Back to Journals » Journal of Inflammation Research » Volume 15
Novel Insights into the Emerging Role of Neat1 and Its Effects Downstream in the Regulation of Inflammation
Authors Pan Y, Wang T, Zhao Z, Wei W , Yang X, Wang X, Xin W
Received 3 December 2021
Accepted for publication 16 January 2022
Published 26 January 2022 Volume 2022:15 Pages 557—571
DOI https://doi.org/10.2147/JIR.S338162
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Yongli Pan,1,2,* Ting Wang,3,* Zhiqiang Zhao,4,* Wei Wei,2,5,* Xinyu Yang,6 Xianbin Wang,7 Wenqiang Xin2,6
1Department of Neurology, Weifang Medical University, Weifang, Shandong, People’s Republic of China; 2Georg-August-University of Göttingen, Göttingen, Lower Saxony, Germany; 3Department of Radiology, The Second Affiliated Hospital of Baotou Medical College, Baotou, 014030, People’s Republic of China; 4Department of Neurosurgery, Heji Hospital affiliated Changzhi Medical College, Shanxi, People’s Republic of China; 5Department of Neurology, Mianyang Central Hospital, Mianyang, Sichuan, People’s Republic of China; 6Department of Neurosurgery, Tianjin Medical University General Hospital, Tianjin, People’s Republic of China; 7Department of Emergency Medicine, The Second Affiliated Hospital of Baotou Medical College, Baotou, 014030, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wenqiang Xin
Department of Neurosurgery, Tianjin Medical University General Hospital, Anshan Road No. 154, Tianjin, 300052, People’s Republic of China
Tel +86– 18526201182, Fax +86– 2260362062, Email [email protected]
Xianbin Wang
Department of Emergency Medicine, The Second Affiliated Hospital of Baotou Medical College, Baotou, 014030, People’s Republic of China
Email [email protected]
Abstract: Nuclear paraspeckle assembly transcript 1 (Neat1) located at chromosome 11 is a long non-coding RNA that is widely expressed in mammalian cell types, and which is overexpressed in several inflammation-related disorders. Inflammation implies a plethora of mutual interactions between both soluble factors and cells due to various stimuli including tissue injury. Although there is no doubt that inflammation is critically involved in multiple biological and pathological processes alike, the precise mechanisms being involved are still open for debate. In this context, the role of Neat1 as a regulator of inflammation, microglial activation, and lipid accumulation under various inflammatory conditions remains elusive. Herein, we review the regulation of Neat1 and how it modulates the expression of its target genes. Thereafter, we will review the impact of Neat1 on inflammation by activating or inhibiting various signaling pathways, such as microRNAs, AKT, TLR4, TRAF6, and NF-κB.
Keywords: Neat1, inflammation, microglia, lipids, microRNA, NF-κB
Introduction
Inflammation is an adaptive pathological response initiated after bacterial or viral infection,1,2 and it is a normal defense mechanism that the body uses to protect itself from tissue injury. In response to these stimuli, immune cells migrate to the site of infection and/or injury prompting tissue repair/wound healing.3 Thus, it is necessary to understand the mechanisms under the inflammation so that we can seek effective anti-inflammatory resolutions.4 However, up to now, our understanding of inflammation is largely based on the central dogma of molecular biology.5 Long non‐coding RNAs (lncRNAs) are defined as a class of RNA transcripts with a length of longer than 200 nucleotides, by far the largest fraction of non-coding transcripts,6,7 which can be transcribed from different genomic regions including intergenic regions, mitochondrial regions, specific chromosomal regions, and intergenic.8,9 Nuclear paraspeckle assembly transcript 1 (Neat1), a nuclear lncRNAs, a vital component of paraspeckles, plays critical roles in multiple cellular physiology and pathophysiology,10,11 such as the innate immune reaction,12 organ development,13,14 cancer15–17 and neurodegenerative diseases.18–21 There are two isoform transcripts of Neat1, Neat1_1 (3.7 kb) and Neat1_2 (23 kb), respectively, both of which are localized to nuclear paraspeckles.22 Paraspeckles are a type of subnuclear body built on the Neat1 and enriched with various proteins, RNAs, which are involved in regulating gene expression by a process called nuclear retention.23 Accumulating evidence has shown that Neat1is dysregulated during the progression of a variety of diseases. For example, Zhou et al and Zhang et al found that Neat1 is aberrantly expressed in glioma and colorectal cancer, while its expression is closely related to tumor cell viability, migration, and invasion, as well as for the overall patient survival.24,25 Neat1 dysregulation has also been shown to be involved in non-cancerous diseases, for instance, Huntington’s Disease (HD), Parkinson’s disease (PD), multiple sclerosis and Hantavirus infection.26 Sunwoo et al identified upregulation of Neat1 in the human HD brain and revealed the neuroprotective function without the precise mechanism in HD pathogenesis.20 Likewise, 1-Methyl-4-phenylpyridinium (MPP+) induced PD was protected by the knockdown of Neat1, and the protect mechanism might relate to α-synuclein expression.27 Notably, the role and mechanism of Neat1 in disorders related to the inflammatory cytokines and mediators are poorly understood and have not been systematically reviewed. Herein, in this study, we discuss and summarize the function and mechanism of Neat1 specific to inflammation and propose potential signaling pathways.
