Back to Journals » OncoTargets and Therapy » Volume 13
Novel lncRNA SFTA1P Promotes Tumor Growth by Down-Regulating miR-4766-5p via PI3K/AKT/mTOR Signaling Pathway in Hepatocellular Carcinoma
Authors Huang G, Yang Y, Lv M, Huang T, Zhan X, Kang W, Hou J
Received 6 February 2020
Accepted for publication 22 July 2020
Published 30 September 2020 Volume 2020:13 Pages 9759—9770
DOI https://doi.org/10.2147/OTT.S248660
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr XuYu Yang
Guohong Huang,* Yimei Yang,* Mengxin Lv, Tian Huang, Xiaoyan Zhan, Wei Kang, Jianghou Hou
Clinical Research Center of Kunming Maternal and Child Health Hospital, Kunming 650031, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianghou Hou
Clinical Research Center of Kunming Maternal and Child Health Hospital, Kunming 650031, People’s Republic of China
Email [email protected]
Background: Hepatocellular carcinoma (HCC) is a common malignancy worldwide with a high mortality rate. lncRNA SFTA1P is highly expressed in HCC. We aimed to study the role of SFTA1P in HCC and its relationship with miR-4766-5p.
Materials and Methods: The levels of SFTA1P in HCC tissues and cell lines were determined. Relationship between SFTA1P and clinical features and prognosis was studied. The influence of SFTA1P on HCC cell viability, migration, invasion and apoptosis was studied in vitro. Rescue experiments were conducted after the binding site between SFTA1P and miR-4766-5p confirmed by dual-luciferase assay. The protein expression of AKT, p-AKT, mTOR and p-mTOR in HCC cells with knockdown of SFTA1P was determined by Western blotting. A tumor study in nude mice was conducted in order to assess the effects of SFTA1P on tumor growth characteristics.
Results: SFTA1P was up-regulated in HCC tissues and cell lines. SFTA1P expression was closely related to tumor size, vascular invasion and TNM stage. Knockdown of SFTA1P inhibited HCC cell viability, migration and invasion and promoted cell apoptosis. MiR-4766-5p was a target of SFTA1P and knockdown of SFTA1P could decrease the protein expression of p-AKT and p-mTOR. Rescue experiments showed that miR-4766-5p mimics could attenuate the promoting role of SFTA1P on HCC cell viability, invasion and migration, and inhibiting role on cell apoptosis. Moreover, we used nude mice models and also found that the knockdown of SFTA1P reduced tumor volume and weight.
Conclusion: lncRNA SFTA1P could promote tumor development in HCC by down-regulating miR-4766-5p expression via PI3K/AKT/mTOR signaling pathway. It may be a potential therapeutic target for HCC.
Keywords: hepatocellular carcinoma, SFTA1P, miR-4766-5p, PI3K/AKT/mTOR signaling pathway, tumor growth
Introduction
Hepatocellular carcinoma (HCC) is a common malignancy worldwide, and its mortality rate is the second highest in tumor-related deaths.1 It is characterized by complex pathophysiological processes, insidious clinical manifestations, low specificity and poor sensitivity of biochemical indicators.2 Therefore, a thorough study of the molecular events in the development of HCC will play an important role in the diagnosis and treatment of HCC patients. As we all know, hundreds of candidate tumor genes and tumor suppressor genes related to HCC have been discovered by sequencing technology in the past few years.3 Some of the genes have been fully confirmed by follow-up experiments, while function of most genes remains unclear. Recent studies have shown that the majority of human genomes are actively transcribed into non-coding RNAs (ncRNAs), of which only 1–2% genomes encode proteins.4 These non-protein-coding RNAs play important roles in cell growth, differentiation and metabolism.5 Long-chain non-coding RNA (lncRNA) is one of those RNAs and is a promising biomarker for treatment or prognosis and a potential target of cancer treatment.
In recent years, more and more studies have shown that lncRNA is closely related to the occurrence and development of HCC, and affects the progress and prognosis of HCC in many ways.6 lncRNA-HOXD-AS1 is markedly up-regulated in HCC tissues and reported to be a prognostic marker for HCC patients.7 lncRNA DIHCC was found down-regulated in HCC and could regulate HCC stem cells by IL-6/STAT3 axis.8 lncRNA SFTA1P is 693 nts long and is located on 10p.9 It is a specific lncRNA in lung tissues, and its expression is different in lung cancer tissues and adjacent tissues.10 In gastric cancer, lncRNA SFTA1P can inhibit cancer cell proliferation and migration via down-regulating TP53.11 Moreover, Liao et al reported HCC patients having higher SFTA1P expression were more likely to have liver fibrosis.12 However, there have no studies about the function of SFTA1P in HCC.
