Back to Journals » OncoTargets and Therapy » Volume 13
Novel Nanocomplexes Targeting STAT3 Demonstrate Promising Anti-Ovarian Cancer Effects in vivo
Authors Zhang X, Lu T, Ma Y, Li R, Pang Y, Mao H, Liu P
Received 27 January 2020
Accepted for publication 6 May 2020
Published 8 June 2020 Volume 2020:13 Pages 5069—5082
DOI https://doi.org/10.2147/OTT.S247398
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Xiaolei Zhang, Tao Lu, Yanhui Ma, Rui Li, Yingxin Pang, Hongluan Mao, Peishu Liu
Department of Obstetrics and Gynecology, Qilu Hospital of Shandong University, Jinan 250012, People’s Republic of China
Correspondence: Peishu Liu; Yanhui Ma
Department of Obstetrics and Gynecology, Qilu Hospital of Shandong University, Jinan 250012, People’s Republic of China
Tel +86-531-82169570
Fax +86-531-86927544
Email [email protected]; [email protected]
Background: Cationic solid lipid nanoparticles (SLN) have attracted intensive interest as an effective gene delivery system for its high biocompatibility, stability and low cytotoxicity. In our previous study, we successfully prepared SLN-STAT3 decoy ODN complexes and made a primary study on its antitumor behavior in ovarian cancer cells in vitro. However, there is little information available so far about the effect of SLN-STAT3 decoy ODN complexes on ovarian cancer in vivo, either little information about the pharmacological toxicology in vivo.
Material and Methods: We applied nanotechnology to improve the gene delivery system and synthesize SLN-STAT3 decoy ODN complexes. Xenograft mouse models were established to assess the antitumor effects of SLN-STAT3 decoy ODN on the tumor growth of ovarian cancer in vivo. To analyze the mechanisms of SLN-STAT3 decoy ODN, we investigated apoptosis, autophagy, epithelial–mesenchymal transition (EMT) in tumor tissues of nude mice and investigated the effects and toxicology of SLN-STAT3 decoy ODN complexes on the vital organs of nude mice.
Results: The results showed that SLN-STAT3 decoy ODN complexes markedly inhibited tumor growth in vivo. SLN-STAT3 decoy ODN complexes could induce cell apoptosis through downregulating Bcl-2, survivin and pro caspase 3, but upregulating Bax and cleaved caspase 3. These complexes could also regulate autophagy through upregulating LC3A-II, LC3B-II and beclin-1, but downregulating p-Akt and p-mTOR. Moreover, these complexes could inhibit cancer cell invasion through reversing EMT. Besides, SLN-STAT3 decoy ODN complexes showed no obvious toxicity on vital organs and hematological parameters of nude mice.
Conclusion: The molecular mechanisms that SLN-STAT3 decoy ODN complexes inhibit tumor growth involved activating the apoptotic cascade, regulating autophagy, and reversing EMT program; and these complexes showed no obvious toxicity on nude mice. Our study indicated that the nanocomplexes SLN-STAT3 decoy ODN might be a promising therapeutic approach for ovarian cancer treatment.
Keywords: STAT3, decoy ODN, SLN, ovarian cancer, in vivo
Introduction
Ovarian cancer is the fifth most common cause of cancer-related deaths among women in 2019, and it would affect about 22,530 new cases and cause approximately 13,980 deaths in the United States.1 Despite developments in cancer therapeutics, the survival rates have not been significantly improved over the past 30 years.2 Surgical cytoreduction and adjuvant chemotherapy are still the standard treatment for ovarian cancer, while the majority will recur due to resistance to chemotherapy and metastasis.3 The five-year survival rate remains low.4 Targeted therapies for cancer have made great progress, including anti-angiogenic drugs, poly ADP-ribose polymerase (PARP) inhibitors. They are of great significance and provide new hope for the treatment of ovarian cancer; however, the responses of most patients to them are transient, further development and advances are needed.5 Therefore, the research and development of targeted drugs for ovarian cancer are demanded.
In recent years, gene therapy has received enormous attention as a new therapeutic strategy in the treatment of neoplastic, genetic and infectious diseases.6–8 The main objective of gene therapy is transferring the gene of interest into the target tissues successfully in vivo.9 There are two major challenges of gene therapy: the target gene and the gene delivery system.
STAT3, as a crucial member of the transcription factor family, can transfer from the cytoplasm to the nucleus and vice versa, regardless of its phosphorylation status.10 Constitutive activation of STAT3 has been found in nearly 70% of solid and hematological tumors,11,12 such as ovarian cancer,13 breast carcinoma,14 malignant melanoma,15 prostate cancer,16 and so on. It is reported that the constitutive activation of STAT3 plays an important role in cell proliferation, differentiation, survival and metastasis.17 STAT3 has been recognized as an oncogene,18 giving good reason for regarding STAT3 as a potential molecular target. The decoy oligodeoxynucleotides (ODN) strategy is a promising approach to inhibit STAT3. The STAT3 decoy ODN is a 15-mer double-strand oligodeoxynucleotides which mimics the STAT3 binding sequences of DNA, acting primarily by competing for binding with the activated STAT3 dimers. The STAT3 decoy ODN has shown growth suppression in human ovarian cancer, glioma cells, hepatocellular carcinoma cells, colorectal carcinoma, and non-small cell lung cancer.19–23 The STAT3 decoy ODN brings hope for ODN-based therapy.
