Back to Journals » Drug Design, Development and Therapy » Volume 11
Novel production method of innovative antiangiogenic and antitumor small peptides in Escherichia coli
Authors Setrerrahmane S, Yu J, Hao J, Zheng H, Xu H
Received 14 March 2017
Accepted for publication 15 September 2017
Published 8 November 2017 Volume 2017:11 Pages 3207—3220
DOI https://doi.org/10.2147/DDDT.S136957
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Frank Boeckler
Sarra Setrerrahmane,1 Jian Yu,1 Jingchao Hao,1,2 Heng Zheng,3 Hanmei Xu1,3
1The Engineering Research Center of Peptide Drug Discovery and Development, China Pharmaceutical University, Nanjing, Jiangsu, 2College of Pharmacy & the Provincial Key Laboratory of Natural Drug and Pharmacology, Kunming, Yunnan, 3State Key Laboratory of Natural Medicines, Ministry of Education, China Pharmaceutical University, Nanjing, Jiangsu, People’s Republic of China
Background: Developing innovative drugs with potent efficacy, specificity, and high safety remains an ongoing task in antitumor therapy development. In the last few years, peptide drugs have become attractive agents in cancer therapy. HM-3, mainly with antiangiogenic effect, and AP25, with an additional antiproliferative effect, are two peptides designed in our laboratory targeting αvβ3 and α5β1 integrins, respectively. The low molecular weight of the two peptides renders their recombinant expression very difficult, and the complicated structure of AP25 makes its chemical synthesis restricted, which presents a big challenge for its development.
Methods: Bifunctional peptides designed by the ligation of HM-3 and AP25, using linkers with different flexibility, were prepared using recombinant DNA technology in Escherichia coli. The fusion peptides were expressed in a modified auto-induction medium based on a mixture of glucose, glycerol, and lactose as carbon substrates and NH4+ as nitrogen source without any amino acid or other elements. Subsequently, the antiangiogenic, antiproliferative, and cell adhesion assays were conducted to evaluate the bioactivity of the two fusion peptides.
Results: The peptides were successfully expressed in a soluble form without any induction, which allows the culture to reach higher cell density before protein expression occurs. Human umbilical vein endothelial cell migration assay and chick embryo chorioallantoic membrane assay showed, at low doses, a significantly increased antiangiogenic effect (>75%) of the purified products compared with the single molecules. Meanwhile, MTT assay confirmed their enhanced antitumor activity against gastric cancer cell line MGC-803; however, no significant effect was observed on hepatoma HepG2 cells and no cytotoxicity on normal human lens epithelial cell SRA01/04 and human epithelial esophageal cells.
Conclusion: Bifunctional molecules with antiangiogenic and antiproliferative effects were obtained by using this technique, which presents an alternative for small peptide production, instead of the conventional chemical method. The increased molecular weight facilitates the peptide expression with a simultaneous improvement in their stability and biological activity.
Keywords: innovative drugs, bifunctional peptides, linkers, auto-induction, antiangiogenic, antitumor
Introduction
Peptides are recognized for being, simultaneously, highly selective, efficacious, relatively safe, and well tolerated. Consequently, they have gained a wide range of applications in medicine and biotechnology during the past decade, and ~140 peptide therapeutics are currently being evaluated in clinical trials.1 Therefore, peptide production in a highly purified and well-characterized form has become a major task in pharmaceutical research and industry.