lncRNA could regulate the expression and biological functions of microRNA. MiR-4766-5p is a newly discovered microRNA and knockdown of miR-4766-5p could markedly promote cell growth and metastasis.13 Liang et al reported miR-4766-5p can be regulated by lncRNA-PRLB in breast cancer, which is related to the progression of breast cancer.14 Phosphoinositide 3-kinase (PI3K) is a key downstream of growth factor receptor tyrosine kinases. It can promote the formation of lipid second messenger PIP3 at cell membrane. PIP3 in turn activates downstream targets, such as Akt.15 PI3K signaling pathway plays an important role in many aspects of cell growth.16 PI3K/Akt signaling is often broken in human cancers. It could regulate essential cell function, such as proliferation, migration and transcription.17 It can regulate epithelial–mesenchymal transition (EMT) in HCC cells.18
In this study, we determined the expression of lncRNA SFTA1P in HCC tissues and the cell lines. Then, we explored the role of SFTA1P expression on HCC cell viability and migration. At last, the molecular mechanism about how SFTA1P regulates HCC was studied. Our study also found SFTA1P may play as an endogenous sponge via inhibiting the expression of miR-4766-5p.
Materials and Methods
Tissues Collection
HCC tissues and the adjacent normal liver tissues were collected from the pathology laboratory of Clinical Research Center of Kunming Maternal and Child Health Hospital between April 2015 and March 2018. HCC tissues and the adjacent normal tissues were obtained from 75 HCC patients. All tissues kept in the refrigerator at −80°C. The use of human tissues was approved by the Ethics Committee of Clinical Research Center of Kunming Maternal and Child Health Hospital and was conducted in accordance with the Declaration of Helsinki. We obtained written informed consent from every patient.
Cell Culture and Transfection
Human HCC cell lines BEL-7402, Hep G2, HCC-9204, Hep 3B, BEL-7405 and normal hepatocyte L-02 were obtained from Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. Cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS) and 1% mycillin. Then, all cells were incubated in cell incubator at 37°C containing 5% CO2. Hep G2 and Hep 3B were transfected with si-SFTA1P or/and SFTA1P pcDNA3.1 or/and miR-4766-5p mimic. For epithelial-like Hep G2, which expresses 3-hydroxy-3-methylglutaryl-CoA reductase and hepatic triglyceride lipase activities, the STR result shows Amelogenin: X, Y; CSF1PO: 10, 11; D13S317: 9, 13; D16S539: 12, 13; D5S818: 11, 12; D7S820: 10; TH01: 9; TPOX: 8, 9; vWA: 17; and there is no evidence of a Hepatitis B virus genome in this cell line. For epithelial-like Hep 3B, the STR result shows Amelogenin: X; CSF1PO: 8; D13S317: 12, 14; D16S539: 10; D5S818: 13; D7S820: 8, 10; TH01: 6, 7; TPOX: 9; vWA: 17, and this line contains an integrated hepatitis B virus genome.
2ʹ-O-methyl (2ʹ-O-Me) oligonucleotides containing miR-4766-5p (UCUGAAAGAGCAGUUGGUGUU) were synthesized by Shanghai Jima Biotechnology Co. LTD (Shanghai, China). They were transfected to cells by Lipofectamine 2000 based on the instructions. SFTA1P pcDNA3.1 (SFTA1P) and the negative control pcDNA3.1 (pcDNA3.1) were synthesized by JRDUN Biotechnology (Shanghai, China). They were co-transfected with the miR-4766-5p or NC into cells using Lipofectamine 2000.
Real-Time Polymerase Chain Reaction (RT-PCR)
Trizol lysis method was used to extract total RNA from HCC tissues and cells. After lysing, trichloromethane was added and mixed, and then centrifugation was performed at 10,000 rpm for 15 min. The watery layer was collected and isopropanol was added to precipitate RNA. RNase Free dd H2O was used to resolve RNA and NANO DROP was used to determine the purity and concentration of RNA. RNA is reversely transcribed into cDNA. QRT-PCR was performed by using the LightCycler and the FastStart DNA Master SYBR Green 1 kit (Roche Applied Sciences). The sequences of the primers were shown as follows: SFTA1P, forward primer: 5ʹ-TTGTGGCACGAGTAAGCCAA-3ʹ and reverse primer: 5ʹ-TCAAGGGCAATATTCCGGGT-3ʹ; GAPDH, forward primer: 5ʹ-AGGTCGGTG TGAACGGATTTG-3ʹ and reverse primer: 5ʹ-TGTAGACCATGTAGTTGAGG TCA-3ʹ.