The Course of Inflammation
Inflammation is programmed progress initiated to protect tissue from injury stimuli, which can cause the release of pathogen or danger-associated molecular patterns (PAMP), and promote tissue recovery via suppressing bacteria, viruses, toxins, and infections by fighting pathogens.28–30 Inflammatory responses have the potential to initially inhibit the release of inflammatory mediators, which appear when with an insult and elapse once the injury is eliminated.30,31 Upon the initial stage of the inflammatory response, PAMPs can be identified by macrophages or mast cells, which in turn promote the release of a pro-inflammation-related mediator such as cytokines, chemokines, and eicosanoids, therefore, enhancing vascular permeability and activating the immune response.1,31,32 Besides that, the neutrophils and monocytes immediately move to the lesion region by a series of pathways and molecules like cytokines and adhesion molecules.32 With the signs of progress of inflammation, monocytes and lymphocytes are recruited and migrate to the lesion site to eliminate the harmful substances, which are finally cleared by macrophages through the cell apoptosis pathway.31 When it comes to the final stage of inflammation, the inflammatory process is replaced by the tissue repair process. In addition, inflammatory reaction normally involves the rapid induction of pro-inflammatory response, closely followed by an anti-inflammatory process.33 To regain homeostasis, resolution of inflammation represents a turning point, since the progression from acute-resolving to persistent-chronic inflammation can appear due to a failure in resolving inflammatory processes.33 Herein, for the sake of promoting tissue to restore homeostasis, the inflammatory reaction must be immediately inhibited to prevent the organ from further damage,31 and a great deal of signal pathways is included in the inflammatory process, providing novel insight into the treatment of inflammation-related disorders.
The Course of Neat1
Although ncRNAs do not encode for proteins, they participate in various biological processes and thus play a vital role in transcriptional and post-transcriptional modulation.34,35 They can be divided into two categories according to the length of the RNA chain, involving lncRNA with lengths of more than 200 nucleotides and small ncRNAs with lengths of less than 200 nucleotides.35,36 Discovered in 2007 by Hutchinson et al,37 Neat1 lncRNA is––as indicated in the name, nuclear enriched abundant transcript–– one of the most abundant lncRNAs localized within paraspeckles in the mammalian nucleus and abundantly expressed in a variety of cell types.38–40 Additionally, it is suggested that Neat1 can target numerous genomic regions in various cells, mainly involving active genes, revealing that it is a regulator for plenty of active genes and signal pathways.41 Structured illumination microscopy detecting RNA and protein components of paraspeckles simultaneously revealed the structural arrangement of Neat1 within paraspeckles; that is, the paraspeckle compositions are located at the core-shell spheroidal structure; however, Neat1’, 5’ and 3’ ends are in paraspeckle around, and the central sequence is present at the core site.42,43 Paraspeckle is a complex consisting of subnuclear ribonucleoprotein within interchromatin, which plays a vital role in modulating transcription and RNA processing mainly via regulating RNA and protein editing and serving as a sponge targeting to various microRNAs.19 There are two subtypes of mono-exonic isoforms transcribed from the Neat1 locus from chromosome 11, known as shorter Neat1_1 with 3684 nucleotides and