcinoma (3.7%) and a mixture of both (1.2%). Most tumors were histological grade 2 (85%), and 11.25% presented with grade 1. We identified T1, T2, T3, and T4 tumors in 11.3%, 66.9%, 9.4%, and 0 patients based on the sixth American Joint Committee on Cancer TNM staging system. Axillary nodal metastases were identified in 58.1% of patients. TNM stage 2 and 3 were the most common stages (58.8% and 30%), followed by 8.1% cases of stage 1 and none of stage 4. There were more hormone receptor (ER/PR)-positive cases than negative cases (ER 67.5% versus 30%, PR 59% versus 38%). Only 32.5% cases were HER2-positive and 65.6% HER2-negative.
Evaluation of XBP1s expression by immunohistochemistry
For each spot, the regions of most intense and/or predominant staining pattern were scored by eye. Nuclear and cytoplasmic staining intensity and positive percentage were determined separately for each specimen. Staining intensity was graded on a scale of 0–3. Positive percentage was classified into five categories: 0 (negative), 1 (1%–25%), 2 (26%–50%), 3 (51%–75%), and 4 (76%–100%). Total scores were calculated from the product of staining intensity and positive percentage that was used to divide all specimens into two groups: a low-expression group (score 0–5) and a high-expression group (score 6–12). For specimens that were uninterpretable or not infiltrating carcinoma, a designation of “not applicable” was given. A sample of immunohistochemistry results is displayed in Figure 1. According to the evaluation standard of immunohistochemical staining, 9.4% and 4.4% of cells showed high XBP1s expression in the nucleus and cytoplasm (XBP1s-C).
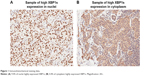  | Figure 1 Immunohistochemical staining data. |
Survival analysis
By July 2013, the median survival of all 160 patients was 101 months (2–131 months). Kaplan–Meier survival analysis was performed with respect to clinical stage. Survival rates from stage 1 to stage 3 were 88.9%, 79.4%, and 59.2%, respectively, and differences were statistically significant (log-rank P=0.005) (Figure 2).
Further survival analyses was performed for XBP1s-C and XBP1s-N, as well as other clinicopathological parameters: age, pathology grade, T stage, N stage, TNM stage, ER, PR, HER2, and HER2 gene amplification (HER2 fluorescence in situ hybridization). The statistical significance of differences was tested via log-rank analysis (Table 2). In the XBP1s-N-expression group, survival rates in patients with low and high XBP1s expression were 75% (108 of 144) and 46.7% (eight of 15), respectively. The log-rank test showed that the difference between survival rates had statistical significance (P=0.018), ie, higher XBP1s-N expression corresponded strongly to poorer survival of patients with BC. In contrast, in the XBP1s-C-expression group, 71.7% (109 of 152) patients with low XBP1s expression and 85.7% (six of seven) patients with high XBP1s expression survived; however, the difference was not significant (P=0.471). Survival curves are shown in Figure 3. Other factors significantly affecting survival included pathological grade (P=0), clinical N stage (P=0.042), TNM stage (P=0.012), ER status (P=0.004), and PR status (P=0.009), correlations that have been widely verified by prior researchers.
  | Table 2 Kaplan–Meier survival analysis and log-rank test |
  | Figure 3 Survival curves of (A) nuclear XBP1s (XBP1s-N) expression and (B) cytoplasmic XBP1s (XBP1s-C) expression. |
Correlation of XBP1s-N expression with various clinicopathological parameters
Because XBP1s-N expression had a significant effect on survival, but was not an independent prognostic factor, we examined the correlation of XBP1s-N expression with other clinicopathological parameters. Under Kruskal–Wallis and Mann–Whitney analyses, XBP1s-N-expression levels showed no significant differences among age–groups <35, 35–50, or >50 years (P=0.477). No differences were seen for pathology grades 1–3, T stages 1–3, N stages 0–3, TNM stages 1–4 (P=0.112, 0.144, 0.071, and 0.241, respectively), for ER status, PR status, HER2 status, or the luminal subgroup (0.718, 0.315, and 0.578, respectively). The same results were obtained from Spearman’s correlation analysis. We can conclude that expression of XBP1s-N was not correlated with any of the clinicopathological parameters analyzed in this study. Detailed data are shown in Table 3.
