Back to Journals » Journal of Inflammation Research » Volume 15
Nrf2 Inhibits the Progression of Parkinson’s Disease by Upregulating AABR07032261.5 to Repress Pyroptosis
Authors Zhong Y, Cai X, Ding L, Liao J , Liu X, Huang Y, Chen X, Long L
Received 29 October 2021
Accepted for publication 16 January 2022
Published 2 February 2022 Volume 2022:15 Pages 669—685
DOI https://doi.org/10.2147/JIR.S345895
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Yunxiao Zhong,1,* Xiaodong Cai,2,* Li Ding,3,* Jinchi Liao,1 Xu Liu,1 Yiying Huang,1 Xiaohong Chen,1 Ling Long1
1Department of Neurology, The Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong Province, 510600, People’s Republic of China; 2Department of Neurology, The Sixth Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong Province, 510655, People’s Republic of China; 3Department of Pathology, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ling Long; Xiaohong Chen
Department of Neurology, The Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong Province, 510600, People’s Republic of China
, Tel +86-20-85253275
Email [email protected]; [email protected]
Objective: Parkinson’s disease (PD) is associated with dysregulated neural cell death, such as pyroptosis, but its regulatory mechanisms are poorly understood. This study investigated roles of nuclear factor E2-related factor 2 (Nrf2) in regulating pyroptosis and PD development.
Methods: Cellular and rat PD models established by 6-OHDA exposure were subjected to Nrf2 overexpression. Neurobehavioral functions were assessed by the traction test, Morris Water Maze, and open field test. Cell proliferation was analyzed by MTS assay, while flow cytometry was applied to quantify levels of reactive oxygen species (ROS) and apoptosis. Nissl bodies in rat brains were detected by Nissl staining, and cell apoptosis in brain tissues was assessed by terminal deoxynucleotidyl transferase dUTP nick-end labeling. Differential expression of lncRNA and mRNA was characterized by deep sequencing.
Results: A cellular PD model was successfully established by inducing PC12 cell differentiation with nerve growth factor-β and exposing differentiated cells to 6-OHDA. Cells exhibited significantly increased ROS levels, enhanced pyroptosis, and inhibited Nrf2 phosphorylation. The rat PD model exhibited impaired muscle strength, increased pyroptosis, and repressed Nrf2 phosphorylation. Nrf2 overexpression effectively repressed pyroptosis in both cellular and rat PD models. Marked alterations of lncRNA and mRNA profiles were induced by Nrf2 overexpression in the cellular PD model, which involved multiple signaling pathways. Silencing of the lncRNA AABR07032261.5 significantly promoted pyroptosis in the cellular PD model.
Conclusion: Nrf2 suppressed PD pathogenesis in cellular and animal models by promoting AABR07032261.5, which repressed pyroptosis.
Keywords: Nrf2, lncRNAs, AABR07032261.5, oxidative stress, pyroptosis, Parkinson’s disease
Introduction
Parkinson’s disease (PD), a common and severe neurological disorder characterized by neurodegenerative progression resulting from early death of dopaminergic neurons in the substantia nigra pars compacta, results in dopamine deficiency in the basal ganglia and movement disorders.1,2 First described in detail nearly two centuries ago, PD remains the second most prevalent neurodegenerative disorder after Alzheimer’s disease, with an annual incidence greater than 160 per 1×105 persons in the aged population.1,3 Recently available methods for PD clinical management, including levodopa, dopamine agonists, monoamine oxidase type B inhibitors, and deep brain stimulation, are mainly used to treat motor symptoms or motor complications of patients of PD rather than be curative.1,4 Previous epidemiological investigations reported that the initiation of PD was associated with multiple risk factors including dairy products, exposure to hazardous chemicals such as pesticides, amphetamine and methamphetamine use, as well as other pathogenic conditions including cancer, traumatic brain injury, diabetes, and hypertension.3 Moreover, research progress during the past decades has shown that the development and progression of PD are closely linked with multiple genetic mutations in genes encoding α-synuclein (α-syn), leucine-rich repeat kinase 2, Parkin, and PTEN-induced putative kinase 1.1,5 Nevertheless, molecular mechanisms driving PD initiation and progression are still poorly understood.
Chronic oxidative stress in the substantia nigra was recently reported to be an essential cellular event closely linked to the pathogenesis of PD.6 Under normal physiological conditions, homeostatic production of cellular reactive oxygen species (ROS) in mitochondria is maintained by the cooperative actions of multiple antioxidant proteins, such as glutathione and superoxide dismutase.7,8 However, mitochondrial depolarization, α-syn accumulation, endoplasmic reticulum stress, abnormal elevation of cytosolic dopamine content, and age-dependent antioxidant dysfunctions contribute to high cellular oxidative stress. Ultimately, this leads to oxidation of proteins, lipids, and other macromolecules, damage to cellular structures, and irreversible neural cell death during the development and progression of PD.7 Furthermore, excessive ROS accumulation and oxidative stress activate various cell-death pathways during PD pathogenesis including apoptosis, autophagic cell death, and necroptosis.6,9 Pyroptosis, a newly discovered inflammatory type of cell death, is characterized by activation of the Nod-like receptor protein 3 (NLRP3) inflammasome, caspase-1 processing, and release of inflammatory factors including damage-associated molecular patterns and inflammatory cytokines including interleukin-1β (IL-1β) and IL-18.10 Although pyroptosis has critical roles in PD development,11 its interplay with oxidative stress and upstream regulatory factors in PD pathogenesis remain largely unexplored.