Western Blotting
Total protein was isolated using RIPA lysis buffer (Beyotime). Equivalent amounts of proteins were fractionated by SDS-PAGE gel and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). The membranes were then incubated with specific primary antibodies against AKT (ab179463; 1:10,000; Abcam, Inc.), p-AKT (Ser473, #4060; 1:2000; Cell Signaling Technology, Inc.), mTOR (2972; 1:1000; Cell Signaling Technology, Inc.), p-mTOR (phospho S2448; ab109268; 1:2000; Abcam, Inc.), GAPDH (#5174; 1:1000; Cell Signaling Technology, Inc.) overnight at 4°C, followed by incubation with HRP-conjugated secondary antibody (1:10,000; Santa Cruz Biotechnology, Inc.) at room temperature for 1 hr. The immunoreactive bands were detected using the enhanced chemiluminescence detection kit (Millipore). GAPDH was used as the control.
Establishment of SFTA1P Overexpressed and Knockdown Cell Lines
lncRNA SFTA1P overexpression lentivirus was obtained from Shanghai Gemma Gene Biology Co. LTD. Cells of logarithmic growth phase were inoculated into the culture plates. Culture was performed for 48–72 h to achieve a density of about 70%. The lentivirus containing overexpressed plasmids is melted on ice and a specific amount of virus is added. After infection for 72 h, observation of green light GFP under fluorescence microscope indicated successful cell infection. RT-PCR was used to determine the expression of lncRNA SFTA1P. In the same way, we use siRNA plasmids of SFTA1P to build knockdown cell lines.
Luciferase Reporter Assay
Starbase 3.0 and AnnoLnc were used to explore the relationship between lncRNA SFTA1P and miR-4766-5p. Then, luciferase reporter assay was applied to verify the target gene. lncRNA-SFTA1P with WT or MUT miR-4766-5p binding sites were generated and fused to the luciferase reporter vector pmirGLO (Promega, Madison, WI, USA). HEK293T cells were placed on a 96-well plate and cotransfected with luciferase plasmids and miR-4766-5p mimic or control miRNA. After 48 h transfection, luciferase activity was measured with the dual-luciferase reporter assay system (Promega). Firefly luciferase activity was normalized against Renilla luciferase activity.
Cell Viability
CCK-8 kit (Beyotime Institute of Biotechnology, China) was used to determine the viability situation of HCC cells in different groups. Cells were inoculated to 96-well plates for 5×103 cells/well. After incubation for 0, 24, 48, 72, 96 h, CCK-8 reagent of 10 ul was added to each well and incubation was continued for 2 h. Micrometer detected the optical density of each hole at the wavelength of 450nm.
Cell Colony Formation Assay
The Hep G2 and Hep 3B cell lines were prepared into a cell suspension and counted. The Hep G2 and Hep 3B cell lines in each group were seeded into a 60 mm plate at a density of 1000 cells. Subsequently, 3 mL of cell culture medium was added to the cell lines at 37°C, and cultured in a 37°C incubator. The culture was terminated upon observation of the colony. Cells were fixed using 5 mL of 4% paraformaldehyde for 15 minutes and then stained with Crystal violet staining for 10–30 minutes.
Flow Cytometry
Cells were washed with PBS and pancreatin was added to digest cells. After digestion at room temperature, FBS was added to stop digestion until the adherent cells could be blown down by light blowing. Cell suspension was transferred into a centrifuge tube and centrifuged at 1000g for 5 min. The cells were collected and the gently suspended with PBS and counted. Cells were centrifuged at 1000g for 5 min and Annexin V-FITC solution was added to resuspend cells. Propyl iodide (PI) was added and mixed uniformly. After incubation in dark for 20 minutes at room temperature, flow cytometer was used for detection.
Transwell Assay for Migration and Invasion
BD matrigel, which was frozen in a refrigerator at −80°C, was turned into a liquid at 4°C overnight. After being diluted 5 times, it was placed in the upper chamber of Transwell chamber and incubated at 37°C for 4–5 h, and the appearance of a “white layer” indicated that it has become solid. Two groups of cell suspensions were prepared and the cell concentration was adjusted to 1×106/mL. Cell suspension of 150 µL was added to each well and 750 µL 1640 medium containing 10% FBS was added to the lower chamber, and then they were incubated at 37°C for 24 h. The Transwell chamber was washed twice with PBS and the cell fixation solution was used to fix for 20min. Crystal violet (0.1%) was added to stain for 10min and cells on the upper surface were washed twice with PBS. Cells on the upper surface were wiped off with a cotton ball and observe under a microscope. For the cell migration experiment, BD matrigel was not used and the cell concentration was adjusted to 5×105/mL. The rest of the steps were the same.
