Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 15
Novel lncRNA SNHG16 Promotes the Growth and Metastasis of Malignant Melanoma by Regulating miR-205-5p/PAK2 Axis
Authors Xia Y, Guan J, Lu X, Liu Y, Luan W
Received 22 May 2022
Accepted for publication 2 August 2022
Published 11 August 2022 Volume 2022:15 Pages 1615—1625
DOI https://doi.org/10.2147/CCID.S374404
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Yun Xia,1,* Jing Guan,1,* Xu Lu,2,* Yifan Liu,2 Wenkang Luan2
1Department of Plastic Surgery, The People’s Hospital of Danyang, Affiliated Danyang Hospital of Nantong University, Danyang, Jiangsu, People’s Republic of China; 2Department of Plastic Surgery, Affiliated People’s Hospital of Jiangsu University, Zhenjiang, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yun Xia, Department of Plastic Surgery, The People’s Hospital of Danyang, Affiliated Danyang Hospital of Nantong University, Xinmin West Road, Danyang, Jiangsu, 212000, People’s Republic of China, Email [email protected] Wenkang Luan, Department of Plastic Surgery, Affiliated People’s Hospital of Jiangsu University, 8 Dianli Road, Zhenjiang, Jiangsu, 212000, People’s Republic of China, Email [email protected]
Background: Long non-coding RNAs (lncRNAs) play an key role in the biological processes of various malignant tumors. SNHG16 has been confirmed to be associated with the progression of various cancers. However, function and molecular mechanism of SNHG16 in melanoma have not been studied by scholars.
Methods: The expression of SNHG16 in melanoma tissues were detected by using qRT-PCR. Melanoma cases from The Cancer Genome Atlas (TCGA) and GEO#GSE15605 were included in this study. CCK-8 assay, EdU assay, transwell and scratch wound assay were used to explore the role of SNHG16 in melanoma cells. Luciferase reporter assays and RNA pull-down assay were used to explore the molecular mechanism of SNHG16 in melanoma.
Results: Here, we found that SNHG16 was up-regulated in melanoma. SNHG16 enhances the growth and metastasis of melanoma. SNHG16 can promote the expression of P21-activated kinases 2 (PAK2) by sponging miR-205-5p. PAK2 is the target gene of miR-205-5p. We demonstrated that SNHG16 promotes the metastasis and growth of melanoma through miR-205-5p/PAK2 axis.
Conclusion: This study firstly confirmed the role and mechanism of SNHG16 in the occurrence and development of melanoma. Therefore, SNHG16 may become a key point in the diagnosis, prognosis and treatment of melanoma patients in the future.
Keywords: melanoma, growth and metastasis, SNHG16, miR-205-5p, PAK2
Introduction
Melanoma is a malignant tumor originated from skin melanocytes.1 Malignant melanoma has a poor prognosis, and is the leading cause of skin tumor related death.2 The incidence rate of melanoma has increased rapidly in in recent decade years.3 In melanoma patients with distant metastasis, despite the use of surgery, chemotherapy, targeted therapy and immunotherapy, their five-year survival rate is still very low.1,4,5 Therefore, it is very necessary to explore the molecular mechanism of the occurrence and development of melanoma and identify new molecular markers for melanoma.
Most DNA can be transcribed into RNA, but very little RNA can be translated into protein.6 This kind of RNA without coding ability is called no-ncoding RNA. lncRNA is a member of no-ncoding RNA, with a length of more than 200 nucleotides.7 It has been showed that lncRNA play an key role in the biological processes of various malignant tumors.8–10 Some specific lncRNA have been identified to be involved in the malignant progression of melanoma.11,12 lncRNA Small Nucleolar RNA Host Gene 16 (SNHG16), located on chromosome 17q25.1, was first described as a potential oncogene leading to poor prognosis in patients with neuroblastoma.13 SNHG16 has been confirmed to be associated with the progression of various cancers, such as colorectal cancer, gastric cancer, non-small cell lung cancer and so on.14–16 However, the expression, function and molecular mechanism of SNHG16 in melanoma have not been studied by scholars.