Nuclear factor E2-related factor 2 (Nrf2), a critical transcription factor belonging to the leucine zipper subfamily, is involved in various cellular processes including antioxidant responses, redox homeostasis, metabolic modulation, proliferation, DNA repair, autophagy, and mitochondrial biology. Nrf2 exerts these varied effects by mediating transcriptional regulation of an array of functional genes.12 Cellular abundance of Nrf2 protein is negatively regulated by ubiquitination catalyzed by the E3 ubiquitin ligase complex of Kelch ECH-associated protein 1 (Keap1) and cullin 3, and subsequent proteosome-mediated degradation.13 However, under certain stresses or pathological conditions, inactivation of Keap1 stabilizes Nrf2 protein, allowing it to enter the nucleus and bind antioxidant response elements to promote target gene expression.13 Specifically, the effects of Nrf2 during oxidative stress are mediated by its transcriptional modulation of heme oxygenase-1, NADH-quinone oxidoreductase 1, and other antioxidant genes.14 Importantly, recent research showed that Nrf2 signaling regulates mitochondrial functions and oxidative stress associated with PD pathogenesis, indicating Nrf2 activation might be a new method to prevent neurodegeneration during PD progression.15 Consistent with this notion, chlorpyrifos repressed Nrf2 signaling to promote oxidative stress and pyroptosis in a cellular PD model.16 However, how Nrf2 affects oxidative stress and pyroptosis in patients with PD requires further investigation.
In this study, we validated the pathological functions of Nrf2 signaling in modulation of oxidative stress and pyroptosis in cellular and animal models of PD. We also investigated potential roles of long non-coding RNAs (lncRNAs) in Nrf2-regulated oxidative stress and pyroptosis during PD development and progression. This study provides new perspectives on the molecular events driving neurodegeneration and cell death in PD pathogenesis, which might aid the development of new diagnostic and therapeutic methods for patients with PD.
Materials and Methods
Cellular PD Model
The cellular PD model was established by inducing the differentiation of PC12 cells into neural cells with nerve growth factor-β (NGF-β) as previously described,17 and then exposing differentiated cells to 6-hydroxydopamine hydrobromide (6-OHDA) according to a previous report.18 In brief, the PC12 cell line (CC9120) was obtained from Cellcook Company (Guangzhou, China), identified by the short tandem repeat profiling, and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% horse serum (Sigma-Aldrich, St Louis, USA), 5% fetal bovine serum (Gibco, Waltham, MA, USA) and 100 T/mL penicillin/streptomycin. Differentiation of PC12 cells in 12-well plates (105 cells/well) was subsequently induced by culturing cells in DMEM containing 1% horse serum and 50 ng/mL NGF-β continuously for 7 d, with the renewal of culture medium every 2 d. Microscopic observation, as well as the detection of tyrosine hydroxylase (TH) mRNA and protein levels, were used to assess PC12 differentiation. Subsequently, differentiated cells were exposed to 100, 300, 500, 700, or 900 μM 6-OHDA (H116; Sigma-Aldrich) for 24 h to induce neurotoxicity mimicking PD development.
Cell Transfection
To overexpress Nrf2 in cells, the coding sequence of rat Nrf2 was amplified by quantitative RT-PCR, ligated into the LV003 plasmid (ForeverGen, Guangzhou, China), and transfected into cultured PC12 cells using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. To silence AABR07032261.5 expression in cells, the si-AABR07032261-1 (5’- GATCCTCTGGGAATGGAAA-3’), si-AABR07032261-2 (5’-GGGAGGAGAGACAAAGGAA-3’), and the si-NC (5’-UUCUCCGAACGUGUCACGUTT-3’) were synthesized by GenePharma (Shanghai, China) and introduced into cultured PC12 cells as described above. Forty-eight hours after transfection, Nrf2 and AABR07032261-1 expression were validated by Western blot and quantitative RT-PCR, respectively.
Rat PD Model
Establishment of the rat PD model was performed by unilateral destruction of the nigral system with a 6-OHDA injection as previously described with minor modifications.19 In brief, male Wistar rats weighing 200–220 g and aged 10–12 w were purchased from Experimental Animal Center of Southern Medical University (Guangzhou, China) and maintained in cages at 20–23°C under a 12:12 light: dark cycle. All experiments were carried out in accordance with the Guide For the Care and Use of Laboratory Animals and were approved by the Ethics Committee for Animal Research of Third Affiliated Hospital of Sun Yat-Sen University. All rats were anesthetized by an intraperitoneal injection of xylazine hydrochloride and ketamine hydrochloride. Rat skulls were exposed by first making an incision and then drilling a hole in the left side of the substantia nigra. Subsequently, 4.8 µL of 6-OHDA solution (2.5 µg/µL; dissolved in saline containing 2 mg/mL ascorbic acid) was injected into the left substantia nigra (5.52 mm posterior to the anterior fonticulus, 1.4 mm on the left side of the center line, and 8.1 mm subdural) at a rate of 1 µL/min. Rats injected with the same volume of saline containing 2 mg/mL ascorbic acid were used as the sham group. Rat brain tissues were collected for assays 4 weeks after the model was established.
Nrf2 Overexpression in Rat Model
Eighteen male Wistar rats were randomly divided into three groups (n = 6 per group): Sham+Vector, PD+Vector, and PD+Nrf2. A recombinant adeno-associated virus vector (AAV) used to overexpress Nrf2 (AAV9-Nrf2) was constructed by WZ Biosciences (Jinan, China). Rats in the PD+Nrf2 group were injected with 1 µL of AAV9-Nrf2 (1.25×1014 vg/mL) in the left substantia nigra. Three weeks after virus injection, rats were subjected to PD model establishment as described above. Rats in the PD+vector group were injected with the same volume of empty AAV9 vector (AAV9-hSyn; 2×1013 vg/mL) and then subjected to PD model establishment. Rats in the Sham+Vector group were injected with the same volume of empty AAV9 vector and subjected to sham operation.
Neurobehavioral Functions
The muscle strength and equilibrium of rats were evaluated by the traction test as previously described.20 The spatial memory capability of rats was assessed by Morris Water Maze test as previously reported.21 Rat motor dysfunction was analyzed with an open field test (OFT) by recording the total distance traveled as described previously.21 The behaviors and spatial memory of rats were observed at days 3 and 5 after injection of 6-OHDA during PD model establishment. Morris Water Maze training was initiated on five days after 6-OHDA injection. Space navigation, space exploration, and OFT were performed on days 5, 6, and 7, respectively, after the initiation of Morris Water Maze training.