longer Neat1_2 with 22,743 nucleotides.19,44 The shorter Neat1_1 isoform (3.7 kb in human) is polyadenylated,23 whereas the longer 23 kb subtype of Neat1_2 is not polyadenylated. Neat1_1 is high abundance and exhibits a high expression in a wide range of tissues.19 Nonetheless, in contrast to the Neat1_2 isoform, the shorter Neat1_1 isoform has no an integral element of paraspeckles; however, it appears as microspeckles and thus may contain other effects that have not been discovered.43 Additionally, Sasaki et al45 indicated that the knockdown of the production of Neat1_2 can lead to paraspeckle elimination even in the presence of intact Neat1_1. Likewise, Neat1_2 defect female mice have a loss function of reproductive tissue development; however, no external or histological abnormalities are observed in Neat1_1 knockout mouse.13,46
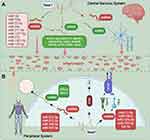
NEAT1 Directly Regulates Inflammation in the Central Nervous System
Inflammatory response, in the central nervous system (CNS), is generally recognized as an essential component of the CNS to restrain abnormal protein aggregates, infection, ischemia, and toxins.47,48 Neat1 has been identified as dysregulated in cell growth, differentiation, apoptosis, cells migration, metastasis, and invasion in neurological disorders,26,49 where regulation of inflammation plays a vital role. Although the fundamental role of Neat1 in terms of inflammation in CNS is becoming increasingly evident,50,51 it is urgent to summarize the inflammatory effect of Neat1 upon inflammatory conditions of the CNS owing to diverse interaction pathways in various neurological disorders with disparate efficiency. The electronic databases, including Cochrane Library, Medline, PubMed, Embase, ScienceDirect, and other databases were retrieved to identify the literature exploring the regulation of Neat1 on inflammation upon various neurological diseases from the inception of electronic databases to September 2021. We retrieved studies using the following keywords in accordance with Boolean logic: (“Neat1” OR “nuclear paraspeckle assembly transcript 1”) AND (“inflammatory” OR “inflammation” OR “microglia”). Beyond this, research on the appraisal reference list was manually checked to determine other potential qualification trials that may have been missed by the database searches. The process was iterated until no more publications would be obtained. A total of 15 publications were identified in this section,50–64 which were performed between 2017 and 2021. The most widely used species and related inflammatory models are mice and middle cerebral artery occlusion (MCAO), respectively. The most common identification method and inflammatory markers are quantitative real-time polymerase chain reaction and interleukin, respectively. Of these 15 studies, 5 on stroke, 6 on PD, 2 on spinal cord injury (SCI), and 1 on traumatic brain injury with the last one on epilepsy study, suggesting that it is sufficient to illustrate that Neat1 can modulate inflammation in the CNS disorders. Up on stroke condition, an upregulation of Neat1 was uncovered in ischemic stroke patients’ blood,61 animals with an MCAO,60 and in vitro cells with hypoxia.50 Similarly, the upregulation of Neat1 has a detrimental effect and accelerates disease progression in PD via regulating inflammation.19 Moreover, the knockdown of Neat1 could inhibit microglia polarization towards the M1 phenotype in various neurological disorders.62 Nevertheless, concerning the differences between diseases (dosage and delivery time), the studies used heterogeneous experimental paradigms meeting their own applied study purposes. More details are shown in Table 1.