Traditional technologies, including conventional chemical synthesis and recombinant DNA technology, have been utilized for innovative peptide drug development. The first approach is labor-intensive and has a relatively high manufacturing cost. Nevertheless, the recombinant approach remains the most attractive due to low cost, high productivity, and rapid process.2 Bacterial, especially Escherichia coli–based systems remain the preferable choice for recombinant protein production both on laboratory scale and in industry because of their ability to grow rapidly and at a high density on inexpensive substrates, the well-characterized genetics, and the availability of an increasingly large number of cloning vectors and mutant host strains.3
HM-3 and AP25 are two polypeptides designed in our laboratory by introducing some modification on endostatin-derived peptide fragments. HM-3 is an 18 amino-acid arginine–glycine–aspartic acid (RGD)-modified polypeptide targeting the αvβ3 integrin. Previous experimental results in vitro indicated that HM-3 could significantly inhibit the migration of endothelial cells but had no inhibitory effect on tumor cells; moreover, in vivo data showed that it could inhibit tumor growth in several animal models. These results indicate that its antitumor effect was accomplished by inhibiting the migration of endothelial cells and angiogenesis.4,5 HM-3 became an attractive innovative peptide for clinical application in cancer therapy, and it has been approved by the China Food and Drug Administration as one of the Chinese national category 1.1 antitumor drugs for phase 1 clinical experiments. AP25 is a 25-amino acid antitumor peptide; due to the cyclic RGD motif, this peptide presents an improved targeting capability of α5β1 and αvβ3 integrins-expressing tumor cells and endothelial cells. Previous in vitro and in vivo experiments revealed that this integrin antagonist peptide has a remarkable antitumor effect on different cancer types;6 however, the structure of this attractive peptide makes it difficult to be synthesized, and this became a challenge in its research development.
Herein, we added the antiproliferative effect to the antiangiogenic one of the HM-3 peptide (a novel antiangiogenic peptide, but it lacks the antiproliferative effect) by its fusion to the AP25 (difficult to be synthesized) peptide using two different linkers to test their contribution in the bioactivity of the final product (Figure 1). In addition, we developed a novel alternative approach for efficient and cost-effective production of small peptides using the fusion technique and recombinant DNA technology in E. coli instead of the traditional chemical synthesis.
Materials and methods
Strains, plasmids, cell lines, and reagents
E. coli strains DH5α and BL21 (DE3) were used as hosts for gene cloning and protein expression, respectively. pGEX-4t-1 and Glutathione Sepharose™ 4B beads were purchased from GE Healthcare Life Sciences (Logan, UT, USA) and used for gene expression and protein purification, respectively. Human umbilical vein endothelial cells (HUVECs; primary cells) were purchased from AllCells (Shanghai, People’s Republic of China); all other cell lines were obtained from the American Type Culture Collection (Shanghai, People’s Republic of China) and maintained in DMEM with 10% fetal bovine serum (FBS; SiJiQing, Hangzhou, People’s Republic of China) and 1% of penicillin–streptomycin solution (Hyclon; GE Healthcare Life Sciences). Peptides HM-3 and AP25 were synthesized by GL Biochem Ltd. (Shanghai, People’s Republic of China) with a purity of >95%. All other biochemicals and chemicals used in the experiment were of analytical grade.
Peptide preparation
Plasmids pGEXF and pGEXR were constructed to encode the two fusion peptides as follows. First, DNA sequences encoding the two peptides were designed based on their amino acid sequences and E. coli codon bias and amplified by a series of polymerase chain reactions (PCRs) using eight primers. Then, the obtained fragments were verified by DNA sequencing and inserted between EcoRI and BamH1 sites of pGEX-4t-1. The obtained plasmids were transformed into E. coli BL21 (DE3) for peptides expression using different media. Harvested cell pellets (8 g/L for each one) were resuspended in 80 mL of lysis buffer (PBS, pH =7.4 with 0.1% [v/v] Triton X-100) and sonicated on ice using JY92-IIN ultrasonic homogenizer (Ningbo Scientz Biotechnology, Ningbo, People’s Republic of China) at 200 W for 5 minutes (30 rounds of 5-second sonication with 5-second intervals). The soluble fractions were isolated by centrifugation at 13,000× g at 4°C for 30 minutes and loaded onto Glutathione Sepharose™ 4B beads pre-equilibrated with equilibration buffer (PBS). After washing with lysis buffer and thrombin cleavage at room temperature for 6 hours, the desired peptides were eluted from the column and further purified to be characterized by tricine–sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis under nonreducing (DL-Dithiothreitol [-DTT]) conditions using anti-HM-3 antibody.