Immunocytochemistry
PC12 cell slides were rinsed with phosphate-buffered saline (PBS), fixed at room temperature with 4% paraformaldehyde in PBS (pH 7.4) for 13 min, and washed twice with pre-chilled PBS. Permeabilization was achieved by incubation with PBS solution containing 0.2% Triton X-100 for 8 min. After washing with PBS, PC12 cell slides were blocked by incubation with 1% bovine serum albumin (BSA) for 35 min, followed by incubation with diluted antibodies recognizing TH (25859-1-AP; Proteintech, Rosemont, IL, USA) for 1 h at room temperature. Following three washes with PBS, slides were incubated with the diluted Peroxidase-AffiniPure goat Anti-Rabbit IgG (H+L) (111-035-003; Jackson Immunoresearch, West Grove, PA, USA) for 1 h at room temperature, rinsed three times with PBS for 2 min, and incubated with DAB substrates. Finally, slides were counterstained with hematoxylin for 20 sec, rinsed twice with distilled water for 2 min, and dehydrated for 2 min each with 50%, 70%, 80%, 90%, 95% and 100% ethanol solutions. Subsequently, slides were immersed in xylene for 10 min, mounted, sealed, and observed by microscopy.
MTS
Rates of PC12 cell proliferation were determined quantitatively with an MTS Assay kit (Colorimetric; ab197010; Abcam, Cambridge, UK) according to the manufacturer’s protocol. Briefly, PC12 cells cultured in 96-well plates (5 × 103 cells/well; 100 μL) for 24 h were mixed with 10 μL of MTS reagent and cultured under normal conditions for another 2–3 h at 37°C. Finally, 96-well plates were briefly placed on a shaker and the absorbance of each well was measured at 490 nm (OD490). Three biological replicates were used for the quantitation and comparison of cell proliferation rates.
Flow Cytometry
The apoptosis of cultured PC12 cells was detected by flow cytometry using an Annexin V-APC/7-AAD Cell Apoptosis Kit (AT105; Multi Sciences Biotech, Hangzhou, China) according to the manufacturer’s instructions. Briefly, approximately 1×106 PC12 cells were collected, washed twice with pre-chilled PBS solution, and resuspended in 500 μL of Positive Control Solution. After incubation on ice for 30 min, cells were washed with PBS, resuspended in 1× binding buffer, stained in the dark with 5 μL of Annexin V-APC and 10 μL 7-AAD solutions for 6 min, and analyzed on a flow cytometer. ROS content of cultured PC12 cells were determined using a Reactive Oxygen Species Assay Kit (50101ES01; Yeason Biotech, Shanghai, China) according to the manufacturer’s instructions. Briefly, cultured PC12 cells at or near 60% confluency were incubated in the dark with DMEM containing 10 μM DCFH-DA at 37°C for 35 min, washed twice with serum-free medium, and evaluated by flow cytometry.
LDH Release
Lactate dehydrogenase (LDH) release by PC12 cells was evaluated using an LDH Cytotoxicity Assay Kit (C0017; Beyotime Biotech, Shanghai, China) according to the manufacturer’s instructions. Briefly, PC12 cells were added to 96-well plates and grown under normal conditions until they reached 80% confluency. Next, cells were washed with PBS and cultured with fresh medium for another 24 h. Subsequently, plates were centrifuged at 400 × g for 5 min and the resulting supernatants were collected. After placing supernatants in a new 96-well plate, 60 μL of fresh LDH detection solution was added and plates were incubated for 30 min at room temperature in the dark. Finally, plates were subjected to measurement of OD490 with a microplate reader.
Histological Evaluation
Distribution of neural cells in the rat hippocampus was assessed by microscopy following standard hematoxylin-eosin (H&E) staining. Nissl bodies in rat brain tissues were detected by a previously described Nissl staining method.22 Briefly, rat brain tissues were fixed in phosphate-buffered 10% formalin, dehydrated, embedded in paraffin, and sliced into sections (10 μm thick). After staining with Nissl dyes using Nissl Staining Solution (G1430; Solarbio Life Science, Beijing, China) according to the manufacturer’s instructions, sections were observed by microscopy. Apoptotic cells in the brain tissues of rats were evaluated using a One Step TUNEL Apoptosis Assay Kit (C1086; Beyotime Biotech, Shanghai, China) according to the manufacturer’s instructions. Briefly, rat brain tissue sections embedded in paraffin were dewaxed with xylene and ethanol, and incubated with 20 μg/mL Protease K solution for 25 min at 37°C. After washing three times with PBS, sections were incubated in the dark with 50 μL of terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) solution for 1 h at 37°C, washed with PBS, mounted, and observed on a fluorescence microscope.
Immunofluorescence
Levels of Nrf2 proteins in rat brain tissues were detected by immunofluorescence using antibodies targeting Nrf2 (ab137550; Abcam). Briefly, rat brain tissue slides were dewaxed, incubated with Protease K, blocked by incubation with 4% BSA for 50 min at room temperature, and incubated with Nrf2 antibodies (1:500) at 4°C overnight. Following three washes with PBS, rat brain sections were incubated in the dark with green fluorophore-conjugated secondary antibodies for 1.5 h at room temperature. After counterstaining with DAPI, sections were mounted and observed by fluorescence microscopy.
ELISA
The levels of interleukin (IL)-18 and IL-1β in cell cultures or rat serum samples were analyzed by enzyme-linked immunosorbent assay (ELISA) kits (IL-18: CSB-E04610r; IL-1β: CSB-E08055r; Cusabio Technology, Houston, TX, USA) according to the manufacturer’s instructions. Briefly, cell culture supernatants were collected, added into 96-well plates (100 μL per well), and incubated at 37°C for 2 h. Following the removal of any liquid, 100 μL of biotin antibody was added to each well for 1-h incubation at 37°C. After three washes, samples were incubated with 100 μL of horseradish peroxidase-conjugated avidin for 1 h at 37°C, washed five times, and incubated in the dark with 90 μL of TMB Substrate for 30 min at 37°C. Following incubations, samples were mixed with 50 μL of Stop Solution and the OD450 of each well was measured with a microplate reader.