In this paper, we found that the expression of SNHG16 was up-regulated in melanoma. SNHG16 enhances the growth and metastasis of melanoma. It has been confirmed that some lncRNA is involved in tumor progression as a competitive endogenous RNA (ceRNA).17 It can sponge miRNA to increase the expression of target genes.18 SNHG16 has also been proved that it acts as a ceRNA in many tumors.19,20 We demonstrated that SNHG16 can promote the expression of P21-activated kinases 2 (PAK2) by sponging miR-205-5p. PAK2 is the target gene of miR-205-5p. More and more evidence shows that the over-expression of PAK2 participate in many signal pathways related to malignant progression of tumors, including melanoma.21 We demonstrated that SNHG16 promotes the metastasis and growth of melanoma through miR-205-5p/PAK2 axis. Thus SNHG16 may become a key point in the diagnosis, prognosis and treatment of melanoma patients in the future.
Materials and Methods
Human Tissues and Cells
The human melanoma tissues comes from the Cancer Genome Atlas (TCGA, 461 melanoma tissues), and 558 normal skin tissue comes from GTEx database, We analyzed the relevant data by using the GEPIA website (http://gepia.cancer-pku.cn/index.html). The published datasets (GEO#GSE15605, 58 melanoma tissues vs 16 normal skin tissue) and melanoma tissues we collected (18 melanoma tissues vs 18 normal skin tissue) also included in the analysis. Melanoma tissues and matched normal skin tissue were obtained in the People’s Hospital of Danyang. The study was sanctioned by the ethics committee of the People’s Hospital of Danyang. The study was conducted in accordance with the Declaration of Helsinki principles. The informed consent was obtained from each participant. We purchased the melanoma cell lines (A375, A875, A2058, MeWo) in the American Type Culture Collection (ATCC, USA). Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA) containing 10% foetal bovine serum (Invitrogen, USA) was used to culture melanoma cell lines. The human epidermal melanocyte (HEMa-LP) were from obtained Invitrogen and incubated in medium 254 (Cascade Biologics). The culture environment was 37 ° C condition with 5% CO2.
Oligonucleotides, Plasmids and Transfection
The mimic and inhibitor of miR-205-5p, small-interfering-RNA (siRNA) of SNHG16 was entrusted to GenePharma (Shanghai, China) to chemical synthesis. The sequences of SNHG16 siRNA: 5′-CCUGGGUAUAAUCUCACAATT-3′, and 5′-GGAACAUACUGCUAUCAUATT-3′. For constructing PAK2 plasmid, pcDNA3.1 vector (Invitrogen, USA) having been inserted the full length of PAK2. The lipofectamine 3000 (Invitrogen, USA) was used to transfect the related oligonucleotides and plasmids to melanoma cells according to the experimental steps.
Cell Proliferation Assay
For the CCK-8 (Beyotime, China) assay, the corresponding melanoma cells were implanted in 96 well plates with 5000 cells per well. CCK-8 reagent was added to the cells at different times (12, 24, 48 and 72 hours), followed by culture at 37 ° C for 2 hours. Using the microplate reader of Thermo Scientific Company to measure the absorbance with optical density of 450 nm. For the EdU assay, EdU Imaging Kit (Life Technologies, USA) was used to detect the DNA synthesis in grown melanoma cells, and the experiment is carried out according to the manufacturer’s instructions. We used Leica DMI3000B microscope to observe the immunostained cells and count.
Cell Invasion and Migration Assays
For the transwell assay, melanoma cells were placed in DMEM with serum-free. The cells were placed on the top of the chamber (BD Biosciences) and the chamber was coated with matrix gel. DMEM containing 10% fetal bovine serum was added to the down chamber as a chemokine. After 24 hours of culture, the invading cells were stained with 0.1% crystal violet, counted and photographed. For the scratch wound assay, the corresponding melanoma cells were inoculated into 6-well plates, followed by tip of 200 μL pipette to scratch the cell layer to form a gap wound. Wound widths (0 and 48 hours) were recorded. Take pictures of cells with a inverted microscope.