HUVEC migration assay
Transwell® chambers (upper chambers, 8.0 μm pore size) were coated with a thin film of growth factor-reduced Matrigel® matrix diluted with a serum-free extracellular matrix (ECM 1:2; Becton Dickinson Lab Ware, Bedford, MA, USA), and after gel polymerization, transwells were placed in a 24-well plate containing invasion medium (ECM with 10% FBS and 1% endothelial cell growth supplement [ECGS]). HUVECs were digested and resuspended in a serum-free ECM and were added to the upper compartment at a density of 1×104 cells/well simultaneously with different concentrations of the tested peptides. The plates were incubated at 37°C for 24 hours. The migrated cells were fixed, stained, and quantified.
Chick embryo chorioallantoic membrane (CAM) assay
CAM assay was performed to evaluate the antiangiogenic activity in vivo.7 Fertilized eggs were kept at 90% of humidity and 37°C. On day 6 of fertilization, a window was made on the top of each egg. Test substances were spotted onto sterilized Whatman® filter paper disks and applied to the surfaces of the growing CAMs above the dense subectodermal plexus on day 8. On day 10 of incubation, CAMs were observed and photographed.
Cell proliferation assay
The cell proliferation assay was conducted on several cancer cell lines, MGC-803 (high expression levels of integrins α5β1 and αvβ3) and HepG-2 (low expression levels of integrins β1 and β3), or on normal cell lines, the human lens epithelial cell (HLEC) SRA01/04 and the human epithelial esophageal cells (HEECs) to test the cytotoxicity. Cells were plated at a density of 3×103 cells/well in 96-well plates and then treated with various concentrations of peptide drugs for 48 hours; 20 μL of MTT solution (0.5 mg/mL in PBS [w/v]) was added to each well, and the plates were incubated for 4 hours at 37°C. After the aspiration of the supernatant, formazan crystals were solubilized in 150 μL dimethyl sulphoxide (DMSO), and the absorbance at 570 nm was measured by using a Multiskan Plus microplate reader (Thermo Electron Corporation, Waltham, MA, USA). The results were expressed as the percentage of the control, considered as 100%.
Cell adhesion of HUVECs
The cell adhesion test was conducted as described previously.8 A 96-well plate was coated with 100 μL of peptides solution (150 μg/mL dissolved in PBS) overnight at 4°C. The solution was then removed, and the wells were blocked with 1% bovine serum albumin (BSA) in PBS for 6 hours at 22°C. At the same time, HUVECs were serum-starved for 4 hours, then were collected, and incubated with integrins αv, β3, α5, β1, αv + β3, and α5 + β1 antibodies (Cell Signaling Technology, Danvers, MA, USA) at 1:400 (v/v) at 4°C 2 hours. The BSA was then removed, and 5×105 cells were added to each well and incubated for 90 minutes. After incubation, nonadherent cells were removed, and adherent cells were washed with PBS, resuspended in 1% SDS solution, and the absorbance at 620 nm was measured. Adhesion was reported as the percentage relative to the amount of cells that adhered in wells bound with BSA.
Three-dimensional modeling
The three-dimensional structure of the peptides was modeled using the homology model method of Molecular Operating Environment (MOE) 2009 (provided by Chemical Computing Group Inc., Montreal, QC, Canada). The structure of residues 1 to 11 of the peptides was taken from 1FUL.pdb9,10 and combined with the modeled structure of the remaining parts using chain A of human endostatin (PDB ID: 1BNL) as template. The force field was set to MMFF94x, and C-terminal outgap modeling was carried out. Rough models were refined with MOE/Minimization program using MMFF94x force field.11
Four steps of energy minimization were performed with a gradual relaxation of restrictions for appropriate atoms. In the first and second steps, the nonhydrogen non–lone pair atoms and the backbone atoms were frozen, respectively. In the third step, the backbone atoms were restricted with a force constant of 300, which was comparable to the strength of a bond in MOE. In the last step, all the restrictions were removed. In all the steps, energy minimization was set to be terminated when the root mean square gradient was <0.01. The resulting structures were evaluated using MOE/protein geometry.