RNA Sequencing and Bioinformatics
Differential expressions of lncRNA and mRNA in differentiated PC12 cells induced by Nrf2 overexpression were characterized by deep RNA sequencing. Briefly, total RNA was prepared from cultured PC12 cells using Trizol (15596026; Thermo Fisher Scientific) according to the manufacturer’s instructions. After RNA quality validation and removal of rRNA components from total RNA with a Human/Mouse/Rat rRNA Removal Kit (Illumina, San Diego, CA, USA), cDNA libraries were constructed using approximately 1.2 μg of total RNA sample from each group by reverse transcription with a TruSeq RNA LT Kit (Illumina). Subsequently, the obtained cDNA library of each group was subjected to sequencing using an Illumina HiSeq 2000 sequencing platform. Following adapter trimming and other processing, reads with a length of >35 bases were mapped to the reference genome database. Differential expressions of lncRNAs and mRNAs between the two groups were defined as the combination of a log2 Ratio (Nrf2/NC) value of ≥0.6 and a P-value ≤0.05 based on reads per kilobase million using Cuffdiff software. Hierarchical clustering and scatter plots of differentially expressed lncRNAs and mRNAs were constructed using R language. Enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for differentiation expressed lncRNAs and mRNAs was performed using the Database for Annotation, Visualization, and Integrated Discovery system. Finally, the interactions between lncRNA, microRNA, and mRNAs were predicted using miRanda 3.3a software (https://anaconda.org/bioconda/miranda).
Quantitative RT-PCR
Total RNA samples were extracted from cultured PC12 cells using Trizol according to the manufacturer’s instructions. Concentrations of total RNA samples were measured using a NanoDrop 2000 instrument (Thermo Fisher Scientific). cDNA was synthesized from 1.5 μg of RNA from each group by reverse transcription using a miScript II RT Kit (218160; QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. A QuantiTect SYBR Green PCR Kit (204143; QIAGEN) was used to quantitate expression levels by quantitative PCR. Relative expressions of mRNAs, microRNAs and lncRNAs were calculated using the standard 2−ΔΔCt method for at least three biological replicates, and calibrated using GAPDH or U6 levels. Primer sequences are listed in Table 1.
 |
Table 1 A List of Primers Used for qRT-PCR Assay |
Western Blot
Total proteins from cultured PC12 cells or rat brain tissues were extracted using cell lysis buffer for Western blotting (P0013; Beyotime Biotech) according to the manufacturer’s instructions. Protein concentrations were analyzed using the BCA method. About 25 μg of protein from each group was boiled in loading buffer for 5 min, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, and blotted onto polyvinylidene fluoride membranes. Subsequently, membranes were blocked by incubation with 5% BSA at room temperature for 2 h, incubated with diluted primary antibodies for 1–2 h at room temperature. After washing twice with Tris-buffered saline containing Tween, membranes were incubated with diluted secondary antibodies for 2 h at room temperature and developed using BeyoECL Plus Substrates (P0018S; Beyotime Biotech). The abundance of GAPDH was used as an internal standard. Antibodies used for Western blotting were anti-TH (25859-1-AP; Proteintech), anti-NRF2 (ab137550; Abcam), anti-p-NRF2 (#bs2013R; BIOSS, Beijing, China), anti-α-syn (4179; Cell Signaling Technology, Danvers, MA, USA), anti-caspase 1 (p10; 22915-1-AP, Proteintech), anti-NLRP3 (ab214185, Abcam), and anti-NLRP1 (4990, Cell Signaling Technology).
Statistical Analysis
Quantitative data from at least three biological replicates were presented as the mean ± standard deviation and analyzed using SPSS 20.0 software (Chicago, IL, USA). Differences between two or more groups were evaluated using the Student’s t-test and ANOVA, respectively. Significant differences were defined when P < 0.05.
Results
Enhanced Oxidative Stress and Pyroptosis in Cellular PD Model
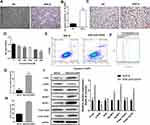
To investigate the association of oxidative stress and pyroptosis with PD pathogenesis, we established a cellular PD model by inducing PC12 cell differentiation and exposing differentiated cells to 6-OHDA. Following induction with NGF-β, we observed significant neurite outgrowth in PC12 cells compared with the negative control group (Figure 1A). Moreover, we observed that TH mRNA levels in PC12 cells treated with NGF-β were significantly higher compared with the negative control group (Figure 1B). Immunocytochemistry indicated that NGF-β treatment markedly increased the TH protein levels in PC12 cells (Figure 1C). These results indicated that treatment with NGF-β induced the differentiation of PC12 cells into neural cells. Next, we exposed differentiated PC12 cells to 100, 300, 500, 700, or 900 μM 6-OHDA to generate a cellular PD model. Our results show that 6-OHDA exposure significantly repressed the proliferation of NGF-β-treated PC12 cells in a concentration-dependent manner (Figure 1D). In addition, 6-OHDA exposure markedly increased the apoptosis of NGF-β-treated PC12 cells, as shown by flow cytometry (Figure 1E). Collectively, these results indicate the successful establishment of a cellular PD model using PC12 cells.
Flow cytometry indicated that ROS content in the cellular PD model (NGF-β+6-OHDA group) were substantially increased compared with the NGF-β group (Figure 1F). Levels of IL-1β in cultures of the cellular PD model group were also markedly elevated compared with the NGF-β group (Figure 1G). In addition, we observed that LDH in the cellular PD model group was substantially increased compared with the NGF-β group (Figure 1H). Western blotting demonstrated increased levels of the PD marker α-syn, as well as pyroptosis markers Nod-like receptor protein 1 (NLRP1), NLRP3, and caspase-1 (p10) in the cellular PD model group compared with the NGF-β group, indicating enhanced oxidative stress and pyroptosis (Figure 1I). In addition, the phosphorylation of Nrf2 proteins was markedly suppressed in the cellular PD model compared with the NGF-β group, suggesting Nrf2 regulates oxidative stress and pyroptosis during PD development (Figure 1I).