However, the actual effect of ODN could only be achieved when it reaches the target tissues and cells with therapeutically efficient concentration. Upon administration, ODN is suffering from nucleases, immune surveillance and endosome-lysosome trafficking before reaching the target tissues and cells.24 There is a pressing need to develop a delivery system that protects ODN from being degraded by nucleases and promotes the efficient translocation across the membrane barriers, leading to intracellular delivery.25
Nanocarriers have been used for the treatment of various diseases including cancer.26 Cationic solid lipid nanoparticles (SLN) are promising delivery systems due to the biocompatibility, safety, less immunogenic, ease of mass production and capable of surface modified.27 Many groups have done research in SLN, and various reports have revealed that cationic SLN are promising systems for transfection; indeed, SLN are the most investigated non-viral vectors.28,29
In our previous work, we incorporated STAT3 decoy ODN into SLN to form SLN-STAT3 decoy ODN complexes. SLN-STAT3 decoy ODN complexes could be efficiently taken up by human ovarian cells and significantly suppressed cancer cell growth. It could induce cell death and inhibit invasion in ovarian cancer cells in vitro as well.30 However, there is little information available so far about the effect of SLN-STAT3 decoy ODN complexes on ovarian cancer in vivo, and either little information about the pharmacological toxicology in vivo. Herein, we studied the antitumor behavior and potential mechanisms of SLN-STAT3 decoy ODN complexes in ovarian cancer on xenografted nude mice, and investigated the effects and safety of SLN-STAT3 decoy ODN complexes on the vital organs of nude mice.
Materials and Methods
Materials
Injectable soya lecithin was purchased from Shanghai Pujiang Phospholipids Co. Ltd (China). Glyceryl monostearate was purchased from Shanghai Chemical Reagent Co. Ltd (China). Cetyltrimethylammonium bromide (CTAB) was purchased from Ameresco Incorporation (USA). STAT3 decoy ODN (sequence: 5ʹ-CATTTCCCGTAAATC-3ʹ, 3ʹ-GTAAAGGGCATTTAG-5ʹ) and STAT3 scrambled ODN (sequence: 5ʹ-CATCTTGCCAATATC-3ʹ, 3ʹ-GTAGAACGGTTATAG-5) was synthesized by Sangon Biotech (Shanghai, China) as used in the previous study.31 McCoy’s 5A medium was purchased from Sigma-Aldrich (St-Louis, MO, USA). Fetal bovine serum (FBS) was obtained from Haoyang Biological manufacture Co., Ltd, Tianjin, China. The primary antibodies against STAT3, p-STAT3 (Ser727, Tyr705), Bcl-2, Bax, caspase 3, LC3A, LC3B, E-cadherin, N-cadherin, mTOR, p-mTOR, AKT, p-AKT, Beclin-1, Snail, MMP-9, VEGR, Vimentin, were obtained from Cell Signaling Technology (Cell Signaling Technology, USA) and Survivin (R&D Systems Inc.). The anti-β-actin primary antibodies and horseradish peroxidase-conjugated second antibody were purchased from Beijing Zhong Shan Biotech Co., Ltd. Beijing, China. The enhanced chemiluminescence (ECL) Western blot reagent was obtained from Merck Millipore. The fluorometric TUNEL system was obtained from KeyGen Biotech, Nanjing, China.
Cell Line and Cell Culture
Human ovarian epithelial cancer cell line SKOV3 was obtained from Qilu Hospital Biotechnology Center, Shandong University, Jinan, China. The use of these cells was approved by the Experimental Ethics Committee of Qilu Hospital of Shandong University. SKOV3 cells were cultured in McCoy’s 5A medium (Sigma-Aldrich, USA) supplemented with 10% FBS and 1% penicillin-streptomycin (Invitrogen, USA), cells were incubated in a humidified atmosphere at 37°C and 5% CO2.
Preparation of SLN and SLN-STAT3 Decoy ODN Complexes
The SLN and SLN-STAT3 decoy ODN complexes were prepared by the solvent diffusion method as previously reported.32 Briefly, acetone (2 mL) containing glyceryl monostearate (20 mg) and soya lecithin (15 mg) was injected into deionized water (20 mL) containing CTAB (15 mg) at 12 mL/h under magnetic stirring at 400 rpm at room temperature. After stirring for 12 hrs to evaporate the organic solvents, the obtained nanoparticles were collected by centrifugation at 15,000 rpm. The pellets were washed three times, filtered through a membrane with a 0.45-μm pore size, and the PH was adjusted to 7.2–7.4. Then, the obtained SLN suspensions were stored at 4°C.
STAT3 decoy ODN solution was mixed with the previously prepared SLN suspension at a fixed ratio of 1:20 (w/w) as we previously used. Then, the mixed suspensions were gently vortexed for 20 s and incubated for 20 mins at room temperature. The SLN-STAT3 decoy ODN complexes were formed finally. Transmission electronic microscopy (TEM) was performed to observe the morphology of the complexes (JEM-1200EX, Japan). The particle size and zeta potential were analyzed by photon correlation spectroscopy using a Zetasizer 3000 (Malvern Instruments, Malvern, England).
Establishment of SKOV3 Xenograft Nude Mice Models
Female nude mice (BALB/c-nu, 4–6 weeks old) were purchased from the Animal Center of China Academy of Medical Sciences (Beijing, China). SKOV3 cells (2 × 107/mL) in 0.2 mL phosphate-buffered saline (PBS) were injected subcutaneously on the right dorsal flank of the nude mice. Fourteen days after injection, the mice were randomly divided into four groups with five mice in each group and independently intratumorally multi-point injected with a total volume of 100 μL of one of the following for each group: (1) PBS alone. (2) SLN-STAT3 scrambled ODN. (3) naked STAT3 decoy ODN. (4) SLN-STAT3 decoy ODN. The injections were performed every 2 days for 30 days. The tumor volume was measured by length (L) and width (W) every 4 days, and calculated from the formula: V (tumor volume) = 0.5 × L × W2. Blood was drawn from the mice’s tail vein for the examination of blood routine, liver and renal function at 12 hrs after the final injection. Then, all of the mice were sacrificed. The tissues of tumors, heart, liver and kidney were collected. Each tumor was divided into two parts, one part frozen at −80°C for Western blot and the other part fixed in formalin and embedded in paraffin for hematoxylin and eosin staining (H&E). The research was approved by the Experimental Ethics Committee of Qilu Hospital of Shandong University. The care of laboratory animals and animal experimentation were in compliance with the recommended procedures of the National Institutes of Health guide for the care and use of laboratory animals.