Quantitative RT-PCR
Trizol reagent (Invitrogen, USA) was used to extract the corresponding RNA. Then, reverse transcription was performed by using transcription Kit (Applied Biosystems, CA) according to the experimental process. Finally, we set the reaction conditions and used ABI StepOnePlus system (Applied Biosystems, CA) for amplification reaction according to the instructions. The primer sequences are as follows: SNHG16 forward 5′-TACTCTGTTGGAAGAGCCTAA-3′ and SNHG16 reverse 5′-GGGTGTTGGTAACGAAA-3′; PAK2 forward 5′-GACTGCTCCTCCCGTTATTG-3′ and PAK2 reverse 5′-ACTTGGCAGCACCATCAA-3′; miR-205-5p forward 5′-ACACTCCAGCTGGGTCCTTCATTCC-3′ and RT 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAGACT-3′. For detecting SNHG16 and PAK2, GAPDH was used as control. U6 was used as control for miR-205-5p. The relative expression were expressed using 2–ΔΔCt.
Western Blot
Firstly, we use the bicinchoninic acid method to quantify the extracted protein. The protein was then separated, transferred to polyvinylidene fluoride membrane and sealed with 5% skimmed milk. Then, it was incubated with primary antibody overnight. Finally, the second antibody was incubated and the signal was detected on the membrane. Antibodies against PAK2 were used (Abcam, UK, 1:1000).
Luciferase Reporter Assay
We first predicted the corresponding binding sites of miR-205-5p. PAK2 3 ‘- UTR and SNHG16 fragments containing miR-205-5p binding sites were inserted into the reporter plasmid. Compared with the wild-type reporter plasmid, we used the mutant as the control and transfected the related oligonucleotides and luciferase reporter plasmid. Finally, the Luciferase Report Assay System (Promega, USA) was used to detect the activity of luciferase.
RNA Pull-Down Assay
Biotinylated miRNA can prove the binding relationship between miR-205-5p and PAK2. Guangzhou Ruibo synthesize biotinylated miR-205-5p, mutants and NC as the control, and then cultured the cells with M-280 streptavidin magnetic beads (Invitrogen). Finally, qRT PCR was used to measure the level of SNHG16 in bound RNA.
Statistical Analysis
All data were analyzed by SPSS13. 0 (mean ± standard deviation). Whether the data have the statistically significant was estimated by t-test or one-way ANOVA. The Spearman correlation analysis was performed. P value less than or equal to 0.05 is considered that this data is statistically significant.
Results
SNHG16 Expression is Increased in Melanoma
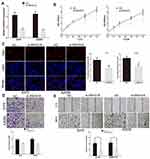
First, we analysed the level of SNHG16 in 461 melanoma tissues and 558 normal skin tissues in TCGA and GTEx datasets by using the GEPIA website (http://gepia.cancer-pku.cn/index.html). We discovered that the expression of SNHG16 in melanoma tissues was higher than that in normal skin tissues (Figure 1A). Published datasets (GEO# GSE15605) were also used to analyse the expression of SNHG16 in melanoma tissues, which confirmed the overexpression of SNHG16 in melanoma (Figure 1B). The same results were demonstrated by detecting the SNHG16 level in 18 melanoma tissues and adjacent normal skin tissues collected for this study (Figure 1C). Then, the level of SNHG16 was measured in cells, and a significant increase was found in melanoma cells (A375, A875, A2058, MeWo) compared to human epidermal melanocytes (HEMa-LP) (Figure 1D). These results indicated that SNHG16 plays a critical role in the progression of melanoma.