Increased Oxidative Stress and Pyroptosis in a Rat PD Model
To further explore the roles of oxidative stress and pyroptosis in PD pathogenesis, we established a rat PD model by inducing unilateral destruction of the nigral system with 6-OHDA. Rat muscle strength and equilibrium in the PD model group were markedly impaired compared with the sham group, as indicated by traction test scores (Figure 2A). Furthermore, numbers of H&E-stained neural cells and Nissl-stained neurons in the hippocampus were significantly reduced in the rat PD model group compared with the negative control group (Figure 2B). Moreover, TUNEL staining showed that numbers of apoptotic cells in the hippocampus of PD model group rats were markedly increased compared with the sham group (Figure 2C). These observations confirmed successful establishment of the rat PD model. Immunofluorescence demonstrated that nuclear distribution of Nrf2 proteins was significantly repressed in the rat PD model compared with the Sham group (Figure 2D). Furthermore, abundances of α-syn, NLRP1, NLRP3, and caspase-1 (p10) proteins were markedly increased, and phosphorylated Nrf2 proteins were markedly downregulated in the rat PD model compared with the sham group (Figure 2E and F). ELISA results demonstrated that IL-1β and IL-18 contents in the rat PD model group were markedly higher compared with the sham group (Figure 2G and H). Furthermore, LDH release in the rat PD model group was higher compared with the sham group (Figure 2I). These results demonstrated enhanced oxidative stress and pyroptosis in the rat PD model.
Nrf2 Overexpression Repressed Pyroptosis in a Cellular PD Model
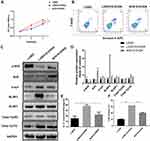
To verify the role of Nrf2 in PD development, we overexpressed Nrf2 in the cellular PD model established by 6-OHDA exposure as described above. Using an MTS assay, we observed that the proliferation rates of PC12 cells exposed to 6-OHDA were markedly increased by Nrf2 overexpression (Figure 3A). In addition, apoptosis of PC12 cells induced by 6-OHDA exposure was partially repressed by Nrf2 overexpression (Figure 3B). Moreover, the decrease of phosphorylated Nrf2 protein and increase of α-syn, NLRP1, NLRP3, and caspase-1 (p10) proteins in PC12 cells induced by 6-OHDA exposure were partially mitigated by Nrf2 overexpression (Figure 3C and D). Similarly, IL-1β contents of the cultured PC12 cells exposed to 6-OHDA were significantly upregulated by Nrf2 overexpression (Figure 3E). In addition, Nrf2 overexpression significantly suppressed LDH release by PC12 cells exposed to 6-OHDA (Figure 3F). These results indicated that Nrf2 overexpression inhibited pyroptosis in the cellular model of PD.
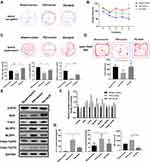
Nrf2 Overexpression Alleviated Motor Dysfunction and Repressed Pyroptosis in a Rat PD Model
To further validate the pathogenic roles of Nrf2, we analyzed the influence of Nrf2 overexpression on motor dysfunction and pyroptosis in the rat PD model. Spatial memory ability was evaluated using the Morris Water Maze test (Figure 4A–C). Escape latencies in the PD model were significantly increased compared with the sham group, and Nrf2 overexpression remarkably shortened escape latencies in the PD model group (Figure 4A and B). In terms of space exploration, the PD model group showed marked decrease in time spent in the target quadrant, mean speed, and numbers of platform crossings compared with the sham group; however, Nrf2 overexpression rescued decreases in time spent in the target quadrant and numbers of platform crossings in the PD model group (Figure 4C). OFT results demonstrated a significant decrease in total distances traveled by rats in the PD model group compared with the sham group, but Nrf2 overexpression increased the total distance traveled in the PD model (Figure 4D). Compared with the sham group, expression of α-syn, NLRP3, NLRP1, and Casp-1 (p10) proteins were upregulated and Nrf2 phosphorylation was repressed in the rat PD model, but these features were significantly mitigated by Nrf2 overexpression (Figure 4E and F). Levels of IL-1β, IL-18, and LDH in the serum of PD model rats were markedly increased compared with sham group rats, but Nrf2 overexpression downregulated the IL-1β, IL-18 and LDH levels in the PD model (Figure 4G). These observations demonstrate that Nrf2 overexpression significantly inhibited motor dysfunction and pyroptosis in the rat PD model.
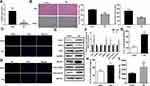
Nrf2 Overexpression Promoted Differential lncRNA Expression in the Cellular PD Model
To analyze the molecular events that mediating Nrf2-regulated pyroptosis in PD, we characterized differentially expressed lncRNAs and mRNAs in the cellular PD model induced by Nrf2 overexpression. Overall, 53 differentially expressed lncRNAs, including 26 upregulated and 27 downregulated lncRNAs, were identified in the Nrf2-overexpressing cellular PD model group compared with the model group (Figure 5A). Furthermore, the expressions of 1907 mRNAs in the cellular PD model were significantly altered by Nrf2 overexpression, including 889 upregulated and 1018 downregulated genes (Figure 5B). KEGG analysis demonstrated that differentially expressed genes related to Nrf2 in the cellular PD model group were significantly enriched in multiple signaling pathways, including the tumor necrosis factor α (TNF-α), Ras, mitogen-activated protein kinase (MAPK), apoptosis, and Toll-like receptor signaling pathways (Figure 5C). Of these, the MAPK and Toll-like receptor signaling pathways are closely involved in pyroptosis, further supporting regulation of pyroptosis by Nrf2 (Figure 5D and E).
We found that differentially expressed lncRNAs were predicted to form interaction networks with multiple microRNAs and functional genes involved in NF-κB, apoptosis, Toll-like receptor, and neurotrophin signaling pathways (Figure 5F). Validation of expression of multiple lncRNAs by quantitative RT-PCR confirmed significantly increased expressions of AABR07032261.5 and AC108574.2 in the cellular PD model following Nrf2 overexpression, consistent with the RNA sequencing results (Figure 5G). We next validated altered expression of eight microRNAs predicted to interact with AABR07032261.5 and AC108574.2 in the PD models. Our results revealed significant decreases in the expressions of rno-miR-1-3p, rno-miR-124-3p, and rno-miR-34a-5p, and an increase in rno-miR-98-5p in the cellular PD model following Nrf2 overexpression (Figure 5H). These results suggest that differential expressions of lncRNAs and their interactions with microRNAs and mRNAs might have a role in Nrf2-regulated motor dysfunction and pyroptosis during PD development.