Scratch Wound Healing Assay
Cells were seeded in 12-well plates at a density of 1.0×105 cells per well. We transfected cells and incubated the dishes at 37°C until cells reached 100% confluence. An artificial gap was scratched using a 100 μL Pipette nozzle. Wells were observed and photographed under an inverted phase-contrast microscope. The software program HMIAS-2000 was used to measure the cell migration distance (μm).
Tumor Formation in Nude Mice
BALB/c nude mice (male, 6–8 weeks, weight 12–13g) were adaptively raised for 2 weeks under SPF conditions. The bedding material and water used for nude mice were firstly sterilized by high-temperature steam, and the edible feed was SPF grade. The survival condition of nude mice was observed every day, and feed and water were timely supplemented. All operations are conducted in the biosafety cabinet. Mice were randomly divided into two groups: NC group and si-SFTA1P group. The left dorsal upper limb skin of nude mice was sterilized, and 200 μL 107/mL Hep G2 cell suspension with si-SFTA1P or si-NC was injected after the skin was dry, respectively. After 8 weeks, nude mice were killed by dislocation and the tumor rate was calculated. The tumor tissue was completely separated, the mass of the tumor was measured, and the tumor volume was calculated. Tumor volume (mm3) = 1/2 (long diameter × short diameter × short diameter). The experimental procedures involving animals were performed with the approval of the Ethics Committee of Clinical Research Center of Kunming Maternal and Child Health Hospital (IACUC Issue No. 2,019,002). All efforts were made to reduce the suffering of animals, according to the Guidelines for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006).
Statistical Analysis
GraphPad Prism 6.0 software (GraphPad Software Inc., San Diego, CA, USA) and SPSS 22.0 software (IBM Corporation, Armonk, NY, USA) were used. The t-test was used for comparison between SFTA1P expression and other variables of patients, and P<0.05 was taken as the difference was statistically significant. We take the average expression level of SFTA1P in liver cancer patients as the cut-off value. Data was presented as mean ± standard deviation (SD) and compared by Student’s t-test or one-way ANOVA as appropriate. The survival curves were plotted by Kaplan–Meier analysis, and analyzed by Log-rank test. Categorical variables were compared by chi-square test.
Results
lncRNA SFTA1P is Up-Regulated in Hepatocellular Carcinoma
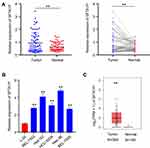
To explore the expression of lncRNA SFTA1P in HCC tissues, we used RT-PCR to determine the relative mRNA levels of lncRNA SFTA1P in 75 pairs of HCC tissues and the adjacent normal liver tissues. As Figure 1A showed, compared with the adjacent normal liver tissues, the mRNA level of SFTA1P in HCC tissues was significantly increased (P < 0.01). The expression levels of SFTA1P in HCC cell lines BEL-7402, BEL-7405, HCC-9204, Hep 3B, Hep G2 and Huh-7, and normal liver cell lines L-02 were also determined by qRT-PCR. As Figure 1B showed, SFTA1P expression was highly in HCC cells at different levels compared with normal liver cells. Hep G2 and Hep 3B cells were selected for further study. In addition, SFTA1P expression in tumor database was retrieved by TCGA visualization software and the results also showed that SFTA1P was highly expressed in HCC tissues (Figure 1C). The relationship between differential expression of SFTA1P and clinical features is shown in Table 1. As shown, SFTA1P expression was closely related to tumor size (P<0.05), vascular invasion (P<0.05), and TNM stage (P<0.05). Nevertheless, SFTA1P expression was not associated with the gender, age, liver cirrhosis, PVTT and differentiation of patients.