Statistical analysis
One-way analysis of variance with Tukey posttest was used to compare all groups with the control groups. Data are expressed as means ± SD and analyzed with Prism software (GraphPad Software, Inc., La Jolla, CA, USA), and P-values <0.05 were considered to be statistically significant (*P<0.05; **P<0.01; ***P<0.001).
Results
Preparation of soluble recombinant fusion peptides F and R
HM-3 and AP25 encoding sequences, together with the linker sequences, were inserted into the multicloning site of the pGEX-4t-1 vector to construct pGEXF and pGEXR plasmids (F referring to the flexible linker (GGGGS)3 and R to the rigid one [A(EAAAK)4A]); their expression was under the control of the inducible T7 promoters.
To select the optimal expression conditions, several fermentation media12 were tested to compare their effects on cell growth and peptide production at shake flask level (Table 1). For Luria–Bertani medium, the expression was induced by isopropyl β-D-thiogalactopyranoside (IPTG) when the bacterial OD at 600 nm (OD600) reached 0.6~0.7, for the auto-induction media (Media 2 and 3), and the bacterial cells were cultured at 37°C for 12 hours.
  | Table 1 Expression medium composition |
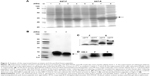
SDS-PAGE analysis of the supernatants indicated that the fusion proteins were successfully expressed in a soluble form in the cytoplasm of BL21 (DE3). The highest expression level was observed when using the modified auto-induction medium (Medium 3), and it accounted for ~40% of the total cellular protein by Gel Image System (Tanon Co., Shanghai, People’s Republic of China) scanning analysis, as shown in Figure 2A. Tricine–SDS-PAGE of the final product showed a single band (Figure 2B) revealing the high purity of the obtained peptides that was further verified by reversed-phase high-performance liquid chromatography analysis and accounted for >95% (Figure S1). Running the purified fusion peptides on tricine–SDS–PAGE under nonreducing conditions showed only one band. Meanwhile, two bands were observed when reducing the samples with DTT; the lower band corresponded to the fusion peptide with disulfide bond that makes the structure more compact and induces the higher mobility on gel, and the upper band corresponded to the fusion peptide after the destruction of disulfide bonds by DTT. The purified peptides were also analyzed by using Western blot analysis using the HM-3 antibody, and the results indicated that the recombinant peptides have preserved their structural characteristics.
Purified peptides exhibited an antiangiogenic effect in vitro and in vivo
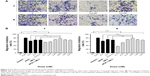
Transwell migration assay was conducted to explore the effects of the recombinant peptides at various doses on the migration of endothelial cells in vitro. The results indicated that the purified peptides, compared with the negative control (blank), exhibited an obvious inhibitory effect on the ECGS-induced migration of HUVECs in a dose-dependent manner in the concentration range between 0.1 and 0.8 nM, as shown in Figure 3. However, the increased dose of 1.6 nM resulted in the gradual decrease in the inhibitory activity of the two peptides on cell migration, and this effect had already been reported for several antiangiogenic drugs.13 Meanwhile, the best inhibitory effect was observed at the dosage of 0.8 nM with 77.07% and 79.43% for peptides F and R, respectively, while the avastin positive control showed an inhibitory effect of 81.19%. Moreover, at the most effective dosage, the two peptides showed increased inhibitory effect compared with the single HM-3 or AP25 or their mixture (P<0.05).