AABR07032261.5 Knockdown Promoted Pyroptosis in the Cellular PD Model
To confirm the role of AABR07032261.5 in PD pathogenesis, we silenced AABR07032261.5 expression in the cellular PD model by transfection with siRNAs targeting AABR07032261.5. Compared with the negative control, AABR07032261.5 expression in the cellular PD model was significantly downregulated (Figure 6A). In the cellular PD model with silenced AABR07032261.5 expression, we observed significantly suppressed proliferation rates compared with the negative control group (Figure 6B). In contrast, flow cytometry indicated that apoptosis in the cellular PD model was markedly increased by the silencing of AABR07032261.5 expression (Figure 6C). Furthermore, secretion of IL-1β and IL-18 in the cellular PD model was markedly increased by silencing of AABR07032261.5 (Figure 6D and E). Western blotting revealed that α-syn, NLRP1/3, and Casp-1 (p10) protein levels in the cellular PD model were markedly increased by AABR07032261.5 knockdown (Figure 6 and G). Furthermore, expression of the predicted target microRNA rno-miR-124-3p was significantly increased by AABR07032261.5 knockdown in the cellular PD model (Figure 6H). In addition, the expression level of Stat3, a target gene of rno-miR-124-3p, was repressed in the cellular PD model by AABR07032261.5 knockdown (Figure 6I). These results indicated that AABR07032261.5 repressed pyroptosis during PD pathogenesis, potentially by modulating the downstream rno-miR-124-3p/Stat3 axis.
Discussion
Clinical management of patients with PD greatly depends on elucidation of underlying molecular pathological processes. The pathological progression of PD is closely controlled by ROS biosynthesis and oxidative stress in the substantia nigra, and mediated by various cell death pathways including apoptosis, necroptosis, autophagic cell death, and pyroptosis.6,9,11 However, the molecular mechanisms regulating pyroptosis during PD development are poorly understood. In this study, we investigated roles of elevated oxidative stress and pyroptosis accompanying marked suppression of Nrf2 in cellular and animal models of PD. Moreover, we identified critical roles of Nrf2 in repressing pyroptosis and suppressing motor dysfunction during PD pathogenesis, by overexpressing Nrf2 in cellular and animal PD models. Furthermore, we used deep sequencing to evaluate how Nrf2 modulated expression of various lncRNAs in the PD model. Our results indicate that lncRNA AABR07032261.5 repressed pyroptosis during PD development, possibly by targeting microRNAs to regulate Stat3 and other signaling pathways. These findings reveal new insights into the molecular mechanisms of pyroptosis regulation underlying PD initiation and progression.
Pyroptosis is a critical cellular pathogenic mechanism associated with the development of multiple neurodegenerative disorders, including PD.11,23 We observed a significant increase in pyroptosis marker gene expression in the cellular and animal PD models, confirming the involvement of pyroptosis in PD pathogenesis. As observed for other types of cell death, initiation of pyroptosis was closely associated with disturbances in cellular redox homeostasis and resultant oxidative stress.24,25 Recent research has implicated pyroptosis in various pathogenic conditions including myocardial ischemia‑reperfusion injury,26 osteogenic dysfunction and periodontitis,24 cancer development,25 cardiomyocyte injury,27 and renal ischemia/reperfusion injury.28 For example, oxidative stress-mediated pyroptosis promoted the effect of enhancer of zeste homolog 2 in regulating renal ischemia/reperfusion injury, which was associated with its ability to inhibit NADPH oxidase 4-dependent ROS production.29 However, little is known about the association between pyroptosis and PD development. In this study, we observed that ROS content in the cellular PD model was greatly increased and positively correlated with enhanced pyroptosis, suggesting oxidative stress is a major causative factor driving pyroptosis during PD pathogenesis. In addition, we discovered that upregulation of caspase-1 (p10) could be rescued by Nrf2 overexpression in PC12 cells exposed to 6-OHDA and PD model rats. Silencing of AABR07032261.5 could also regulate caspase-1 (p10) in the cellular PD model, although total caspase-1 protein (p45) was unchanged. Caspase‑1 can be activated by the NLRP3 inflammasome formation during NLRP3-regulated pyroptosis,30 whereas cleaved caspase‑1 (p10) is generated by hydrolysis during activation of caspase-1.31 Although total expression of caspase-1 (P45) did not change, regulation of caspase-1 (p10) levels indicates a change in pyroptosis.
Nrf2 is a major regulator of cellular redox status and oxidative stress, which are implicated in PD pathogenesis.12,15 Furthermore, the Nrf2 signaling pathway is an essential modulator of pyroptosis under various contexts.28,32 Activation of the Nrf2 signaling pathway inhibited pyroptosis underlying the development of renal ischemia-reperfusion injury in a mouse model.28 In addition, enhancement of Nrf2 signaling with phenolic acid suppressed pyroptosis and alleviated Gouty arthritis development.32 Moreover, inhibition of Nrf2 signaling by chlorpyrifos enhanced oxidative stress and pyroptosis in a cellular PD model.16 The relationship between Nrf2 signaling and pyroptosis in PD development requires further validation using animal models or clinical investigations. In the present study, we showed that Nrf2 expression was repressed in the cellular and animal PD models evaluated, but Nrf2 overexpression inhibited pyroptosis marker gene expression in these models. These findings confirm a regulatory role of Nrf2 in pyroptosis during PD pathogenesis.