  | Table 3 Correlation of nuclear XBP1s expression with various clinicopathological parameters |
Multivariate Cox regression analysis
Multivariate Cox regression analyses were performed with factors that have previously been proven to affect survival. The results are shown in Table 4. In the Cox model, the P-value of XBP1s-N expression was 0.083, and RR was 2.074 (95% CI 0.909–4.736), indicating that XBP1s-N expression was not an independent prognostic factor. P-values for pathological level and TNM stage were 0.247 and 0.155, respectively. We concluded from these results that pathological grade and TNM stage are not independent prognostic factors of BC. Clinical N-stage, ER-status, and PR-status correlation coefficients were negative (−0.079, −0.409, and −0.607, respectively); however, P-values were all greater than 0.05 (0.799, 0.36, and 0.177, respectively), indicating no statistical significance. Therefore, N stage, ER status, and PR status were not independent prognostic factors either in the present study.
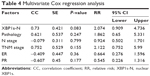  | Table 4 Multivariate Cox regression analysis |
Discussion
Our survival analysis showed that elevated expression of XBP1s-N is associated with worse clinical outcome. The overall survival rate of patients with low XBP1s-N expression was 1.6 times higher than patients with high XBP1s-N expression. However, the same was not true for the XBP1s-C-expression group. XBP1s is an important participant in the UPR, and has been studied as a novel protein involved in BC in recent years. Researchers have observed that the expression level of XBP1s in BC tissue is correlated with prognosis; however, these studies were mostly performed in the context of gene expression.12,21–25 In this study, using tissue-microarray methods, we showed that a higher expression of XBP1s protein in nuclei corresponded to poorer survival in BC. This finding suggested that XBP1s affects survival in patients via its role as a nuclear transcription factor.
Scientists have explored the mechanism of XBP1s in tumor progression. The low sugar, anoxic, and acidic microenvironment in malignant tumors can lead to an increase in proteins unfolding/misfolding, causing higher ER stress.21,26,27 In response to ER-stress signals, IRE1 in the endoplasmic reticulum membrane dimerizes and becomes autophosphorylated, resulting in its activation and the unconventional splicing of XBP1 pre-mRNA. XBP1-splicing creates transcriptionally active XBP1s.8 XBP1s enters the nucleus and activates a variety of genes involved in protein maturation, degradation, or ER expansion, which enables cells to produce and secrete proteins efficiently, helping to resolve ER stress and promote cell survival.28–30 However, if the UP level exceeds a threshold, affected cells are committed to cell death.31,32 This mechanism gives us a rough understanding of why XBP1s expression in nuclei affects BC survival. Similarly to the discoveries about hormone receptors and consequent endocrine treatment in BC,7 XBP1s may have potential value in prognostication and targeted therapy in the future.10
Our multivariate Cox regression analysis showed that it is too early to conclude that XBP1s is an independent prognostic factor. Other factors, such as pathology grade, N stage, TNM stage, ER, and PR, had no significance in our model either. The generally accepted biomarker HER2, which has been shown to have a detrimental effect on relapse-free survival and risk of death,6,33 likewise showed no effect on survival in this study. This result is not so surprising when considering that the sample was not very large and the majority of our samples were from middle- to late-stage patients. More importantly, ER status was reported to be related to the induction of PR.34,35 This may be the main reason that even though ER and PR are both important biomarkers that obviously affect BC survival,36–38 they do not always show significance in multivariate-regression models,39 which is consistent with our results. It is noteworthy that the P-value of XBP1s-N expression in our model was 0.083, which was the lowest P-value and very close to 0.05. Further studies with a larger samples and detailed sample stratification are needed to revise our conclusion.