 |
Table 1 Preclinical and Clinical Studies Assessing the Effect of NEAT1 on the Regulation of Inflammation in Central Nervous System |
Neat1 Directly Regulates Inflammation in the Peripheral System
Peripheral inflammation is a non-specific, adaptive response to infections or noxious stimuli aiming to limit injury and enhance recovery.65 Systemic inflammation is characterized by an up-regulated expression of circulating cytokines, chemokines, and acute-phase proteins, as can be seen in multiple chronic disorders, such as obesity, diabetes, and metabolic syndrome, can lead to serious health risks.66 It is of interest that peripheral inflammatory stimuli that circulate in the blood may lead to the release of cytokines in the CNS mainly via regulating the integrity of the blood–brain barrier and activation of microglia,67,68 suggesting that peripheral and central inflammation can be mutually modulated.65 Indeed, Neat1 can regulate inflammation in CNS; therefore, the effect of Neat1 on peripheral inflammation deserves more attention. The aforementioned literature search strategy with the keywords of “Neat1” OR “nuclear paraspeckle assembly transcript 1” was also applied in this section and yielded a total of 53 studies,69–121 suggesting broad regulatory effect of Neat1 on peripheral inflammation. Sepsis is a systemic inflammatory response syndrome triggered by infection, with a global incidence of approximately 437 per 100,000 person-years.70 Ten of these included studies, respectively, demonstrated that circulating Neat1 correlated with an increased risk of sepsis, elevated severity, and unfavorable prognosis, as well as higher expression of pro-inflammatory cytokines in sepsis patients, and knockdown of Neat1 promoted viability and reduced the expression of inflammatory cytokines in lipopolysaccharide (LPS)-induced cells.69–78 Additionally, inflammation is a vital driver of atherosclerosis and myocardial infarction, and besides various proteins and microRNAs, Neat1 has been involved in inflammation regulation.79–85 Fu et al,83 Chen et al,84 and Wang et al85 indicated that silencing of Neat1 can suppress lipid uptake inflammation-related molecules including interleukin (IL)‐6, reactive oxygen species, IL‐1β, and tumor necrosis factor (TNF)‐α. Meanwhile, Gast et al79 applied a Neat1 -/- mice and showed the first direct illustration that Neat1 was involved in the regulation of T cell and monocyte-macrophage lineage differentiation and functions via serving as a novel lncRNA-type immunoregulator in myocardial infarction. The characters and primary mechanism of some of these studies are summarized in Table 2, demonstrating Neat1 may offer a promising strategy to treat the disorders with peripheral inflammation.
 |  |  |
Table 2 Preclinical and Clinical Studies Assessing the Effect of NEAT1 on the Regulation of Inflammation in Peripheral System |
Neat1 Can Regulate the Activation of Microglia
Microglia cells serve as “brain-resident macrophages” that comprise approximately 10% of all the cells in the CNS,122 which play a key role in phagocytic elimination of influencing brain development and maintenance of the neural environment. Normally, the microglia are in the rest state to keep homeostasis in the fields around microglia cells.123 Upon an inflammatory condition, the activation of microglia is the first step of response to inflammation, and then other immune cells like neutrophils, T cells, natural killer cells, and so on were activated in the brain.123–125 Stimuli results in the activation of microglia, which represents the first step of an inflammatory response, followed by four distinct phenotypes, such as ramified, intermediate, amoeboid, and round phenotypes.123,126 Based on the morphological progression and its substance secretion, microglia are characterized as M1-type and M2-type.123 M1-type, the pro-inflammatory phenotype, produces various pro-inflammatory factors such as IL-1β, IL-6, interferon-α, motif chemokine ligand, cyclooxygenase-2, and inducible nitric oxide synthase.123,127–130 M2, anti-inflammatory phenotype, microglia involve the states of both alternative activation and acquired deactivation, which are accompanied by IL-4/IL-13 and IL-10/transforming growth factor (TGF)-β, respectively.123,131,132 Ni et al62 exposed Neat1 knockdown BV-2 cells to oxygen–glucose deprivation/reoxygenation (OGD/R) and then detected the various levels of markers of M1 (CD16, CD32, and CD86) and M2 (BDNF, PDGF, and Arg-1) microglia, demonstrating that Neat1 knockdown suppresses M1microglial polarization but does not promote M2 microglial polarization. Jin et al60 revealed that microglia had shorter ramifies and larger cell bodies after MCAO; however, the antisense oligonucleotides (ASO)-Neat1 group (Neat1 knockdown) was associated with significantly fewer activated microglia. Also, pyknotic nuclei and vacuolated cytoplasm were markedly reduced in the cortex and striatum of ASO-Neat1 mice. Similarly, Han et al50 suggested that Yin Yang 1 (YY1)-induced upregulation of Neat1 contributed to the OGD/R injury and neuroinflammation damage to microglial cells. Neat1 knockdown might therefore contribute to suppressing microglia activation, and information regarding this aspect, however, is scarce.