Detection of Apoptosis and Autophagy
TUNEL assay was carried out using a Fluorometric TUNEL system (KeyGen Biotech, Nanjing, China) according to the manufacturer’s instructions to evaluate the effects of SLN·STAT3 decoy ODN complexes on apoptosis in the tumor tissues. The tissue sections were rinsed with PBS for 5 min following incubation with proteinase K (18 µg/mL) for 20 min, and then blocked with fetal bovine serum for 15 min at room temperature. TdT reaction mix (50 µL) was added to the sections followed by incubation in a humidified chamber for 60 min at 37°C. The sections were then rinsed with PBS and observed under a fluorescence microscope. Cells that nucleus were stained with green fluorescent were defined as positive. To quantify the positive cells, cells were counted under a microscope at ×200 magnification in five random fields. The intracellular autophagosomes were observed by TEM, according to the previously described methods.33,34
Western Blot Assay
The tumor tissues were lysed in lysis buffer for Western blot assay. Equal amounts of protein were separated by SDS-PAGE and transferred onto nitrocellulose membranes (Millipore, Bedford, MA, USA). The membranes were blocked in Tris-buffered saline with 5% (w/v) non-fat dry milk and then incubated with a primary antibody (1:1000) at 4°C. After washing with TBST 3 times, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody. Immunoreactive proteins were visualized using an enhanced chemiluminescence (ECL) detection system (Pierce, Rockford, IL, USA). Band density was quantified by Image J software. The quantity of β-actin was used for internal control. The density value of each sample was normalized to its β-actin density value to get its relative quantity value.
Statistical Analyses
All experiments were independently repeated at least three times. All data were analysed using GraphPad Prism version 7.0. The data with normal distribution were presented as means ± standard deviation (SD). One-way analysis of variance (ANOVA) was used to estimate differences among the four groups. Differences between the two groups were determined by the least significant difference t (LSD-t) test. Statistical differences were marked by the following symbols: * P < 0.05; # P > 0.05; and P < 0.05 was considered statistically significant.
Results
Characterization of SLN-STAT3 Decoy ODN Complexes
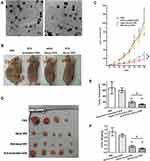
SLN and STAT3 decoy ODN were successfully synthetized in our study. SLN and STAT3 decoy ODN were mixed at weight ratios (w/w), and the optimal ratio for preparing the complexes was 20:1. The mean particle size of the complexes was 101.30±11.89 nm, with a polydispersity index of 0.24±0.03. The value of the zeta potential was 20.03 ±0.93 mV. As shown in Figure 1A, both SLN and SLN-STAT3 decoy ODN complexes were spherical or ellipsoidal under TEM.
SLN-STAT3 Decoy ODN Complexes Show Significant Antitumor Activities in vivo
To assess the antitumor effect of SLN-STAT3 decoy ODN on the tumor growth of ovarian cancer in vivo, SKOV3 xenograft mouse models were established (Figure 1B). Tumor growth curves as shown in Figure 1C indicated that the tumor growth was obviously inhibited in the SLN-STAT3 decoy ODN complexes treated group, compared with the other three groups. Figure 1D shows the tumor tissues of the nude mice, one mouse in the PBS group died of cachexia before the end of treatment. As presented in Figure 1E and F, STAT3 decoy ODN treatment significantly decreased tumor volume and weight, while SLN-STAT3 decoy ODN treatment led to more obvious inhibition effects on the volumes and weights of tumors, compared to the naked STAT3 decoy ODN group (P < 0.05). The results suggest that SLN-STAT3 decoy ODN markedly inhibited tumor growth in vivo.
SLN-STAT3 Decoy ODN Complexes Induce Cell Apoptosis of Ovarian Cancer in vivo
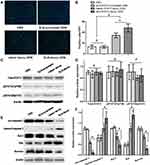
The Results of Fluorometric TUNEL Assay
To detect the cell apoptosis in the tumor tissues of the nude mice, the Fluorometric TUNEL assay was performed. Cell nuclei with green fluorescent staining were defined as apoptotic cells, as shown in Figure 2A. TUNEL assay revealed: there were 13±4 apoptotic cells/high power field (/HPF) in the PBS group, 13±3 apoptotic cells/HPF in the SLN-STAT3 scrambled ODN group, 34±4 apoptotic cells/HPF in the naked STAT3 decoy ODN group, while there were 48±8 apoptotic cells/HPF in the SLN-STAT3 decoy ODN group. The results showed that the green fluorescence in the SLN-STAT3 decoy ODN group and naked STAT3 decoy ODN group were dramatically increased, and apoptotic cells/HPF in SLN-STAT3 decoy ODN group were significantly increased than that in naked STAT3 decoy ODN group (P < 0.05) (Figure 2B). Meanwhile, the tumor cells of the PBS and SLN-STAT3 scrambled ODN groups were observed with little green fluorescence, demonstrating that the two groups had little apoptosis. The results indicated that the SLN-STAT3 decoy ODN promoted apoptosis of ovarian cancer cells in vivo.