In further exploration of the correlation of XBP1s-N expression with other clinicopathological parameters, we found that it had no significant correlation with age, pathology grade, T stage, N stage, TNM stage, ER, PR, HER2 status, or luminal subtype. From this, we can preliminarily conclude that XBP1s-N expression is independent of these clinicopathological factors. Interestingly, XBP1 has been reported to interact with ER in a ligand-independent manner and can also induce transcription from estrogen-responsive elements containing a luciferase-reporter gene, even in the absence of estrogen.40 Further research found large-scale chromatin unfolding associated with XBP1-mediated increases in ER-transcriptional activity.41 Newer studies on ER- and PR-associated genes in BC found a high degree of correlation between ER levels and expression of XBP1.42 These findings seem contradictory with ours; it is unclear whether XBP1s expression is correlated with ER expression. We can only temporarily speculate that XBP1 may not be related to the hormone-signal pathway on a macroscopic level. The relationship of XBP1s with the ER is a largely unclear issue that needs to be further studied.
Conclusion
Our study indicates that XBP1s is an important biomarker in nuclei that is correlated with an adverse effect on BC survival. Although it is too early to say XBP1s is an independent prognostic factor, its prognostic value requires further research. Moreover, XBP1s may be a new prognostic and therapeutic target, which may potentially improve the survival of patients.10,28
Acknowledgment
This work was supported financially by the National Natural Science Foundation of China (81202094).
Disclosure
The authors report no conflicts of interest in this work.
References
Early Breast Cancer Trialists’ Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. | ||
Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9(1):45–53. | ||
Early Breast Cancer Trialists’ Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 1998;351(9114):1451–1467. | ||
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. | ||
Peto R, Davies C, Godwin J, et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100 000 women in 123 randomised trials. Lancet. 2012;379(9814):432–444. | ||
Krishnamurti U, Silverman JF. HER2 in breast cancer: a review and update. Adv Anat Pathol. 2014;21(2):100–107. | ||
Jensen EV, Jordan VC. The estrogen receptor: a model for molecular medicine. Clin Cancer Res. 2003;9(6):1980–1989. | ||
Back SH, Schröder M, Lee K, Zhang K, Kaufman RJ. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods. 2005;35(4):395–416. | ||
Romero-Ramirez L, Cao H, Nelson D, et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64(17):5943–5947. | ||
Shajahan AN, Riggins RB, Clarke R. The role of X-box binding protein-1 in tumorigenicity. Drug News Perspect. 2009;22(5):241–246. | ||
Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol Rev. 2011;91(4):1219–1243. | ||
Cao SS, Kaufman RJ. Unfolded protein response. Curr Biol. 2012;22(16):R622–R626. | ||
Koumenis C. ER stress, hypoxia tolerance and tumor progression. Curr Mol Med. 2006;6(1):55–69. | ||
Maurel M, Chevet E, Tavernier J, Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014;39(5):245–254. | ||
Bertucci F, Houlgatte R, Benziane A, et al. Gene expression profiling of primary breast carcinomas using arrays of candidate genes. Hum Mol Genet. 2000;9(20):2981–2991. | ||
Gu Z, Lee RY, Skaar TC, et al. Association of interferon regulatory factor-1, nucleophosmin, nuclear factor-κB, and cyclic AMP response element binding with acquired resistance to Faslodex (ICI 182,780). Cancer Res. 2002;62(12):3428–3437. | ||
Scriven P, Coulson S, Haines R, Balasubramanian S, Cross S, Wyld L. Activation and clinical significance of the unfolded protein response in breast cancer. Br J Cancer. 2009;101(10):1692–1698. | ||
Chen X, Iliopoulos D, Zhang Q, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508(7494):103–107. | ||