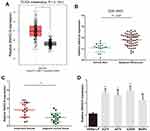 |
Figure 1 SNHG16 expression is increased in melanoma. (A) GEPIA website (http://gepia.cancer-pku.cn/index.html) was used to detect the expression of SNHG16 in TCGA melanoma dataset (461 melanoma tissues and 558 normal skin tissue). (B) The level of SNHG16 was analyzed in published melanoma datasets (GEO#GSE15605). (C) The overexpression of SNHG16 was analyzed in 18 melanoma tissues and adjacent normal skin tissue. (D) The expression profile of SNHG16 in melanoma cell lines (A375, A875, A2058, MeWo) and human epidermal melanocytes (HEMa-LP). *P < 0.05, **P < 0.01, ***P < 0.001. |
SNHG16 Promotes the Growth and Metastasis of Melanoma Cells
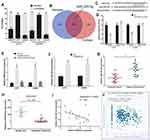
To further explore the effect of SNHG16 in melanoma cells, SNHG16 siRNA was transfected into A375 and A2058 cells (Figure 2A). CCK8 and EdU assays were used to analyse the proliferation ability of melanoma cells. We found that the proliferation of A375 and A2058 cells was inhibited by SNHG16 siRNA (Figure 2B). The number of EdU-positive cells in SNHG16 siRNA-transfected melanoma cells was significantly reduced compared to that in control cells (Figure 2C). Meanwhile, the migration and invasion abilities of melanoma cells were estimated by wound healing and transwell assays. The knockdown of SNHG16 caused a significant decline in the migration and invasion of melanoma cells (Figure 2D and E). These results suggested that SNHG16 enhances the growth and metastasis of melanoma cells.
SNHG16 Actes as a Sponge of miR-205-5p in Melanoma
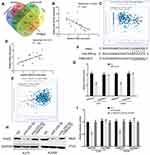
lncRNAs have been proven to act as ceRNAs that can mediate mRNA by sponging miRNAs in many tumors.9,22 First, we extracted RNA from the nucleus and cytoplasm and conducted qRT‒PCR experiments. It was found that SNHG16 was located in the cytoplasm and the nucleus (Figure 3A). Bioinformatics prediction software (lncBase V3.0 and StarBase 3.0) was used to identify the miRNAs that bind to SNHG16, and 53 potentially binding miRNAs were found (Figure 3B and Supplementary Table 1). Three miRNAs were expressed at low levels in melanoma and were negatively correlated with the expression of SNHG16. Among them, miR-205-5p had the highest correlation with SNHG16. For the luciferase reporter assay, we constructed SNHG16 luciferase reporter plasmids that included the binding sites of miR-205-5p and mutant vectors as the control (Figure 3C). The luciferase activity of wild-type SNHG16 luciferase reporter plasmids was repressed by the miR-205-5p mimic, but no changes were found in the mutant plasmid (Figure 3D). An RNA pull-down assay was used to further confirm the direct binding relationship between SNHG16 and miR-205-5p in melanoma. We synthesized biotinylated-miR-205-5p and found that SNHG16 was pulled down by biotinylated-miR-205-5p (Figure 3E). Meanwhile, the miR-205-5p levels increased along with the knockdown of SNHG16 in melanoma cells (Figure 3F). We also found that the expression of miR-205-5p was decreased in the melanoma tissues we collected, and the same results were confirmed by analysing published datasets (GEO# GSE24996) (Figure 3G and H). SNHG16 had a negative correlation with miR-205-5p in both the melanoma tissues we collected and TCGA melanoma database (Figure 3I and J). All results suggested that miR-205-5p and SNHG16 show a direct binding state in melanoma.