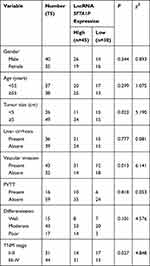 |
Table 1 Relationship Between lncRNA SFTA1P and Clinical Characteristics |
Knockdown of SFTA1P Inhibited HCC Cell Viability and Promotes Apoptosis
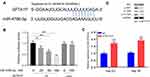
We used siRNA transfection method to interfere lncRNA SFTA1P expression in Hep G2 and Hep 3B cells. After transfection, the mRNA levels of SFTA1P in Hep G2 and Hep 3B cells were determined by RT-PCR. As Figure 2A showed, SFTA1P expression level in si-SFTA1P group was significantly decreased compared with NC group (P<0.01). In order to test the role of SFTA1P on HCC cell viability, CCK-8 assay was used to determine the viability of Hep G2 and Hep 3B cells in si-SFTA1P group and NC group. As Figure 2B showed, the cell viability in NC group was stably higher than the si-SFTA1P group (P<0.05). To further test the influence of SFTA1P on cell viability, clone formation assay was performed. As Figure 2C and D showed, the number of cloning cells in si-SFTA1P group was evidently reduced compared with NC group (P<0.05). Flow cytometry was applied to test the influence of SFTA1P expression on HCC cell apoptosis and results are shown in Figure 2E and F. As shown, the apoptosis rates of Hep G2 and Hep 3B cells in si-SFTA1P group was significantly promoted compared with NC group.
Knockdown of SFTA1P Inhibits HCC Cell Migration and Invasion
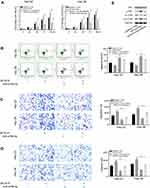
To explore the influence of SFTA1P on cell transfer capacity, Transwell assay was performed and results are shown in Figure 3A. As shown, the number of cells crossed the cell membrane in si-SFTA1P group was significantly lower than the NC group (P<0.05). The invasion experiment was carried out in a chamber containing matrigel and results are shown in Figure 3B. As shown, knockdown of SFTA1P inhibited the migration capacity of Hep G2 and Hep 3B cells. Scratch healing experiment was used to further verify the influence of SFTA1P expression on HCC cell migration and results are shown in Figure 3C. As shown, the migration rate of Hep G2 and Hep 3B cells in si-SFTA1P group was markedly slower than that in NC group.
lncRNA SFTA1P Sponges miR-4766-5p and Influences PI3K/AKT/mTOR Pathway
Starbase 3.0 and AnnoLnc were used to explore the relationship between lncRNA SFTA1P and miR-4766-5p and the result indicated miR-4766-5p was a target gene of lncRNA SFTA1P (Figure 4A). To verify miR-4766-5p was a potential target of lncRNA SFTA1P, luciferase reporter assay was performed. Compared with co-transfection of miR-NC + SFTA1P wt, reporter activity in Hep G2 cells co-transfected with miR-4766-5p plasmid + SFTA1P wt was decreased. However, the reporter activity in miR-NC + SFTA1P mut and miR-4766-5p plasmid + SFTA1P mut had no significant difference (Figure 4B). Then, pcDNA3.1-SFTA1P or pcDNA3.1-NC plasmid was transfected to Hep G2 and Hep 3B cells, and the relative expression levels of miR-4766-5p were determined by RT-PCR. As Figure 4C showed, knockdown of SFTA1P increased miR-4766-5p expression. To verify the influence of SFTA1P on the expression levels of key proteins in the PI3K/AKT/mTOR pathway, Western blotting was applied. As Figure 4D showed, the protein expression of total AKT and mTOR in NC and si-SFTA1P group had no significant difference, while the protein expressions of p-AKT and p-mTOR in si-SFTA1P group were markedly reduced compared to NC group. Those indicated SFTA1P may participate in the progression of HCC via influencing the protein expression of key proteins in the PI3K/AKT/mTOR pathway.
miR-4766-5p Mimic Reverses SFTA1P-Induced Tumor-Promoting Effects
In this study, Hep G2 and Hep 3B cells were co-transfected with pcDNA3.1-miR-4766-5p and pcDNA3.1-SFTA1P plasmid, and then CCK8, Transwell assay, and Flow cytometry were used to explore the combined action of miR-4766-5p and lncRNA SFTA1P on HCC cell viability, invasion, migration and apoptosis. Results of CCK-8 (Figure 5A) showed overexpression of SFTA1P promoted the viability of Hep G2 and Hep 3B cells, while further transfection of miR-4766-5p counteracted the promoting role. Results of FCM (Figure 5B) showed overexpression of miR-4766-5p abolished the inhibiting role of SFTA1P on the apoptosis of SFTA1P. Results of Transwell assay (Figure 5C and D) showed overexpression of SFTA1P promoted the migration and invasion ability of Hep G2 and Hep 3B cells, while overexpression of miR-4766-5p offset the role. Moreover, miR-4766-5p mimics rescued the increased expression of p-AKT and pmTOR by overexpression of SFTA1P (Figure 5E).