Li H, Chen X, Gao Y, Wu J, Zeng F, Song F. XBP1 induces snail expression to promote epithelial-to-mesenchymal transition and invasion of breast cancer cells. Cell Signal. 2015;27(1):82–89. | ||
Ming J, Ruan S, Wang M, et al. A novel chemical, STF-083010, reverses tamoxifen-related drug resistance in breast cancer by inhibiting IRE1/XBP1. Oncotarget. 2015;6(38):40692–40703. | ||
Yadav RK, Chae SW, Kim HR, Chae HJ. Endoplasmic reticulum stress and cancer. J Cancer Prev. 2014;19(2):75–88. | ||
Yoshida H. ER stress and diseases. FEBS J. 2007;274(3):630–658. | ||
Ulianich L, Insabato L. Endoplasmic reticulum stress in endometrial cancer. Front Med (Lausanne). 2014;1:55. | ||
Honma Y, Harada M. New therapeutic strategy for hepatocellular carcinoma by molecular targeting agents via inhibition of cellular stress defense mechanisms. J UOEH. 2014;36(4):229–235. | ||
Zhu J, Chen M, Chen N, et al. Glycyrrhetinic acid induces G1-phase cell cycle arrest in human nonsmall cell lung cancer cells through endoplasmic reticulum stress pathway. Int J Oncol. 2015;46(3):981–988. | ||
Sovolyova N, Healy S, Samali A, Logue SE. Stressed to death: mechanisms of ER stress-induced cell death. Biol Chem. 2014;395(1):1–13. | ||
Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14(9):581–597. | ||
Koong AC, Chauhan V, Romero-Ramirez L. Targeting XBP-1 as a novel anti-cancer strategy. Cancer Biol Ther. 2006;5(7):756–759. | ||
Shuda M, Kondoh N, Imazeki N, et al. Activation of the ATF6, XBP1 and GRP78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol. 2003;38(5):605–614. | ||
Manie SN, Lebeau J, Chevet E. Cellular mechanisms of endoplasmic reticulum stress signaling in health and disease – 3: orchestrating the unfolded protein response in oncogenesis – an update. Am J Physiol Cell Physiol. 2014;307(10):C901–C907. | ||
Vandewynckel YP, Laukens D, Geerts A, et al. The paradox of the unfolded protein response in cancer. Anticancer Res. 2013;33(11):4683–4694. | ||
Wang WA, Groenendyk J, Michalak M. Endoplasmic reticulum stress associated responses in cancer. Biochim Biophys Acta. 2014;1843(10):2143–2149. | ||
Petrelli F, Barni S. Role of HER2-neu as a prognostic factor for survival and relapse in pT1a-bN0M0 breast cancer: a systematic review of the literature with a pooled-analysis. Med Oncol. 2012;29(4):2586–2593. | ||
Harris HA, Katzenellenbogen JA, Katzenellenbogen BS. Characterization of the biological roles of the estrogen receptors, ERα and ERβ, in estrogen target tissues in vivo through the use of an ERα-selective ligand. Endocrinology. 2002;143(11):4172–4177. | ||
Sá SI, Pereira PA, Malikov V, Madeira MD. Role of estrogen receptor α and β in the induction of progesterone receptors in hypothalamic ventromedial neurons. Neuroscience. 2013;238:159–167. | ||
Huber KE, Carey LA, Wazer DE. Breast cancer molecular subtypes in patients with locally advanced disease: impact on prognosis, patterns of recurrence, and response to therapy. Semin Radiat Oncol. 2009;19(4):204–210. | ||
Omlin A, Amichetti M, Azria D, et al. Boost radiotherapy in young women with ductal carcinoma in situ: a multicentre, retrospective study of the Rare Cancer Network. Lancet Oncol. 2006;7(8):652–656. | ||
Provenzano E, Hopper JL, Giles GG, Marr G, Venter DJ, Armes JE. Biological markers that predict clinical recurrence in ductal carcinoma in situ of the breast. Eur J Cancer. 2003;39(5):622–630. | ||
Zhang Y, Gao H, Gao W. The volume and duration of wound drainage are independent prognostic factors for breast cancer. Tumor Biol. 2014;35(4):3563–3568. | ||
Ding L, Yan J, Zhu J, et al. Ligand-independent activation of estrogen receptor α by XBP-1. Nucleic Acids Res. 2003;31(18):5266–5274. | ||
Fang Y, Yan J, Ding L, et al. XBP-1 increases ERα transcriptional activity through regulation of large-scale chromatin unfolding. Biochem Biophys Res Commun. 2004;323(1):269–274. | ||
Andres SA, Wittliff JL. Co-expression of genes with estrogen receptor-α and progesterone receptor in human breast carcinoma tissue. Horm Mol Biol Clin Investig. 2012;12(1):377–390. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