SNHG16 Promotes PAK2 Expression by Binding miR-205-5p in Melanoma Cells
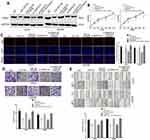
Four bioinformatics software programs (TargetScan, StarBase, miRPathDB and PITA) were used to identify the target gene of miR-205-5p (Figure 4A). Among the candidate genes, PAK2 attracted our attention, mainly due to the negative correlation between the expression of PAK2 and miR-205-5p and the positive correlation between PAK2 and SNHG16 (Figure 4B–E). The 3’-UTR of PAK2 shares the same binding sites of miR-205-5p with SNHG16 (Figure 4F). We constructed the wild-type PAK2 luciferase reporter plasmid, and the mutant plasmid was used as the control. A luciferase reporter assay showed that the luciferase activity of the PAK2 wild-type reporter plasmid was inhibited by the miR-205-5p mimic in melanoma (Figure 4G). The protein expression levels of PAK2 were repressed by the miR-205-5p mimic in melanoma cells (Figure 4H). TCGA database and our melanoma samples also proved the negative correlation between miR-205-5p and PAK2 (Figure 4B and C). These results demonstrated that miR-205-5p targeted the inhibition of PAK2. Moreover, the SNHG16 plasmid reversed the inhibitory effect of miR-205-5p on the protein level of PAK2 (Figure 4H). The luciferase activity of the PAK2 wild-type reporter plasmid showed a significant decrease in the SNHG16 knockdown group, and the effect of SNHG16 was reversed by the miR-205-5p inhibitor (Figure 4I).
The expression of PAK2 was decreased at the protein level when SNHG16 siRNA was transfected, and this effect was abolished by transfection with the miR-205-5p inhibitor (Figure 5A). We then further verify the regulatory effect of SNHG16 on PAK2. We found that the protein expression of PAK2 was increased when SNHG16 plasmid was transfected, and this effect can be reversed by a miR-205-5p mimic (Supplementary Figure 1). All these data demonstrated that SNHG16 promotes PAK2 expression by binding miR-205-5p in melanoma.
SNHG16 Promotes the Growth and Metastasis of Melanoma Cells by Regulating the miR-205-5p/PAK2 Axis
We explored whether SNHG16 promotes the malignant progression of melanoma through the miR-205-5p/PAK2 pathway. The miR-205-5p mimic, SNHG16 siRNA, SNHG16 siRNA plus miR-205-5p inhibitor and miR-205-5p mimic plus PAK2 plasmid were transfected into melanoma cells. Western blotting detected the protein expression of PAK2 in different treatment groups (Figure 5A). CCK8, EdU, wound healing and transwell assays showed that the proliferation and metastasis of melanoma cells were suppressed by the miR-205-5p mimic, and the PAK2 plasmid reversed the effect of miR-205-5p (Figure 5B–E). Therefore, we demonstrated that miR-205-5p acts as a tumor suppressor gene in melanoma by targeting PAK2. Moreover, SNHG16 knockdown caused a decrease in the PAK2 level, metastasis and growth of melanoma cells, and this negative regulatory effect was abolished by transfection with the miR-205-5p inhibitor (Figure 5B–E). In conclusion, these results suggested that SNHG16 has a cancer-promoting effect by regulating the miR-205-5p/PAK2 axis.
Discussion
LncRNA, a kind of non-coding RNA with a length of more than 200 nucleotides, is involved in many biological processes and the malignant progression of tumors.9,10 Many special lncRNAs have been proven to be abnormally expressed and used as specific molecular markers in many human tumors.23,24 SNHG16, a non-coding RNA expressed in aggressive neuroblastoma, is 2435 nucleotides long and located on chromosome 17q25.1.13,25 In recent years, the role of SNHG16 in malignant tumors has been widely studied. It is considered that SNHG16 plays the role of a carcinogenic lncRNA in some tumors.19,25 For instance, SNHG16 promotes the proliferation, metastasis and autophagy of neuroblastoma by affecting the expression of ATG5 through miR-542-3p.26 Exosomal SNHG16, secreted by cancer stem cells, promotes the malignant progression of glioma by regulating TLR7.27 It has been reported that SNHG16 promotes the proliferation and migration of melanoma cells.28 However, there are few studies on the expression, function and mechanism of SNHG16 in melanoma. In this paper, we detected the expression of SNHG16 in TCGA melanoma database and our melanoma tissues. We found that SNHG16 is un-regulated in melanoma tissues compared to normal skin tissues. CCK8, EdU, wound healing and transwell assays showed that SNHG16 promoted the growth and metastasis of melanoma cells. Then, we deeply studied the mechanism of SNHG16 in melanoma.