Effects of SLN-STAT3 Decoy ODN Complexes on the Expression of Apoptosis-Related Proteins
To further explore the possible mechanisms underlying the SLN-STAT3 decoy ODN complexes induce apoptosis, Western blot assay was performed to test the related protein expression. The results showed that the expression of STAT3 and p-STAT3 (Ser727, Tyr705) had no significant difference among the four groups (Figure 2C and D). However, the expression levels of pro caspase 3, Bcl-2 and Survivin were down-regulated in SLN-STAT3 decoy ODN and naked STAT3 decoy ODN groups, while the expression of cleaved caspase 3 and Bax was up-regulated in SLN-STAT3 decoy ODN and naked STAT3 decoy ODN groups, especially in the SLN-STAT3 decoy ODN group (Figure 2E). The expression levels of apoptosis-related proteins were significantly regulated in SLN-STAT3 decoy ODN group (P < 0.05) (Figure 2F). These results indicated that SLN-STAT3 decoy ODN was more effective in regulating the expression level of apoptosis-related proteins, thus promoting the occurrence of tumor cell apoptosis in vivo.
SLN-STAT3 Decoy ODN Complexes Induce Cell Autophagy of Ovarian Cancer in vivo
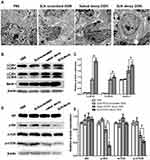
Autophagy Observed Under TEM
TEM images revealed more autophagic cells in the tumor tissues of SLN-STAT3 decoy ODN group than the other three groups, and there are multiple autophagosomes in the cytoplasm around the nucleus. Some autophagosomes had intact envelops and contain organelles and protease particles (Figure 3A).
Effects of SLN-STAT3 Decoy ODN Complexes on the Expression of Autophagy-Related Proteins
Because conversion of cytosolic LC3-I to LC3-II through lipidation by a ubiquitin-like system is a classical marker of autophagosome formation,35 we evaluated SLN-STAT3 decoy ODN effect on the conversion of LC3-I to lipidated LC3-II in four groups. Western blot assay showed that the protein level of LC3A-II and LC3B-II increased in SLN-STAT3 decoy ODN and naked STAT3 decoy ODN groups, especially in SLN-STAT3 decoy ODN group (P < 0.05) (Figure 3B and C). SLN-STAT3 decoy ODN complexes resulted in a noticeable decrease in the expression of p-Akt and p-mTOR, whereas Beclin-1 expression increased. Protein level of total Akt and total mTOR were similar in the four groups (P>0.05) (Figure 3D and E).
Effects of SLN-STAT3 Decoy ODN Complexes on the Invasion of Ovarian Cancer Cells in vivo
In the process of xenograft tumor resection, we found that there was heavy adhesion between tumors and peripheral tissues with no apparent limits in PBS and SLN-STAT3 scrambled ODN treated nude mice (Figure 4Ab and c). The tumor tissue boundaries were relatively clear in SLN-STAT3 decoy ODN group (Figure 4Aa). Western blot showed a downregulation of expression of VEGF and MMP9 in SLN-STAT3 decoy ODN group and naked STAT3 decoy ODN group compared with the other two groups (Figure 4B), and the difference in SLN-STAT3 decoy ODN group was more significant than that in naked STAT3 decoy ODN group (P < 0.05) (Figure 4C). The expression of epithelial marker protein, such as E-cadherin was significantly up-regulated, while the expression of mesenchymal marker protein, such as N-cadherin and Vimentin were significantly down-regulated in SLN-STAT3 decoy ODN group. Similarly, the protein level of EMT related regulators, such as Snail was significantly down-regulated in SLN-STAT3 decoy ODN group (P < 0.05) (Figure 4D and E).
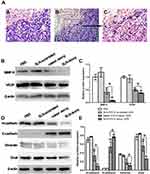
Pharmacological Toxicology of SLN-STAT3 Decoy ODN in Nude Mice
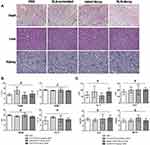
H&E staining of heart, liver and kidney in nude mice showed no obvious histopathological changes among the four treatment groups (Figure 5A). Then, we examined the effects of SLN-STAT3 decoy ODN on hematological parameters of nude mice. Results showed that the counts of white blood cell (WBC), red blood cells (RBC), hemoglobin (HGB) and platelet (PLT) were similar, with no significant statistical difference among the four treatment groups (P > 0.05) (Figure 5B). Additionally, the effects of SLN-STAT3 decoy ODN on liver and kidney function in nude mice were tested. Comparisons of ALT, AST, SCr and BUN levels revealed no statistical difference among the four groups (P > 0.05) (Figure 5C).
Discussion
Studies have provided evidences for persistent activation of STAT3 presents in nearly 70% of tumors,36 and STAT3 is the key node in transcriptional activation downstream of cytokine or kinase action in multiple cancers.37 As reported, STAT3 is the best-studied family member of the JAK-STAT pathway in cancer, and is a known oncogene in various types of solid malignancies, like melanoma or lung cancer.38–40 Overactivation of STAT3 is commonly associated with an upstream oncogenic driver, such as hyperactive mutated tyrosine kinases. The STAT3 activation in epithelial ovarian cancer (EOC) controls the properties of both tumor cells and their microenvironment, making multiple distinct functions during the progression of EOC. Increasing evidence reveals activation of the STAT3 pathway has a significant correlation with reduced survival of recurrent EOC, suggesting that STAT3 emerges as an attractive target for anti-cancer therapy.41 Constitutive STAT3 activation plays an important role in disrupting the normal physiological roles in regulating cell proliferation, survival, angiogenesis and immune function. Blockage of STAT3 was found to inhibit cell proliferation, induce cell death, suppress angiogenesis and stimulate immune responses. One efficient strategy to block the STAT3 pathway employs decoy ODN which mimic DNA binding consensus sequences that are combined with activated STAT3 and inhibits STAT3-responsive gene transcription in vitro and in vivo.42–44 First-in-human trial of a STAT3 decoy ODN in head and neck tumors provides the first demonstration of a successful strategy to inhibit tumor STAT3 signaling, thereby paving the way for broad clinical development.45 In our previous work, we found that STAT3 decoy ODN inhibited ovarian cancer cell growth in vivo.31 However, transfection efficiency and bioavailability were still the problems and challenges we needed to face.