Knockdown of lncRNA SFTA1P Inhibits HCC Development in vivo
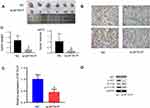
Mice were transfected with Hep G2 cells, which were infected with si-SFTA1P or NC. Tumors were separated and peeled, and then tumor weight and volume were measured. As Figure 6A showed tumor weight and volume in si-SFTA1P group were less than that in NC group (P<0.05). Results of immunohistochemistry showed Ki-67 and PCNA were stably inhibited by si-SFTA1P(Figure 6B). QRT-PCR was used to determine the expression levels of SFTA1P in si-SFTA1P group and NC group. Results (Figure 6C) showed the expression level of SFTA1P in si-SFTA1P group was much lower compared with NC group (P<0.05). Western blotting was used to measure the protein expression of AKT, p-AKT, mTOR and mTOR in si-SFTA1P group and NC group. Results (Figure 6D) showed SFTA1P expression had no influence on the protein expression of AKT, and mTOR, while knockdown of lncRNA SFTA1P decreased the expression of p-AKT and p-mTOR. The results in mice were consistent with those in cells.
Discussion
The high incidence, easy to metastasize and poor prognosis of HCC can affect human life and health. The diagnosis and treatment of HCC are still a worldwide medical problem.19,20 Although previous studies have found many abnormally expressed protein-coding genes in HCC, there is still an urgent need to find new molecular markers that can help early diagnosis and risk assessment. lncRNA can regulate gene expression and protein synthesis in several ways, and can regulate the nuclear development process through gene inhibition or gene activation.21 Many studies have shown that dysregulation of lncRNA is associated with a variety of human diseases. Some lncRNAs are potential biomarkers for HCC diagnosis and may serve as the therapeutic targets.22
SFTA1P is a pseudogene-derived long non-coding RNA and located in 10p14. The length of SFTA1P is 693 nt.23 It was significantly down-regulated in gastric cancer (GC) tissues compared to the normal tissues.11 Zhang et al reported SFTA1P was found down-regulated in lung adenocarcinoma (LAC).23 Xu et al reported downregulation of SFTA1P could inhibit LAC cell migration and invasion.24 In this study, we used RT-PCR to determine the relative mRNA levels of lncRNA SFTA1P in HCC tissues and the adjacent normal liver tissues. Results showed SFTA1P was highly expressed in HCC tissues. The mRNA levels of SFTA1P in 5 kinds of HCC cell lines were also higher compared with the normal liver cell. This suggests that lncRNA SFTA1P may play a role as an oncogene in HCC. The correlation analysis between SFTA1P expression and clinicopathologic indicators of HCC showed high SFTA1P expression indicates increased tumor size in HCC patients. Based on the above studies, we tested the effect of lncRNA SFTA1P on the viability, apoptosis, migration and invasion of HCC cells in vitro. We constructed HCC cell lines Hep G2 and Hep 3B with si-SFTA1P plasmid. CCK-8 and cloning formation assay indicated knockdown of SFTA1P inhibited Hep G2 and Hep 3B cell viability. Results of FCM showed knockdown of SFTA1P promoted the apoptosis. Scratch healing assay and Transwell assay showed knockdown of SFTA1P evidently inhibited Hep G2 and Hep 3B cell migration and invasion.
Similar to lncRNAs, miRNAs, a class of small noncoding RNAs, are also involved in HCC progression.25 Accumulating studies reveal that lncRNAs can serve as a competing endogenous RNA (ceRNA) by sponging miRNAs, thereby causing a loss of miRNA function.26 To further explore the molecular mechanism about how SFTA1P affects HCC viability, migration and invasion, we used Starbase 3.0 and AnnoLnc databases to explore the relationship between lncRNA SFTA1P and miR-4766-5p, and the result indicated miR-4766-5p was a target gene of lncRNA SFTA1P. Luciferase reporter assay also verifies miR-4766-5p was a target of lncRNA SFTA1P. RT-PCR results showed knockdown of SFTA1P up-regulated miR-4766-5p expression. Our study showed knockdown of SFTA1P reduced the protein expression of p-AKT and p-mTOR. Akt is a homologue of the v-Akt, and activated Akt adjusts many processes, such as apoptosis and proliferation.27 mTOR is a large protein kinase and the target of rapamycin.28 mTOR signaling pathway senses and integrates a variety of environmental cues to regulate organismal growth and homeostasis.29 PI3K/AKT/mTOR pathway is activated in many human cancers.30 Deregulation of the PI3K/Akt/mTOR pathway plays an important regulating role in HCC carcinogenesis.31 Those indicated SFTA1P may participate in the progression of HCC via influencing the protein expression of key proteins in the AKT/mTOR pathway. PI3K/Akt/mTOR signaling is one of the most critical intracellular pathways in both normal and cancer cells, and inhibition of this signaling has been shown to lead to regression of many tumor types, including HCC.