To validate the antiangiogenic potential of the purified products in vivo, CAM model was established to explore whether the local administration of the final products has an antiangiogenic activity. Results indicated, as shown in Figure 4, that the two peptides displayed a strong antiangiogenic response in the CAM with 78.86% (F) and 84.14% (R), respectively. Few neovessels were observed in the region where the peptides were applied, indicating that the purified peptides were shown to be potent inhibitors of angiogenesis at the dose of 3 nM in CAM.
Antitumor activity evaluation
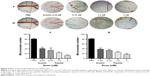
The two peptides showed a significant inhibition of gastric cancer cell MGC-803 proliferation in a dose-dependent manner between 5 and 80 nM and caused ~80% growth inhibition at 80 nM, with an IC50 of 26.24±0.63 nM and 12.80±0.52 nM for the peptides F and R, respectively. However, the purified products up to 80 nM showed no cytotoxicity to normal HLEC SRA01/04 and HEEC and no significant effect on human hepatoma cells, HepG2. In addition, as shown in Figure 5, the fusion peptides exhibited a better inhibition effect than the individual control peptides or their mixture did.
Antitumor effect was also morphologically assessed using Hoechst 33342/propidium iodide (PI) nuclear staining and fluorescence microscopy. MGC-803 was double-stained with Hoechst/PI and visualized using fluorescence microscopy. As shown in Figure 6, at the dose of 80 nM, the majority of the cells were red-stained with PI indicating their death, and this lethality was proportional to the peptide dose.
Two peptides interacted with integrin αvβ3 and α5β1
Previous studies had found that the RGD motif and the RGD cyclic peptide could bind integrin αvβ3 and α5β1, respectively.14,15 Based on these data and the structure of the fusion peptides, which contain an RGD cyclic peptide on the N-terminus and an RGD motif on its C-terminus, HUVEC adhesion test was conducted to verify whether the purified peptides have conserved their properties after being expressed in E. coli. The results indicated that the HUVECs showed an increased binding to the plates coated with the peptides compared with BSA-coated wells. Furthermore, monoclonal antibody against αv or β3 or α5 or β1, individually or altogether, inhibited HUVEC adhesion, and as indicated in Figure 7, the inhibition was higher for the new fusion peptides, indicating that the purified peptides have a higher receptor-binding affinity to the integrin receptors.
Three-dimensional structure modeling
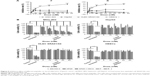
In the modeled structures shown in Figure 8A, the two functional parts were linked by different linkers; the results indicated that the two parts could perform their respective functions. The resulting structures were evaluated using MOE/protein geometry, which provides a variety of stereochemical measurements to inspect the structural quality in a given protein, including Ramachandran φ-ψ dihedral plots, side-chain rotamer quality, and nonbonded contact quality. For the evaluation of the φ-ψ torsion quality, it uses a reference data set as the basis for identifying potentially erroneous protein backbone topologies.9 Figure 8B shows the Ramachandran plot of the conducted peptide models. It was indicated that 98.2% and 98.4% of the plots are located in the “core” and “allowed” regions, while only 1.8% and 1.6% are located in the “disallowed” region for the peptides R and F, respectively.
Discussion
Chemical synthesis remains the most used technique for peptide production; however, its procedures are not ideally suited for the synthesis of long-chain polypeptides because the technical difficulties, the solubility, and the purification become formidable as the number of the amino acid residues increase; the recombinant expression is recently a newly developed approach for peptide synthesis.