It seems that currently applied therapeutics may not be able to effectively modulate the STAT3 signaling pathway and suffer from a variety of drawbacks such as low bioavailability and lack of specific tumor targeting.46 The application of decoy ODN is limited, primarily owing to the lack of efficient and safe carriers to transmit the decoy ODN. For the purpose of obtaining effective decoy ODN therapeutics, the important priority is the development of a delivery system that protects the decoy ODN from degraded by the nuclease and facilitates the cellular uptake. Nanocarriers can be considered as potential candidates in cancer therapy and they can encapsulate the drug to protect it against degradation thus resulting in its enhanced bioavailability for therapeutic application. Solid lipid nanoparticles (SLN) are one of the newest members of the lipid-based nanocarriers family, and many studies demonstrating their usefulness in numerous diseases have been published.47,48 In our previous work, we had synthesized SLN as a carrier for STAT3 decoy ODN and prepared SLN-STAT3 decoy ODN complex. The SLN-STAT3 decoy ODN can produce a series of antitumor effects in ovarian cancer cells in vitro. However, the biological effects of SLN-STAT3 decoy ODN in vivo have not been well illustrated.
In the present study, we studied the antitumor behavior and potential mechanisms of SLN-STAT3 decoy ODN complexes in ovarian cancer on xenografted nude mice. Our results suggested that SLN-STAT3 decoy ODN showed more advantages compared with naked STAT3 decoy ODN. First of all, the cellular uptake of SLN-STAT3 decoy ODN complexes was significantly higher than naked STAT3 decoy ODN,30 this result had been reported in our previous article. Nanotechnology aims to the development of materials and structures with low size (1–1000 nm).49 The mean particle size of the SLN-STAT3 decoy ODN complexes in our study was only 101.30±11.89 nm, which could result in high cell uptake rate and bioavailability. Secondly, SLN-STAT3 decoy ODN had a stronger antitumor effect in vivo than naked STAT3 decoy ODN. The growth curves of xenograft nude mice revealed that the tumor growth treated with SLN-STAT3 decoy ODN was significantly inhibited than treated with naked STAT3 decoy ODN. Statistical comparison of tumor weight and volume was consistent with the above results. Thirdly, the SLN-STAT3 decoy ODN was more capable of inducing tumor cell apoptosis in vivo than naked STAT3 decoy ODN. Fourthly, SLN-STAT3 decoy ODN played a more significant role than naked STAT3 decoy ODN in regulating the target protein of the downstream signal pathway, which involved tumor apoptosis, autophagy, and epithelial-mesenchymal transition (EMT). The antitumor mechanism of SLN-STAT3 decoy ODN is further elaborated as follows:
Apoptosis is an evolutionarily conserved process that plays an essential role in organism development and tissue homeostasis. However, in pathological conditions, particularly cancer, cells lose their ability to undergo apoptosis induced death leading to uncontrolled proliferation.50 It is reported STAT3 decoy ODN can induce tumor cells apoptosis in vitro and in vivo.19,31,51,52 To further explore the underlying apoptosis induction mechanism of SLN-STAT3 decoy ODN in ovarian cancer in the nude mice, apoptotic proteins were determined by Western blot assay (Figure 2). The results showed a down-regulation of Bcl-2, Survivin and pro caspase 3 but an up-regulation of Bax and cleaved caspase 3 in SLN-STAT3 decoy ODN group compared with the other three groups. All the above proteins are pivotal regulators of cell apoptosis.53 The Bcl-2 protein functions through hetero-dimerization with pro-apoptotic members of the BH3 family to prevent mitochondrial pore formation and prevent cytochrome c release and initiation of apoptosis.54 Survivin inhibits apoptosis by inhibiting caspases.55 Caspase-3 is a central effector caspase, and cleaved caspase 3 participates in nuclear changes during apoptosis.56 Bax participate in pro-apoptotic signaling.53 SLN-STAT3 decoy ODN complexes could regulate these apoptosis-related proteins, activate the apoptotic cascade in tumour tissues, and lead to reduced tumour burden.
Autophagy serves to maintain intracellular homeostasis through a process that involves lysosomal degradation and recycling of unnecessary or damaged cellular components, and in turn promotes cell survival. Autophagy plays a complex and controversial role in tumorigenesis and the treatment of malignant tumor.35 Autophagy has been a mechanism of drug resistance in cancer therapy.57 STAT3 has been identified as a new autophagy regulator.58 In our study, the expression of LC3A-II and LC3B-II which are autophagosome-specific markers increased in SLN-STAT3 decoy ODN treatment nude mice. Beclin-1, Akt, and mTOR are factors that can regulate cell autophagy.59 We observed that the complexes resulted in a noticeable decrease in the expression of p-Akt and p-mTOR, whereas Beclin-1 expression increased. The protein level of total Akt and total mTOR was not decreased. The down-regulation of p-Akt and p-mTOR led to the inhibition of the Akt/mTOR pathway. The lack of Akt/mTOR resulted in the transcriptional activation of autophagy genes and mediated phosphorylation of proteins necessary for autophagosome formation. Beclin-1 plays an important part in controlling the initiation of autophagy and autophagosome nucleation.60 These results demonstrated that the SLN-STAT3 decoy ODN complexes played a role in autophagy through the Akt/mTOR pathway and regulating Beclin-1 expression in vivo.