By bioinformatic algorithms, we predicted a novel microRNA, miR-4766-5p, as a potential target of SFTA1P. Dual-luciferase reporter gene assay revealed the direct interaction between SFTA1P and miR-4766-5p. To verify the role of miR-4766-5p in HCC, miR-4766-5p mimic and pcDNA3.1-SFTA1P were co-transfected to Hep G2 and Hep 3B cells. Then, MTT, cloning formation, transwell, scratch healing assay and FCM assay were applied to determine the viability, migration, invasion and apoptosis capacity of cells. Results showed miR-4766-5p promoted the apoptosis of Hep G2 and Hep 3B cells and inhibited the cell viability, invasion and migration ability. Moreover, miR-4766-5p attenuated the promoting role of SFTA1P on HCC cell viability, invasion and migration, and inhibiting role on HCC cell apoptosis. In breast cancer, miR-4766-5p mimics was also reported to promote apoptosis and inhibit migration in tumor cells.32 Moreover, we used nude mice models also found knockdown of SFTA1P reduced tumor growth in vivo. Ki-67 and PCNA were stably inhibited by si-SFTA1P. Ki-67 is a nuclear and nucleolar protein, which is related to cell proliferation.33 The higher the Ki-67 labelling index, the poorer the prognosis of HCC.34 PCNA is an adjunct protein of DNA polymerase, and its level reflects the cell proliferation rate and DNA synthesis rate. It can evaluate the malignancy and proliferation potential of tumor and is a reliable proliferation marker in the current study on tumor proliferation activities.35 PCNA expression was positively correlated with NET-1, whose expression was positively correlated to the TMN stages of HCC.36 Moreover, knockdown of lncRNA SFTA1P also decreased the expression of p-PI3K, p-AKT and p-mTOR in tumorigenic tissue. We speculated upregulation of SFTA1P promoted the development of HCC cells by activating PI3K/AKT/mTOR signaling pathway. MiR-4766-5p is a target gene of SFTA1P and overexpression of miR-4766-5p reduced SFTA1P expression, which further silenced PI3K/AKT/mTOR signaling pathway. Therefore, miR-4766-5p counteracted the effects of SFTA1P as an inducer of tumor progression.
In conclusion, lncRNA SFTA1P was up-regulated in HCC and can promote the tumor growth by targeting down-regulating miR-4766-5p via PI3K/AKT/mTOR signaling pathway.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Granot E, Sokal EM. Hepatitis C virus in children: deferring treatment in expectation of direct-acting antiviral agents. Isr Med Assoc J. 2015;17(11):707–711.
2. Dhar D, Antonucci L, Nakagawa H, et al. Liver cancer initiation requires p53 inhibition by CD44-enhanced growth factor signaling. Cancer Cell. 2018;33(6):1061–1077. doi:10.1016/j.ccell.2018.05.003
3. Hansji H, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front Genet. 2014;5:379. doi:10.3389/fgene.2014.00379
4. Boon RA, Jaé N, Holdt L, Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol. 2016;67(10):1214–1226. doi:10.1016/j.jacc.2015.12.051
5. Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi:10.1016/j.cell.2010.06.040
6. Majem B, Rigau M, Reventós J, Wong D. Non-coding RNAs in saliva: emerging biomarkers for molecular diagnostics. Int J Mol Sci. 2015;16(4):8676–8698. doi:10.3390/ijms16048676
7. Wang H, Huo X, Yang X-R, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. doi:10.1186/s12943-017-0680-1
8. Wang X, Sun W, Shen W, et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol. 2016;64(6):1283–1294. doi:10.1016/j.jhep.2016.01.019
9. Groen JN, Capraro D, Morris KV. The emerging role of pseudogene expressed non-coding RNAs in cellular functions. Int J Biochem Cell Biol. 2014;54:350–355. doi:10.1016/j.biocel.2014.05.008
10. Huang G-Q, Ke Z-P, H-B H, Gu B. Co-expression network analysis of long noncoding RNAs (IncRNAs) and cancer genes reveals SFTA1P and CASC2 abnormalities in lung squamous cell carcinoma. Cancer Biol Ther. 2017;18(2):115–122. doi:10.1080/15384047.2017.1281494
11. Ma H, Ma T, Chen M, Zou Z, Zhang Z. The pseudogene-derived long non-coding RNA SFTA1P suppresses cell proliferation, migration, and invasion in gastric cancer. Biosci Rep. 2018;38(2):BSR20171193. doi:10.1042/BSR20171193
12. Liao H-T, Huang J-W, Lan T, Wang -J-J, Zhu B, Yuan K-F ZY. Identification of the aberrantly expressed LncRNAs in hepatocellular carcinoma: a bioinformatics analysis based on RNA-sequencing. Sci Rep. 2018;8(1):5395. doi:10.1038/s41598-018-23647-1
13. Wei Y, Wang Y, Zang A, Wang Z, Fang G, Hong D. MiR-4766-5p inhibits the development and progression of gastric cancer by targeting NKAP. Onco Targets Ther. 2019;12:8525. doi:10.2147/OTT.S220234
14. Liang Y, Song X, Li Y, et al. A novel long non-coding RNA-PRLB acts as a tumor promoter through regulating miR-4766-5p/SIRT1 axis in breast cancer. Cell Death Dis. 2018;9(5):1–16.
15. Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4(4):257–262. doi:10.1016/S1535-6108(03)00248-4
16. Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988. doi:10.1038/nrd1902
17. Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9(6):667–676. doi:10.1023/B:APPT.0000045801.15585.dd
18. Yan W, Fu Y, Tian D, et al. PI3 kinase/Akt signaling mediates epithelial–mesenchymal transition in hypoxic hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2009;382(3):631–636. doi:10.1016/j.bbrc.2009.03.088
19. Heimbach JK, Kulik LM, Finn RS, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–380. doi:10.1002/hep.29086
20. Balogh J, Victor III D, Asham EH, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41. doi:10.2147/JHC.S61146
21. Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi:10.1038/nature20149
22. Huo X, Han S, Wu G, et al. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol Cancer. 2017;16(1):165. doi:10.1186/s12943-017-0734-4
23. Zhang H, Xiong Y, Xia R, Wei C, Shi X, Nie F. The pseudogene-derived long noncoding RNA SFTA1P is down-regulated and suppresses cell migration and invasion in lung adenocarcinoma. Tumor Biol. 2017;39(2):1010428317691418.
24. Xu N, Qiao L, Yin L, Li H. Long noncoding RNA ROR1-AS1 enhances lung adenocarcinoma metastasis and induces epithelial-mesenchymal transition by sponging miR-375. J BUON. 2019;24(6):2273–2279.
25. Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56(6):1371–1383. doi:10.1016/j.jhep.2011.11.026
26. Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7(39):64148. doi:10.18632/oncotarget.11637
27. Nakanishi K, Sakamoto M, Yasuda J, et al. Critical involvement of the phosphatidylinositol 3-kinase/Akt pathway in anchorage-independent growth and hematogeneous intrahepatic metastasis of liver cancer. Cancer Res. 2002;62(10):2971–2975.
28. Sarbassov DD, Ali SM, Kim D-H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–1302. doi:10.1016/j.cub.2004.06.054
29. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi:10.1016/j.cell.2012.03.017
30. Karar J, Maity A. PI3K/AKT/mTOR pathway in angiogenesis. Front Mol Neurosci. 2011;4:51. doi:10.3389/fnmol.2011.00051
31. Zhou Q, Lui VW, Yeo W. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Future Oncol. 2011;7(10):1149–1167. doi:10.2217/fon.11.95
32. Yang Q. A novel long non-coding RNA-PRLB acts as a tumor promoter through regulating miR-4766-5p/SIRT1 axis in breast cancer. Cell Death Dis. 2018;9(5):563.
33. Endl E, Gerdes J. The Ki-67 protein: fascinating forms and an unknown function. Exp Cell Res. 2000;257(2):231–237. doi:10.1006/excr.2000.4888
34. Daveau M, Scotte M, François A, et al. Hepatocyte growth factor, transforming growth factor α, and their receptors as combined markers of prognosis in hepatocellular carcinoma. Mol Carcinog. 2003;36(3):130–141. doi:10.1002/mc.10103
35. Yuli S, Suyan W, Yulin L, Tian C, Department E. The effects and mechanism of silenced PCNA gene expression on proliferation and apoptosis of thyroid carcinoma cells. Anti Tumor Pharm. 2018.
36. Shen SQ, Li K, Zhu N, Nakao A. Expression and clinical significance of NET-1 and PCNA in hepatocellular carcinoma. Med Oncol. 2008;25(3):341–345. doi:10.1007/s12032-008-9042-6
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.