Product proteolysis degradation, low expression levels, and poor recovery yields are the most important problems in small peptide expression; however, alternative expression strategies have been developed by the fusion of the desired peptide with a partner protein that enhances product stability, facilitates product recovery, and/or aids in the acquisition of biological activity of the recombinant peptide.16
In this study, the desired peptides were successfully expressed and purified by fusion to glutathione-S-transferase (GST) proteins. This affinity tag possesses the capability to be expressed in large amounts in E. coli and the ability to act as a chaperon to facilitate the protein folding, and frequently, the fused protein can be expressed in a soluble form rather than inclusion bodies, avoiding the addition of protein refolding steps, which could affect the protein structure and bioactivity.17 Previous work revealed that GST proteins have some redox activity and can help to get the aimed peptides in the correct oxidation state.18 Purified proteins using the GST fusion system have been used successfully in vaccine production and immunological studies,19,20 protein–protein and protein–DNA interaction studies, and other biochemical analyses.21
On the other hand, bifunctional molecules have been developed as a class of novel therapeutics with multifunctional properties. By genetically fusing two or more domains together, the product may possess functions derived from each of their component moieties, to achieve enhanced therapeutic effects.22 However, direct fusion of functional domains may lead to the product’s misfolding,23 low yield,24 or impaired bioactivity.25 Therefore, linkers have shown increasing importance in the design of bifunctional molecules, to obtain bioactive molecules with an enhanced effect. The selection of a peptide linker with the ability to maintain domain function of the fusion protein is quite essential. Fusion protein characteristics are variously influenced by the linker sequence depending on its flexibility. However, these linker effects have been studied exclusively for large proteins,24,26 and no studies are available about their use in small peptides fusion. The selected flexible spiral linker (GGGGS)327 and the rigid helical linker [A(EAAAK)4A]24,27–29 reported higher efficacy in the bifunctional fusion protein domains separation. The small size of the amino acids composing the first linker (Gly, Ser) provides flexibility and allows for mobility of the connecting functional domains. The incorporation of Ser could maintain the stability of the linker in aqueous solutions by forming hydrogen bonds with the water molecules. However, the rigid linker was reported to form α-helical structure highly stabilized by the Glu−-Lys+ salt bridges with intrasegment hydrogen bonds.30 The linkers mentioned above were used to construct the HM-3-AP25 fusion peptides. Molecular modeling indicated that whatever the type of linker, the distance between the active domains of the two peptides remains long enough to prevent their interaction and keep the molecule biological activity; this prediction was confirmed by bioactivity tests for the proteins, which showed that both the two peptides have approximately the same properties with only a little higher biological activity for the bifunctional fusion peptide with the rigid linker (P>0.05). The two spacers are also long enough to increase the molecular weight of the primer peptide and overcome the proteolysis problem commonly observed in small peptide degradation during the expression.
The fusion proteins were expressed in an auto-induced culture condition that allows cultures to reach very high cell densities before recombinant protein expression occurs.12,31,32 Compared with the IPTG induction, this approach reduces the risk of cell toxicity while enhancing product recovery. We found also that increasing the glucose concentration in the previous reported media could result in increased yields of fusion protein, and this is probably due to the use of alternative basal media together with increased levels of carbon source.
HUVEC migration assay and CAM assay revealed that both the two peptides exhibited a strong antiangiogenic effect in vitro and in vivo by inhibiting the endothelial cell migration and new vessel formation; this effect is due to the blockage of the integrin and the inhibition of endothelial cell attachment to the extracellular matrix. In cancer therapy, angiogenesis is an important hallmark, and reducing the formation of new vessels can inhibit tumor expansion, invasion, and metastasis. Cell viability investigations on human gastric cancer cell line MGC-803 showed that both F and R peptides displayed a significant antitumor activity at a dose up to 80 nM; however, they exhibited no significant effect toward hepatic carcinoma cell line Hep-G2 (low expression level of integrins).33 This assay on normal HLEC SRA01/04 or HEECs revealed that neither F nor R peptide was toxic (cell viabilities ~100%) for all tested concentrations, while the docetaxel positive control showed a strong cytotoxicity for all cells including the normal cells. These results confirmed that peptides are highly selective and safe, especially targeting integrin-expressed cells. This safety makes these antitumor peptides a good choice in cancer therapy, instead of the conventional chemotherapeutic agents, which enter normal tissues in the body with indiscriminate cytotoxicity and do not preferentially accumulate in tumor sites.