lncRNA is a type of large non-coding RNA (>200 nucleotides) that regulates gene transcription.33 A recent study showed that lncRNA-regulated gene transcription is a critical molecular mechanism associated with PD development and progression.34 Specifically, lncRNAs were reported to be implicated in multiple pathological events of PD, including α-syn aggregation, apoptosis of dopaminergic neurons, mitochondrial dysfunction, autophagy, and neuroinflammation.34 In addition, accumulating evidence indicates important roles of Nrf2 as an upstream regulator of lncRNA expression and function.35 Several lncRNAs, such as smoking-and-cancer-associated lncRNA-1 and nuclear-LUCAT1, are regulated by the Nrf2 signaling pathway.35 However, the interplay between Nrf2 and lncRNAs in PD remains poorly understood. In this study, we comprehensively characterized differentially expressed lncRNAs and mRNAs caused by Nrf2 overexpression in a cellular PD model. We found that the lncRNA AABR07032261.5 suppressed pyroptosis during PD development. Also, we found that AABR07032261.5 regulated rno-miR-124-3p to modulate Stat3 expression in the PD model. Recent research demonstrated that targeting of Stat3 expression by miR-124-3 is closely involved in the progression of PD and other neurological disorders.36,37 Collectively, these results indicate that Nrf2-regulated AABR07032261.5 expression plays essential roles in modulating pyroptosis in PD pathogenesis, possibly by regulating downstream players such as rno-miR-124-3p and Stat3.
In summary, we found that Nrf2 could suppress the development and progression of PD by promoting expression of lncRNA AABR07032261.5, which inhibited pyroptosis. These findings provide new insights into the molecular mechanisms underlying PD development and progression, especially in terms of epigenetic modulation, that can be explored as new targets for PD diagnosis and treatment. However, because of the limitations of this study (for instance, 6-OHDA-mediated oxidative stress responses were not confirmed in the rat PD model), it is necessary to further explore the role and molecular mechanism of AC108574.2 in pyroptosis associated with PD.
Abbreviations
6-OHDA, 6-Hydroxydopamine hydrobromide; AAV, adeno-associated virus vector; BSA, bovine serum albumin; DMEM, Dulbecco’s modified Eagle’s medium; ELISA, enzyme linked immunosorbent assay; H&E, hematoxylin-eosin; KEEG, Kyoto Encyclopedia of Genes and Genomes; IL-1β, interleukin-1β; Keap1, Kelch ECH-associated protein 1; LDH, lactate dehydrogenase; lncRNAs, long non-coding RNAs; LRRK2, leucine-rich repeat kinase 2; MAPK, mitogen-activated protein kinase; NGF-b, nerve growth factor b; NLRP1, Nod-like receptor protein 1; NLRP3, Nod-like receptor protein 3; Nrf2, nuclear factor E2-related factor 2; PBS, phosphate-buffered saline; PD, Parkinson’s disease; ROS, reactive oxygen species; TH, tyrosine hydroxylase; TNF-α, tumor necrosis factor α; TUNEL, terminal deoxynucleotidyl transferase dUTP nick-end labeling; α-syn, alpha-synuclein.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of Guangdong Province (2018A0303130307), Guangdong Basic and Applied Basic Research Foundation (2019A1515011739) and the Fundamental Research Funds for the Central Universities (19ykpy27). We thank Liwen Bianji (Edanz) (www.liwenbianji.cn/) for basic language editing of a draft of this manuscript.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015;386:896–912. doi:10.1016/s0140-6736(14)61393-3
2. Raza C, Anjum R, Shakeel NUA. Parkinson’s disease: mechanisms, translational models and management strategies. Life Sci. 2019;226:77–90. doi:10.1016/j.lfs.2019.03.057
3. Ascherio A, Schwarzschild MA. The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol. 2016;15:1257–1272. doi:10.1016/s1474-4422(16)30230-7
4. Emamzadeh FN, Surguchov A. Parkinson’s disease: biomarkers, treatment, and risk factors. Front Neurosci. 2018;12:612. doi:10.3389/fnins.2018.00612
5. Puschmann A. New genes causing hereditary Parkinson’s disease or Parkinsonism. Curr Neurol Neurosci Rep. 2017;17:66. doi:10.1007/s11910-017-0780-8
6. Trist BG, Hare DJ, Double KL. Oxidative stress in the aging substantia nigra and the etiology of Parkinson’s disease. Aging Cell. 2019;18:e13031. doi:10.1111/acel.13031
7. Puspita L, Chung SY, Shim JW. Oxidative stress and cellular pathologies in Parkinson’s disease. Mol Brain. 2017;10:53. doi:10.1186/s13041-017-0340-9
8. Zhong H, Hao L, Li X, Wang C, Wu X. Anti-inflammatory role of trilobatin on lipopolysaccharide-induced acute lung injury through activation of ampk/gsk3β-nrf2 pathway. Signa Vitae. 2020;16:160–166.