Epithelial-mesenchymal transition (EMT) is a significant mechanism involved in the progression of cancer, and EMT plays an important role in cancer cell migratory and invasive behaviours.61 STAT3 activation exerts a critical influence on establishing cell polarity during directed cancer cell progression, processes significant for EMT programmes.62 E-cadherin is an epithelial marker, the down-regulation of E-cadherin is characteristic of the EMT process.63 In our study, the expression of E-cadherin was significantly up-regulated in SLN-STAT3 decoy ODN treatment group, which suggested that SLN-STAT3 decoy ODN could inhibit EMT process. Snail is a zinc-finger transcription regulator that inhibits the transcription of E-cadherin and initiates EMT.64 The protein level of EMT related regulators Snail was down-regulated in SLN-STAT3 decoy ODN group. Moreover, the expression of mesenchymal marker proteins such as N-cadherin and Vimentin were also significantly down-regulated in SLN-STAT3 decoy ODN group. It is well known that VEGF and MMP9 are closely related to cancer cell invasion and metastasis, our results showed the expression of VEGF and MMP9 was down-regulated in SLN-STAT3 decoy ODN treatment group. All the results indicated that the SLN-STAT3 decoy ODN complexes could reverse the EMT program, thereby inhibiting the invasion and metastasis of ovarian cancer cells in vivo.
EOC patients have a 5-year survival rate as low as 30%, because of resistance and toxicity of anti-cancer drugs. Treatment for EOC remains challenging, and there is an urgent need to develop more feasible agents exhibiting low toxicity with more distinct molecular target in order to effectively assist in the treatment of ovarian cancers. One advantage of the SLNs is that they reduce the toxicity of the therapeutic molecule that they transfer protecting them. It has been reported that general uncharged SLN have no cytotoxic effects in vitro when the lipid concentration reaches 2.5%.65,66 It is crucial to explore the toxicology of SLN-STAT3 decoy ODN. Our previous study found that naked STAT3 decoy ODN may have side-effects on the livers of the mouse.31 It is reported that rifampicin encapsulated into SLN resulting in low or no hepatotoxicity.67 In the present study, we detected the morphology of the mouse heart, liver and kidney by H&E staining. No obvious cardiac, hepatic and renal toxicity was induced by SLN-STAT3 decoy ODN. Further research showed that SLN-STAT3 decoy ODN had no significant toxic effects on hematological parameters (such as RBC, WBC, HGB, PLT, ALT, AST, SCr and BUN) of nude mice. The results indicated a value for future SLN-based gene delivery in vivo biomedical research.
Conclusion
Our study demonstrated that STAT3 decoy ODN delivered by SLN can effectively suppress ovarian cancer cell growth in xenograft nude mice in vivo. SLN-STAT3 decoy ODN have a more obvious therapeutic effect than the naked STAT3 decoy ODN. The molecular mechanisms that SLN-STAT3 decoy ODN inhibits tumor growth involve activating the apoptotic cascade; regulating autophagy; and reversing EMT program. SLN-STAT3 decoy ODN showed no obvious toxicity on vital organs and hematological parameters of nude mice. Our study provided evidence for the gene therapy of ovarian cancer and the data indicated that SLN-STAT3 decoy ODN might be a promising therapeutic approach for ovarian cancer treatment.
Ethics Approval
The Experimental Ethics Committee of Qilu Hospital of Shandong University approved this research. The care of laboratory animals and animal experimentation were in compliance with the recommended procedures of the National Institutes of Health guide for the care and use of laboratory animals.
Acknowledgments
We gratefully thank Professor Na Zhang (School of Pharmaceutical Sciences, Shandong University, Jinan, Shandong, China) for the assistance provided in SLN synthesis. This work was supported by the National Natural Science Foundation of China (81101984 and 81702559) and Natural Science Foundation of Shandong Province (ZR2016HM27).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Kim A, Ueda Y, Naka T, et al. Therapeutic strategies in epithelial ovarian cancer. J Exp Clin Cancer Res. 2012;31(1):14. doi:10.1186/1756-9966-31-14
3. Ozols RF, Bookman MA, Du Bois A, et al. Intraperitoneal cisplatin therapy in ovarian cancer: comparison with standard intravenous carboplatin and paclitaxel. Gynecol Oncol. 2006;103(1):1–6. doi:10.1016/j.ygyno.2006.06.026
4. Baldwin LA, Huang B, Miller RW, et al. Ten-year relative survival for epithelial ovarian cancer. Obstet Gynecol. 2012;120(3):612–618. doi:10.1097/AOG.0b013e318264f794
5. Beaver JA, Coleman RL, Arend RC, et al. Advancing drug development in gynecologic malignancies. Clin Cancer Res. 2019;25(16):4874–4880. doi:10.1158/1078-0432.CCR-19-0619
6. Assi H, Candolfi M, Baker G, et al. Gene therapy for brain tumors: basic developments and clinical implementation. Neurosci Lett. 2012;527(2):71–77. doi:10.1016/j.neulet.2012.08.003
7. Keeler AM, Flotte TR. Cell and gene therapy for genetic diseases: inherited disorders affecting the lung and those mimicking sudden infant death syndrome. Hum Gene Ther. 2012;23(6):548–556. doi:10.1089/hum.2012.087