Conclusion
We reported for the first time the use of two kinds of linkers for small peptides ligation to construct two bifunctional antiangiogenic and antitumor bifunctional peptides, and the desired sequences were expressed in a soluble form in E. coli using a cost-effective auto-induction medium. The bioactivity evaluation and the molecular modeling results proved that whatever the flexibility of the linker, it could effectively separate the domains of a bifunctional molecule with a little increase in the biological activity when using the rigid helical linker.
Acknowledgments
This work was supported by 863 High-Technology Development Planning (Number 2012AA020304) and Priority Academic Program Development of Jiangsu Higher Education Institutions and the National Science and Technology Major Projects of New Drugs (Numbers 2012ZX09103301-004 and 2014ZX09508007) in People’s Republic of China.
Disclosure
The authors report no conflicts of interest in this work.
References
Kaspar AA, Reichert JM. Future directions for peptide therapeutics development. Drug Discov Today. 2013;18(17–18):807–817. | ||
Kelland LR. Overcoming the immortality of tumour cells by telomere and telomerase based cancer therapeutics–current status and future prospects. Eur J Cancer. 2005;41(7):971–979. | ||
Sørensen HP, Mortensen KK. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol. 2005;115(2):113–128. | ||
Xu HM, Chen YL. An RGD-modified endostatin-derived synthetic peptide shows antitumor activity in vivo. Bioconjug Chem. 2008;19(10):1980–1986. | ||
Xu H, Li P, Ren Y, Yang Y, Huang X, Liu Z. RGD-modified angiogenesis inhibitor HM-3 dose: dual function during cancer treatment. Bioconjug Chem. 2011;22(7):1386–1393. | ||
Yin R, Zheng H, Xi T, Xu HM. Effect of RGD-4C position is more important than disulfide bonds on antiangiogenic activity of RGD-4C modified endostatin derived synthetic polypeptide. Bioconjug Chem. 2010;21(7):1142–1147. | ||
Dai J, Peng L, Fan K, et al. Osteopontin induces angiogenesis through activation of PI3K/AKT and ERK1/2 in endothelial cells. Oncogene. 2009;28(38):3412–3422. | ||
Dia VP, Gonzalez de Mejia E. Lunasin potentiates the effect of oxaliplatin preventing outgrowth of colon cancer metastasis, binds to α5β1 integrin and suppresses FAK/ERK/NF-κB signaling. Cancer Lett. 2011;313(2):167–180. | ||
Yin R, Zheng H, Xi T, Xu HM. Effect of RGD-4C position is more important than disulfide bonds on antiangiogenic activity of RGD-4C modified endostatin derived synthetic polypeptide. Bioconjug Chem. 2010;21(7):1142–1147. | ||
Assa-Munt N, Jia X, Laakkonen P, Ruoslahti E. Solution structures and integrin binding activities of an RGD peptide with two isomers. Biochemistry. 2001;40(8):2373–2378. | ||
Sun R, Zheng H, Fang Z, Yao W. Rational design of aminoacyl-tRNA synthetase specific for p-acetyl-L-phenylalanine. Biochem Biophys Res Commun. 2010;391(1):709–715. | ||
Li Z, Kessler W, van den Heuvel J, Rinas U. Simple defined autoinduction medium for high-level recombinant protein production using T7-based Escherichia coli expression systems. Appl Microbiol Biotechnol. 2011;91(4):1203–1213. | ||
Huang Y, Stylianopoulos T, Duda DG, Fukumura D, Jain RK. Benefits of vascular normalization are dose and time dependent–letter. Cancer Res. 2013;73(23):7144–7146. | ||
Wickström SA, Alitalo K, Keski-Oja J. Endostatin associates with integrin alpha5beta1 and caveolin-1, and activates Src via a tyrosyl phosphatase-dependent pathway in human endothelial cells. Cancer Res. 2002;62(19):5580–5589. | ||