Neat1 Can Regulate the Expression of Lipids
Lipids are a kind of hydrophobic or amphiphilic small molecules that are insoluble in water and soluble in nonpolar solvents.133 They are an essential structural component of cell membranes and play a vital role in regulating important biological processes that support multiple biological functions, and dysregulation of lipid metabolism has been revealed to be involved in inflammation-related disorders, such as atherosclerosis, cancer, and neurodegenerative diseases.134 Since bioactive lipids emerged as vital factors during all phases of the inflammatory process, we also explored the effect of paraspeckles of Neat1 on lipid uptake. Atherosclerosis has been recognized as a chronic inflammation process induced by lipid accumulation in the vessel wall.135 Neat1 was significantly increased in human macrophages THP‐1 cells incubated with oxidized low-density lipoprotein (ox‐LDL). To investigate the role of Neat1 in foam cells formation, Neat1 was downregulated in THP‐1 cells by transfection of Neat1 small interfering RNA (siRNA), as demonstrated by Wang et al,85 knockdown of Neat1 restrained CD36 protein expression, triglyceride, and oil‐red staining levels, suggesting that Neat1 could modulate the formation of foam cells triggered by ox‐LDL in vitro. Therein, abnormality of triglyceride metabolism is a lipid metabolism disorder and includes hypertriglyceridemia and liver triglyceride accumulation.136 Using rapamycin after transplantation can cause hypertriglyceridemia and liver triglyceride accumulation.137 Fan et al indicated that the down-regulation of the expression of hsa-miR-372–3p contributes to triglyceride accumulation induced by rapamycin, further using dual-luciferase reporter assay and bioinformatics to determine the relationship between hsa-miR-372–3p and Neat1.137 In vivo, Neat1 was upregulated in non-alcoholic fatty liver disease patients, and besides that, knockdown of Neat1 can inhibit lipid accumulation in mice with a high-fat diet.138 Although Neat1 has been illustrated to be collectively effective in regulating lipids and inflammation, information regarding this aspect is scarce. Herein, more evidence-based information is provided in this respect.
Neat1 Correlates with the Level of Inflammatory Cytokines in Clinical Studies
Liquid biopsy is a new diagnostic tool conducted on blood or other biofluids to evaluate the inflammation-derived components and their genomic or proteomic profiles. In clinical practice, it is widely accepted that Neat1 is identified as a potential contributor to the increased secretion of multiple pro-inflammatory factors in diverse inflammatory disorders.103,139 For instance, in patients with systemic lupus erythematosus, the expression level of Neat1 is upregulated in the blood and correlates with the increased expression of pro-inflammatory chemokines and cytokines, such as IL-1β, IL-6, and CXC chemokine ligand 10 (CXCL10), exacerbating the severity of disorder.103 In asthma, Neat1 exhibits the potential to differentiate patients with asthma or not, especially in predicting exacerbation risk and severity of asthma. Neat1 relative expression was positively correlated with TNF-α, IL-1β, and IL-17, while negatively correlated with miR‐124 expression in patients with asthma in exacerbation. Interestingly, miR‐124 relative expression was positively correlated with TNF‐α, IL‐1β, IL‐17, and exacerbation severity as well.87 Likewise, sepsis patients were associated with a higher incidence of plasma Neat1 expression than those in healthy control and further disclosed that Neat1 revealed a good predictive value for sepsis risk and was positively associated with levels of TNF-α, IL-1β, IL-6, and IL-8, while negatively correlated with level of IL-10.74,76 Hence, Neat1 expression is aberrantly-mostly upregulated in inflammatory pathological conditions, indicating that it may serve as a potential diagnostic and prognostic biomarker to monitor disease activity and treatment outcome in multiple inflammatory disorders; however, the potential of Neat1 as a biomarker and as a therapeutic target for clinical CNS inflammation highly warrants further research to elucidate its exact role due to limited clinical data, moreover, different liquid biopsy is deserved to be performed to detect the expression of Neat1 since most studies use blood samples.