The present theory is that lncRNAs can play biological roles as ceRNAs in human tumor.29,30 ceRNAs, RNAs with miRNA binding sites, regulate the expression of oncogenes and tumour suppressor genes by competitively binding miRNAs to liberate mRNAs.17 SNHG16 has also been confirmed to act as a ceRNA in many tumors. SNHG16 contributes to the progression of osteosarcoma by sponging miR-1285-3p20 and promotes colorectal cancer progression by regulating the miR-132-3p/USP22 pathway.31 Here, we used bioinformatics prediction software to identify the potential ceRNA network of SNHG16. SNHG16, miR-205-5p and PAK2 were found to have potential ceRNA correlations in melanoma based on the results of prediction software and TCGA and our melanoma tissue databases. miR-205-5p has been proven to have both tumor suppressing and tumor promoting effects. For example, miRNA-205-5p acts as a tumor suppressor in renal carcinoma cells by regulating VEGFA.32 Exosomal miR-205-5p enhances the metastasis of nasopharyngeal carcinoma by targeting desmocollin-2.33 Recent research has shown that miR-205-5p increases sensitivity to vemurafenib and induces cell apoptosis in melanoma by down-regulating TNFAIP8.34 We further proved that miR-205-5p plays an anti-cancer role in melanoma. PAK2 is the target gene of miR-205-5p, and SNHG16 can directly bind to miR-205-5p. The expression of PAK2 was decreased at the protein level when SNHG16 siRNA was transfected, but the protein expression of PAK2 was increased when SNHG16 plasmid was transfected. We further demonstrated that SNHG16 up-regulates the level of PAK2 by decoying miR-205-5p in melanoma.
PAKs, members of the STE20 family of serine/threonine kinases, play a major role in regulating changes in actin cytoskeleton organization, cell shape, and adhesion dynamics.35 Six PAK isoforms were divided into two families: Group I (PAK1-3) and Group II (PAK4-6).36 Many scholars have proven that PAK2 participates in many signalling pathways related to tumors, including melanoma.21,37 Here, we further showed that co-transfection of a miR-205-5p inhibitor can reverse the effect of SNHG16 siRNA on the metastasis and growth of melanoma cells. In this study, we constructed a ceRNA network of SNHG16/miR-205-5p/PAK2 in the malignant progression of melanoma.
Conclusion
In conclusion, SNHG16 play an vital role in the malignant progression of melanoma. SNHG16 competitively bind to miR-205-5p to cut off the targeted binding between miR-205-5p and PAK2. Thus, SNHG16 can up-regulate the expression of PAK2 by liberating PAK2 mRNA transcripts. SNHG16 exerts its pro-cancer effect through regulating miR-205-5p/PAK2 axis. In order to find novel biological targets for early diagnosis and targeted therapy of melanoma patients, deeply studying the internal mechanism of SNHG16/miR-205-5p/PAK2 regulatory network is necessary.
Disclosure
This study was funded by Zhenjiang Social Development Foundation of Zhenjiang key R & D projects (SH2020044), the People’s Hospital of Danyang scientific research project (DRYK202009). The authors declare that they have no competing interests.