Sudhakar A, Sugimoto H, Yang C, Lively J, Zeisberg M, Kalluri R. Human tumstatin and human endostatin exhibit distinct antiangiogenic activities mediated by αVβ3 and α5β1 integrins. Proc Natl Acad Sci U S A. 2003;100(8):4766–4771. | ||
Arnau J, Lauritzen C, Petersen GE, Pedersen J. Reprint of: Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr Purif. Epub 2011 Sep 2. | ||
Harper S, Speicher DW. Purification of proteins fused to glutathione S-transferase. Methods Mol Biol. 2011;681:259–280. | ||
Nozach H, Fruchart-Gaillard C, Fenaille F, et al. High throughput screening identifies disulfide isomerase DsbC as a very efficient partner for recombinant expression of small disulfide-rich proteins in E. coli. Microb Cell Fact. 2013;12:37. | ||
Yip YL, Smith G, Ward RL. Comparison of phage pIII, pVIII and GST as carrier proteins for peptide immunisation in Balb/c mice. Immunol Lett. 2001;79(3):197–202. | ||
Smyth DR, Mrozkiewicz MK, McGrath WJ, Listwan P, Kobe B. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 2003;12(7):1313–1322. | ||
Vikis HG, Guan KL. Glutathione-S-transferase-fusion based assays for studying protein-protein interactions. Methods Mol Biol. 2004;261:175–186. | ||
Kim BJ, Zhou J, Martin B, et al. Transferrin fusion technology: a novel approach to prolonging biological half-life of insulinotropic peptides. J Pharmacol Exp Ther. 2010;334(3):682–692. | ||
Zhao HL, Yao XQ, Xue C, Wang Y, Xiong XH, Liu ZM. Increasing the homogeneity, stability and activity of human serum albumin and interferon-α2b fusion protein by linker engineering. Protein Exp Purif. 2008;61(1):73–77. | ||
Amet N, Lee HF, Shen WC. Insertion of the designed helical linker led to increased expression of tf-based fusion proteins. Pharm Res. 2009;26(3):523–528. | ||
Bai Y, Ann DK, Shen WC. Recombinant granulocyte colony-stimulating factor-transferrin fusion protein as an oral myelopoietic agent. Proc Natl Acad Sci U S A. 2005;102(20):7292–7296. | ||
Huang Z, Chong Z, Chen S, Ye F, Xing XH. Active inclusion bodies of acid phosphatase PhoC: aggregation induced by GFP fusion and activities modulated by linker flexibility. Microb Cell Fact. 2013;12(1):25. | ||
Bai Y, Shen WC. Improving the oral efficacy of recombinant granulocyte colony-stimulating factor and transferrin fusion protein by spacer optimization. Pharm Res. 2006;23(9):2116–2121. | ||
Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14(8):529–532. | ||
Amet N, Wang W, Shen WC. Human growth hormone–transferrin fusion protein for oral delivery in hypophysectomized rats. J Control Release. 2010;141(2):177–182. | ||
Chen X, Zaro J, Shen W. Fusion protein linkers: effects on production, bioactivity, and pharmacokinetics. In: Schmidt SR (editor). Fusion Protein Technologies for Biopharmaceuticals: Applications and Challenges. John Wiley & Sons, Inc.; 2013:57–73. | ||
Blommel PG, Becker KJ, Duvnjak P, Fox BG. Enhanced bacterial protein expression during auto-induction obtained by alteration of lac repressor dosage and medium composition. Biotechnol Prog. 2007;23(3):585–598. | ||
Setrerrahmane S, Zhang Y, Dai G, Lv J, Tan S. Efficient production of native lunasin with correct N-terminal processing by using the pH-induced self-cleavable Ssp DnaB mini-intein system in Escherichia coli. Appl Biochem Biotechnol. 2014;174(2):612–622. | ||
Yassin S, Hu J, Xu H, Li C, Setrerrahmane S. In vitro and in vivo activities of an antitumor peptide HM-3: a special dose-efficacy relationship on an HCT-116 xenograft model in nude mice. Oncol Rep. 2016;36(5):2951–2959. |
Supplementary material
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.