9. Morris G, Walker AJ, Berk M, Maes M, Puri BK. Cell death pathways: a novel therapeutic approach for neuroscientists. Mol Neurobiol. 2018;55:5767–5786. doi:10.1007/s12035-017-0793-y
10. Frank D, Vince JE. Pyroptosis versus necroptosis: similarities, differences, and crosstalk. Cell Death Differ. 2019;26:99–114. doi:10.1038/s41418-018-0212-6
11. Wang S, Yuan YH, Chen NH, Wang HB. The mechanisms of NLRP3 inflammasome/pyroptosis activation and their role in Parkinson’s disease. Int Immunopharmacol. 2019;67:458–464. doi:10.1016/j.intimp.2018.12.019
12. de la Vega MR, Chapman E, Zhang DD. NRF2 and the Hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi:10.1016/j.ccell.2018.03.022
13. Tonelli C, Chio IIC, Tuveson DA. Transcriptional regulation by Nrf2. Antioxid Redox Signal. 2018;29:1727–1745. doi:10.1089/ars.2017.7342
14. Loboda A, Damulewicz M, Pyza E, Jozkowicz A, Dulak J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell Mol Life Sci. 2016;73:3221–3247. doi:10.1007/s00018-016-2223-0
15. Ammal Kaidery N, Ahuja M, Thomas B. Crosstalk between Nrf2 signaling and mitochondrial function in Parkinson’s disease. Mol Cell Neurosci. 2019;101:103413. doi:10.1016/j.mcn.2019.103413
16. Zhao MW, Yang P, Zhao LL. Chlorpyrifos activates cell pyroptosis and increases susceptibility on oxidative stress-induced toxicity by miR-181/SIRT1/PGC-1alpha/Nrf2 signaling pathway in human neuroblastoma SH-SY5Y cells: implication for association between chlorpyrifos and Parkinson’s disease. Environ Toxicol. 2019;34:699–707. doi:10.1002/tox.22736
17. Rostamian Delavar M, Baghi M, Safaeinejad Z, et al. Differential expression of miR-34a, miR-141, and miR-9 in MPP+-treated differentiated PC12 cells as a model of Parkinson’s disease. Gene. 2018;662:54–65. doi:10.1016/j.gene.2018.04.010
18. Luo Y, Jiang Y, He Y, et al. Vina-ginsenoside R4 from panax ginseng leaves alleviates 6-OHDA-induced neurotoxicity in PC12 cells via the PI3K/Akt/GSK-3β signaling pathway. J Agric Food Chem. 2020;68:15239–15248. doi:10.1021/acs.jafc.0c06474
19. Ghahari L, Safari M, Rahimi Jaberi K, et al. Mesenchymal stem cells with granulocyte colony-stimulating factor reduce stress oxidative factors in Parkinson’s disease. Iran Biomed J. 2020;24:89–98. doi:10.29252/ibj.24.2.89
20. Li J, Dai X, Zhou L, Li X, Pan D. Edaravone plays protective effects on LPS-induced microglia by switching M1/M2 phenotypes and regulating NLRP3 inflammasome activation. Front Pharmacol. 2021;12:691773. doi:10.3389/fphar.2021.691773
21. Boccella S, Iannotta M, Cristiano C, et al. Treatment with 2-pentadecyl-2-oxazoline restores mild traumatic brain injury-induced sensorial and neuropsychiatric dysfunctions. Front Pharmacol. 2020;11:91. doi:10.3389/fphar.2020.00091
22. Gao Y, Cui M, Zhong S, et al. Dihydroartemisinin ameliorates LPS-induced neuroinflammation by inhibiting the PI3K/AKT pathway. Metab Brain Dis. 2020;35:661–672. doi:10.1007/s11011-020-00533-2
23. Li Q, Shi N, Cai C, et al. The role of mitochondria in pyroptosis. Front Cell Dev Biol. 2020;8:630771. doi:10.3389/fcell.2020.630771
24. Liu S, Du J, Li D, et al. Oxidative stress induced pyroptosis leads to osteogenic dysfunction of MG63 cells. J Mol Histol. 2020;51:221–232. doi:10.1007/s10735-020-09874-9
25. Le X, Mu J, Peng W, et al. DNA methylation downregulated ZDHHC1 suppresses tumor growth by altering cellular metabolism and inducing oxidative/ER stress-mediated apoptosis and pyroptosis. Theranostics. 2020;10:9495–9511. doi:10.7150/thno.45631
26. Zhou Y, Li KS, Liu L, Li SL. MicroRNA132 promotes oxidative stress induced pyroptosis by targeting sirtuin 1 in myocardial ischaemia reperfusion injury. Int J Mol Med. 2020;45:1942–1950. doi:10.3892/ijmm.2020.4557
27. Fan X, Zhan E, Yao Y, et al. MiR-599 protects cardiomyocytes against oxidative stress-induced pyroptosis. Biomed Res Int. 2021;2021:3287053. doi:10.1155/2021/3287053
28. Diao C, Chen Z, Qiu T, et al. Inhibition of PRMT5 attenuates oxidative stress-induced pyroptosis via activation of the Nrf2/HO-1 signal pathway in a mouse model of renal ischemia-reperfusion injury. Oxid Med Cell Longev. 2019;2019:2345658. doi:10.1155/2019/2345658
29. Liu H, Chen Z, Weng X, et al. Enhancer of zeste homolog 2 modulates oxidative stress-mediated pyroptosis in vitro and in a mouse kidney ischemia-reperfusion injury model. FASEB J. 2020;34:835–852. doi:10.1096/fj.201901816R
30. Hughes MM, O’Neill LAJ. Metabolic regulation of NLRP3. Immunol Rev. 2018;281:88–98. doi:10.1111/imr.12608
31. Qiu Z, He Y, Ming H, et al. Lipopolysaccharide (LPS) aggravates high glucose- and hypoxia/reoxygenation-induced injury through activating ROS-dependent NLRP3 inflammasome-mediated pyroptosis in H9C2 cardiomyocytes. J Diabetes Res. 2019;2019:8151836. doi:10.1155/2019/8151836
32. Lin Y, Luo T, Weng A, et al. Gallic acid alleviates gouty arthritis by inhibiting NLRP3 inflammasome activation and pyroptosis through enhancing Nrf2 signaling. Front Immunol. 2020;11:580593. doi:10.3389/fimmu.2020.580593
33. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110. doi:10.1126/sciadv.aao2110
34. Lv Q, Wang Z, Zhong Z, Huang W. Role of long noncoding RNAs in Parkinson’s disease: putative biomarkers and therapeutic targets. Parkinsons Dis. 2020;2020:5374307. doi:10.1155/2020/5374307
35. Bhattacharjee S, Li J, Dashwood RH. Emerging crosstalk between long non-coding RNAs and Nrf2 signaling. Cancer Lett. 2020;490:154–164. doi:10.1016/j.canlet.2020.07.011
36. Vuokila N, Aronica E, Korotkov A, et al. Chronic regulation of miR-124-3p in the perilesional cortex after experimental and human TBI. Int J Mol Sci. 2020;21. doi:10.3390/ijms21072418
37. Geng L, Liu W, Chen Y. miR-124-3p attenuates MPP (+)-induced neuronal injury by targeting STAT3 in SH-SY5Y cells. Exp Biol Med. 2017;242:1757–1764. doi:10.1177/1535370217734492
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.