References
1. Ferrone S. Human Melanoma: From Basic Research to Clinical Application. Berlin; New York: Springer-Verlag; 1990.
2. Ahmed B, Qadir MI, Ghafoor S. Malignant melanoma: skin cancer-diagnosis, prevention, and treatment. Crit Rev Eukaryot Gene Expr. 2020;30(4):291–297. doi:10.1615/CritRevEukaryotGeneExpr.2020028454
3. Little EG, Eide MJ. Update on the current state of melanoma incidence. Dermatol Clin. 2012;30(3):355–361. doi:10.1016/j.det.2012.04.001
4. Kozovska Z, Gabrisova V, Kucerova L. Malignant melanoma: diagnosis, treatment and cancer stem cells. Neoplasma. 2016;63(4):510–517. doi:10.4149/neo_2016_403
5. Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10(2):103–121. doi:10.1080/15592294.2014.1003746
6. Elgar G, Vavouri T. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008;24(7):344–352. doi:10.1016/j.tig.2008.04.005
7. Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21(5):542–551. doi:10.1038/s41556-019-0311-8
8. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi:10.4161/rna.20481
9. Cheng JT, Wang L, Wang H, et al. Insights into biological role of LncRNAs in epithelial-mesenchymal transition. Cells. 2019;8(10):1178. doi:10.3390/cells8101178
10. Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15–24. doi:10.1016/j.addr.2015.05.012
11. Ma DM, Sun D, Wang J, Jin DH, Li Y, Han YE. Long non-coding RNA MIR4435-2HG recruits miR-802 from FLOT2 to promote melanoma progression. Eur Rev Med Pharmacol Sci. 2020;24(5):2616–2624. doi:10.26355/eurrev_202003_20530
12. Luan W, Zhou Z, Ni X, et al. Long non-coding RNA H19 promotes glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J Cancer Res Clin Oncol. 2018;144(3):531–542. doi:10.1007/s00432-018-2582-z
13. Yu M, Ohira M, Li Y, et al. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol. 2009;34(4):931–938. doi:10.3892/ijo_00000219
14. Christensen LL, True K, Hamilton MP, et al. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol. 2016;10(8):1266–1282. doi:10.1016/j.molonc.2016.06.003
15. Lian D, Amin B, Du D, Yan W. Enhanced expression of the long non-coding RNA SNHG16 contributes to gastric cancer progression and metastasis. Cancer Biomark. 2017;21(1):151–160. doi:10.3233/CBM-170462
16. Han W, Du X, Liu M, Wang J, Sun L, Li Y. Increased expression of long non-coding RNA SNHG16 correlates with tumor progression and poor prognosis in non-small cell lung cancer. Int J Biol Macromol. 2019;121:270–278. doi:10.1016/j.ijbiomac.2018.10.004
17. Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54(5):766–776. doi:10.1016/j.molcel.2014.03.045
18. Fernandes M, Marques H, Teixeira AL, Medeiros R. ceRNA network of lncRNA/miRNA as circulating prognostic biomarkers in non-hodgkin lymphomas: bioinformatic analysis and assessment of their prognostic value in an NHL cohort. Int J Mol Sci. 2021;23:1. doi:10.3390/ijms23010201
19. Xia W, Liu Y, Cheng T, Xu T, Dong M, Hu X. Extracellular vesicles carry lncRNA SNHG16 to promote metastasis of breast cancer cells via the miR-892b/PPAPDC1A axis. Front Cell Dev Biol. 2021;9:628573. doi:10.3389/fcell.2021.628573
20. Xiao X, Jiang G, Zhang S, et al. LncRNA SNHG16 contributes to osteosarcoma progression by acting as a ceRNA of miR-1285-3p. BMC Cancer. 2021;21(1):355. doi:10.1186/s12885-021-07933-2