8. Deng W, Bivalacqua TJ, Champion HC, et al. Gene therapy techniques for the delivery of endothelial nitric oxide synthase to the lung for pulmonary hypertension. Methods Mol Biol. 2010;610:309–321. doi:10.1007/978-1-60327-029-8_18
9. El-Aneed A. An overview of current delivery systems in cancer gene therapy. J Control Release. 2004;94(1):1–14. doi:10.1016/j.jconrel.2003.09.013
10. Pranada AL, Metz S, Herrmann A, et al. Real time analysis of stat3 nucleocytoplasmic shuttling. J Biol Chem. 2004;279(15):15114–15123. doi:10.1074/jbc.M312530200
11. Frank DA. Stat3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251(2):199–210. doi:10.1016/j.canlet.2006.10.017
12. Bar-Natan M, Nelson EA, Xiang M, et al. Stat signaling in the pathogenesis and treatment of myeloid malignancies. JAK-STAT. 2012;1(2):55–64. doi:10.4161/jkst.20006
13. Yang C, Lee H, Jove V, et al. Prognostic significance of b-cells and pstat3 in patients with ovarian cancer. PLoS One. 2013;8(1):e54029. doi:10.1371/journal.pone.0054029
14. Chen Y, Wang J, Wang X, et al. Stat3, a poor survival predicator, is associated with lymph node metastasis from breast cancer. J Breast Cancer. 2013;16(1):40–49. doi:10.4048/jbc.2013.16.1.40
15. Lee I, Fox PS, Ferguson SD, et al. The expression of p-stat3 in stage iv melanoma: risk of CNS metastasis and survival. Oncotarget. 2012;3(3):336–344. doi:10.18632/oncotarget.475
16. Singh N, Hussain S, Bharadwaj M, et al. Overexpression of signal transducer and activator of transcription (stat-3 and stat-5) transcription factors and alteration of suppressor of cytokine signaling (socs-1) protein in prostate cancer. J Recept Signal Transduct. 2012;32(6):321–327. doi:10.3109/10799893.2012.733885
17. Siveen KS, Sikka S, Surana R, et al. Targeting the stat3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014;1845(2):136–154. doi:10.1016/j.bbcan.2013.12.005
18. Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi:10.1016/S0092-8674(00)81959-5
19. Liu M, Wang F, Wen Z, et al. Blockage of stat3 signaling pathway with a decoy oligodeoxynucleotide inhibits growth of human ovarian cancer cells. Cancer Invest. 2014;32(1):8–12. doi:10.3109/07357907.2013.861471
20. Gu J, Li G, Sun T, et al. Blockage of the stat3 signaling pathway with a decoy oligonucleotide suppresses growth of human malignant glioma cells. J Neurooncol. 2008;89(1):9–17. doi:10.1007/s11060-008-9590-9
21. Sun X, Zhang J, Wang L, et al. Growth inhibition of human hepatocellular carcinoma cells by blocking stat3 activation with decoy-odn. Cancer Lett. 2008;262(2):201–213. doi:10.1016/j.canlet.2007.12.009
22. Souissi I, Najjar I, Ah-Koon L, et al. A STAT3-decoy oligonucleotide induces cell death in a human colorectal carcinoma cell line by blocking nuclear transfer of STAT3 and STAT3-bound NF-κB. BMC Cell Biol. 2011;12(1):14. doi:10.1186/1471-2121-12-14
23. Njatcha C, Farooqui M, Kornberg A, et al. Stat3 cyclic decoy demonstrates robust antitumor effects in non-small cell lung cancer. Mol Cancer Ther. 2018;17(9):1917–1926. doi:10.1158/1535-7163.MCT-17-1194
24. Siddiqui A, Patwardhan GA, Liu YY, et al. Mixed backbone antisense glucosylceramide synthase oligonucleotide (MBO-asGCS) loaded solid lipid nanoparticles: in vitro characterization and reversal of multidrug resistance in NCI/ADR-RES cells. Int J Pharm. 2010;400(1–2):251–259. doi:10.1016/j.ijpharm.2010.08.044
25. de Jesus MB, Zuhorn IS. Solid lipid nanoparticles as nucleic acid delivery system: properties and molecular mechanisms. J Control Release. 2015;201:1–13. doi:10.1016/j.jconrel.2015.01.010
26. Shi J, Kantoff PW, Wooster R, et al. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2016;17(1):20–37. doi:10.1038/nrc.2016.108
27. Severino P, Szymanski M, Favaro M, et al. Development and characterization of a cationic lipid nanocarrier as non-viral vector for gene therapy. Eur J Pharm Sci. 2014;66C:78–82. doi:10.1016/j.ejps.2014.09.021
28. Vighi E, Montanari M, Ruozi B, et al. The role of protamine amount in the transfection performance of cationic sln designed as a gene nanocarrier. Drug Deliv. 2012;19(1):1–10. doi:10.3109/10717544.2011.621989
29. Vighi E, Ruozi B, Montanari M, et al. pDNA condensation capacity and in vitro gene delivery properties of cationic solid lipid nanoparticles. Int J Pharm. 2010;389(1–2):254–261. doi:10.1016/j.ijpharm.2010.01.030
30. Ma Y, Zhang X, Xu X, et al. Stat3 decoy oligodeoxynucleotides-loaded solid lipid nanoparticles induce cell death and inhibit invasion in ovarian cancer cells. PLoS One. 2015;10(4):e0124924. doi:10.1371/journal.pone.0124924
31. Zhang X, Liu P, Zhang B, et al. Inhibitory effects of stat3 decoy oligodeoxynucleotides on human epithelial ovarian cancer cell growth in vivo. Int J Mol Med. 2013;32(3):623–628. doi:10.3892/ijmm.2013.1431
32. Yu W, Liu C, Ye J, et al. Novel cationic sln containing a synthesized single-tailed lipid as a modifier for gene delivery. Nanotechnology. 2009;20(21):215102. doi:10.1088/0957-4484/20/21/215102
33. Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12(9):823–830. doi:10.1038/ncb0910-823
34. Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544.
35. Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15(7):713–720. doi:10.1038/ncb2788
36. Fagard R, Metelev V, Souissi I, et al. Stat3 inhibitors for cancer therapy: have all roads been explored? JAK-STAT. 2013;2(1):e22882. doi:10.4161/jkst.22882
37. Orlova A, Wagner C, de Araujo ED, et al. Direct targeting options for stat3 and stat5 in cancer. Cancers (Basel). 2019;11(12):1930. doi:10.3390/cancers11121930
38. Corvinus FM, Orth C, Moriggl R, et al. Persistent stat3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005;7(6):545–555. doi:10.1593/neo.04571
39. Yu H, Lee H, Herrmann A, et al. Revisiting stat3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. doi:10.1038/nrc3818
40. Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109(9):1139–1142. doi:10.1172/JCI0215617
41. Wu CJ, Sundararajan V, Sheu BC, et al. Activation of stat3 and stat5 signaling in epithelial ovarian cancer progression: mechanism and therapeutic opportunity. Cancers (Basel). 2019;12(1):24. doi:10.3390/cancers12010024
42. Jing N, Tweardy DJ. Targeting stat3 in cancer therapy. Anticancer Drugs. 2005;16(6):601–607. doi:10.1097/00001813-200507000-00002
43. Leeman RJ, Lui VWY, Grandis JR. Stat3 as a therapeutic target in head and neck cancer. Expert Opin Biol Ther. 2006;6(3):231–241. doi:10.1517/14712598.6.3.231
44. Sen M, Tosca PJ, Zwayer C, et al. Lack of toxicity of a stat3 decoy oligonucleotide. Cancer Chemother Pharmacol. 2009;63(6):983–995. doi:10.1007/s00280-008-0823-6
45. Sen M, Thomas SM, Kim S, et al. First-in-human trial of a stat3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov. 2012;2(8):694–705. doi:10.1158/2159-8290.CD-12-0191
46. Ashrafizadeh M, Ahmadi Z, Kotla NG, et al. Nanoparticles targeting stats in cancer therapy. Cells. 2019;8(10):1158. doi:10.3390/cells8101158
47. Feng L, Mumper RJ. A critical review of lipid-based nanoparticles for taxane delivery. Cancer Lett. 2013;334(2):157–175. doi:10.1016/j.canlet.2012.07.006
48. Naseri N, Valizadeh H, Zakeri-Milani P. Solid lipid nanoparticles and nanostructured lipid carriers: structure, preparation and application. Adv Pharm Bull. 2015;5(3):305–313. doi:10.15171/apb.2015.043
49. Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5(3):161–171. doi:10.1038/nrc1566
50. Toit AD. Cell death: balance through a bivalent regulator. Nat Rev Mol Cell Biol. 2013;14(9):546–547. doi:10.1038/nrm3637
51. Zhang X, Liu P, Zhang B, et al. Role of stat3 decoy oligodeoxynucleotides on cell invasion and chemosensitivity in human epithelial ovarian cancer cells. Cancer Genet Cytogenet. 2010;197(1):46–53. doi:10.1016/j.cancergencyto.2009.10.004
52. Shen J, Li R, Li G. Inhibitory effects of decoy-odn targeting activated stat3 on human glioma growth in vivo. In Vivo. 2009;23(2):237–243.
53. Ghobrial IM, Witzig TE, Adjei AA. Targeting apoptosis pathways in cancer therapy. CA Cancer J Clin. 2005;55(3):178–194. doi:10.3322/canjclin.55.3.178
54. Masood A, Azmi AS, Mohammad RM. Small molecule inhibitors of bcl-2 family proteins for pancreatic cancer therapy. Cancers (Basel). 2011;3(2):1527–1549. doi:10.3109/10799893.2012.733885
55. Giraud AS, Menheniott TR, Judd LM. Targeting stat3 in gastric cancer. Expert Opin Ther Targets. 2012;16(9):889–901. doi:10.1517/14728222.2012.709238
56. Kamada S, Kikkawa U, Tsujimoto Y, et al. Nuclear translocation of caspase-3 is dependent on its proteolytic activation and recognition of a substrate-like protein(s). J Biol Chem. 2005;280(2):857–860. doi:10.1074/jbc.C400538200
57. Li YJ, Lei YH, Yao N, et al. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36(1):52. doi:10.1186/s40880-017-0219-2
58. Jonchère B, Bélanger A, Guette C, et al. Stat3 as a new autophagy regulator. JAK-STAT. 2013;2(3):e24353.
59. Salminen A, Kaarniranta K, Kauppinen A. Beclin 1 interactome controls the crosstalk between apoptosis, autophagy and inflammasome activation: impact on the aging process. Ageing Res Rev. 2013;12(2):520–534. doi:10.1016/j.arr.2012.11.004
60. He C, Wei Y, Sun K, et al. Beclin 2 functions in autophagy, degradation of g protein-coupled receptors, and metabolism. Cell. 2013;154(5):1085–1099. doi:10.1016/j.cell.2013.07.035
61. Nieto MA, Huang RY, Jackson RA, et al. Emt: 2016. Cell. 2016;166(1):21–45. doi:10.1016/j.cell.2016.06.028
62. Wendt MK, Balanis N, Carlin CR, et al. Stat3 and epithelial-mesenchymal transitions in carcinomas. JAK-STAT. 2014;3(1):e28975. doi:10.4161/jkst.28975
63. Kim MJ, Lim J, Yang Y, et al. N-myc downstream-regulated gene 2 (ndrg2) suppresses the epithelial-mesenchymal transition (EMT) in breast cancer cells via stat3/snail signaling. Cancer Lett. 2014;354(1):33–42. doi:10.1016/j.canlet.2014.06.023
64. Lee MY, Shen MR. Epithelial-mesenchymal transition in cervical carcinoma. Am J Transl Res. 2012;4(1):1–13.
65. Schubert MA, Müller-Goymann CC. Characterisation of surface-modified solid lipid nanoparticles (sln): influence of lecithin and nonionic emulsifier. Eur J Pharm Biopharm. 2005;61(1–2):77–86. doi:10.1016/j.ejpb.2005.03.006
66. Cortesi R, Campioni M, Ravani L, et al. Cationic lipid nanosystems as carriers for nucleic acids. N Biotechnol. 2014;31(1):44–54. doi:10.1016/j.nbt.2013.10.001
67. Singh H, Jindal S, Singh M, et al. Nano-formulation of rifampicin with enhanced bioavailability: development, characterization and in-vivo safety. Int J Pharm. 2015;485(1–2):138–151. doi:10.1016/j.ijpharm.2015.02.050
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.