21. Gao C, Ma T, Pang L, Xie R. Activation of P21-activated protein kinase 2 is an independent prognostic predictor for patients with gastric cancer. Diagn Pathol. 2014;9:55. doi:10.1186/1746-1596-9-55
22. Lin X, Xiang X, Feng B, et al. Targeting long non-coding RNAs in hepatocellular carcinoma: progress and prospects. Front Oncol. 2021;11:670838. doi:10.3389/fonc.2021.670838
23. Zhao GY, Ning ZF, Wang R. LncRNA SNHG19 promotes the development of non-small cell lung cancer via mediating miR-137/E2F7 axis. Front Oncol. 2021;11:630241. doi:10.3389/fonc.2021.630241
24. Zhou FC, Zhang YH, Liu HT, Song J, Shao J. LncRNA LINC00588 suppresses the progression of osteosarcoma by acting as a ceRNA for miRNA-1972. Front Pharmacol. 2020;11:255. doi:10.3389/fphar.2020.00255
25. Xiao Y, Xiao T, Ou W, et al. LncRNA SNHG16 as a potential biomarker and therapeutic target in human cancers. Biomark Res. 2020;8:41. doi:10.1186/s40364-020-00221-4
26. Wen Y, Gong X, Dong Y, Tang C. Long non coding RNA SNHG16 facilitates proliferation, migration, invasion and autophagy of neuroblastoma cells via sponging miR-542-3p and upregulating ATG5 expression. Onco Targets Ther. 2020;13:263–275. doi:10.2147/OTT.S226915
27. Zhang R, Li P, Lv H, Li N, Ren S, Xu W. Exosomal SNHG16 secreted by CSCs promotes glioma development via TLR7. Stem Cell Res Ther. 2021;12(1):349. doi:10.1186/s13287-021-02393-8
28. Bi LL, Hua XQ, Li WH, Wang L, Li Y, Jia XF. SNHG16 promotes cell proliferation and migration through sponging miR-132 in melanoma. J Biol Regul Homeost Agents. 2020;34(4):1307–1316. doi:10.23812/20-172-A
29. Jia X, Shi Y, Zhu Y, et al. Integrated analysis of mRNA-miRNA-lncRNA ceRNA network in human HR+/Her-2- breast cancer and triple negative breast cancer. J Comput Biol. 2020;27(7):1055–1066. doi:10.1089/cmb.2019.0152
30. Seo D, Kim D, Chae Y, Kim W. The ceRNA network of lncRNA and miRNA in lung cancer. Genomics Inform. 2020;18(4):e36. doi:10.5808/GI.2020.18.4.e36
31. He X, Ma J, Zhang M, Cui J, Yang H. Long non-coding RNA SNHG16 activates USP22 expression to promote colorectal cancer progression by sponging miR-132-3p. Onco Targets Ther. 2020;13:4283–4294. doi:10.2147/OTT.S244778
32. Huang J, Wang X, Wen G, Ren Y. miRNA2055p functions as a tumor suppressor by negatively regulating VEGFA and PI3K/Akt/mTOR signaling in renal carcinoma cells. Oncol Rep. 2019;42(5):1677–1688. doi:10.3892/or.2019.7307
33. Yang W, Tan S, Yang L, et al. Exosomal miR-205-5p enhances angiogenesis and nasopharyngeal carcinoma metastasis by targeting desmocollin-2. Molecular Therapy Oncolytics. 2022;24:612–623. doi:10.1016/j.omto.2022.02.008
34. Ge X, Niture S, Lin M, Cagle P, Li PA, Kumar D. MicroRNA-205-5p inhibits skin cancer cell proliferation and increase drug sensitivity by targeting TNFAIP8. Sci Rep. 2021;11(1):5660. doi:10.1038/s41598-021-85097-6
35. Radu M, Semenova G, Kosoff R, Chernoff J. PAK signalling during the development and progression of cancer. Nat Rev Cancer. 2014;14(1):13–25. doi:10.1038/nrc3645
36. Arias-Romero LE, Chernoff J. A tale of two Paks. Biology of the Cell. 2008;100(2):97–108. doi:10.1042/BC20070109
37. Ni X, Ding Y, Yuan H, et al. Long non-coding RNA ZEB1-AS1 promotes colon adenocarcinoma malignant progression via miR-455-3p/PAK2 axis. Cell Prolif. 2020;53(1):e12723. doi:10.1111/cpr.12723
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.