The Mechanism of Neat1 in the Effect of Regulating Inflammation
Neat1-miRNA-mRNA Axis is the Key Player in Regulating Inflammation
MicroRNAs (miRNAs) are small non-coding RNAs with lengths of approximately 18–24 nucleotides and can post-transcriptionally modulate gene expression through binding to 3′‐untranslated regions of mRNAs, which further induce degradation or suppression of mRNAs, and then inhibit gene translation.85,140,141 MiRNAs can regulate various target genes via serving as mediators, and one target gene may be regulated by multiple miRNAs.141,142 MiRNAs are involved in crucial cellular functions of inflammatory progression, such as miR‐195 and miR‐146b‐5p, which can regulate inflammatory profile in affecting the crosstalk with smooth muscle cells and foam cell formation via targeting different mRNA, respectively.85 lncRNAs play a key role in multiple human diseases by regulating miRNA since they have direct ‘sponging-like effects’ on miRNAs,141,143 which in turn modulate target genes via imperfect complementarity binding to the 3-UTR of mRNA.141 In addition, some lncRNAs can compete with miRNAs directly through targeting mRNA.140,141 That shows the lncRNA–microRNA–mRNA axis, herein, contributes to the regulation of inflammatory disorders, and the emerging evidence has been identified that the anti- or proinflammatory effects of specific miRNAs are highly modulated by Neat1.114,117 As downstream of Neat1, the top two miRNA reported in frequency were miRNA-124 and miRNA-129 as shown in Table 2. MiR-124, a tumor inhibitor, has recently been reported to be involved in the inhibition of inflammatory responses by diverse pathways, for instance, it plays a crucial role in traumatic brain injury and intestine by regulating inflammation processes and reduces skin inflammation by inhibiting the innate immune response.144–146 Chen et al80 reported that the overexpression of Neat1 markedly increased H9c2 cells inflammation, while the miR-124 could reverse the effects by repressing the NF-κB signaling pathway. With regard to miR-129, Li et al111 uncovered that Neat1 was upregulated, while miR-129 was distinctly downregulated in vitro and in vivo in renal fibrosis models, what is more, Neat1 knockdown and miR-129 overexpression can suppress renal fibrosis, as shown by a decreased expression of inflammatory factor release. Likewise, Zhang et al100 performed the luciferase reporter assay and RNA pull-down and illustrated a direct interaction between miR-129-5p and NEAT1, which in turn interacted with NF-kB signaling pathway. Besides that, Table 2 provides more miRNAs involved in the interaction between Neat1 and inflammatory regulation, hence, a plethora of miRNAs is found in the downstream of Neat1 of which the precise signaling cascades that are regulated under inflammatory conditions are also diverse.
Neat1 Regulates Inflammation Through the NF-κB Pathway
Nuclear factor-kB (NF-κB) is present in almost all types of cells and primarily serves as a transcription factor implicated in various biological processes, such as apoptosis, cell proliferation, tumorigenesis, inflammation, and various autoimmune disorders.147 It is revealed to promote multiple proinflammatory mediators, and suppression of NF-κB signaling correlates with beneficial effects in inflammatory conditions. For instance, LncRNA Snhg8 acts as a competitive endogenous RNA (ceRNA) by binding to miR-425–5p, which was revealed to boost microglial inflammation by targeting the sirtuin1 (SIRT1)‐mediated NF‐κB pathway.148 Furthermore, microglia can mediate experimental autoimmune encephalomyelitis, a neuroinflammatory disorder, and pathogenesis progression, therein, T cells activate the microglial noncanonical NF-κB pathway plays a crucial role.149 As aforementioned, LncRNA can act on mRNA indirectly through miRNA or mRNA, and there is such a relationship between Neat1 and NF‐κB. Xiao et al150 demonstrated that Neat1 was up-regulated in rheumatoid arthritis, a chronic inflammatory disease, and the knockdown of it attenuated TNF-α-induced cell proliferation and inflammatory cytokine production while promoting cell apoptosis by targeting miR-204-5p through activating the NF-κB signaling pathway. Likewise, Nong et al117 indicated Neat1 functioned as a ceRNA by sponging to reversed miR-193a-3p overexpression and alleviated inflammatory injury of normal human fibroblast cell-line WI-38 cells induced by LPS via regulating the activation of NF-κB signaling. Of note, Neat1 is able to interact indirectly with NF-κB via mRNA as well as to regulate inflammation, as indicated by Wang et al that Neat1 knockdown by tail vein injection of siRNAs has the potential to inhibit the expression levels of pro-inflammatory factor by inhibiting the Toll-like receptor 2/NF-κB signaling pathway in myocardial injury.81 Hence, Neat1 appears to interact with NF-κB indirectly via miRNA and mRNA, however, the same signaling pathways that are regulated under other inflammatory conditions (except for the disorders described in the Tables) are not fully known due to currently limited evidence, yet.
Neat1 Regulate Inflammation Through AKT, TLR4, and TRAF6 Pathway
Despite the NF-κB pathway being involved downstream of Neat1, the following question remains: how can Neat1 achieve this pro‐inflammatory effect through the NF-κB signaling pathway? The activation of Toll-like receptor 4 (TLR4)/NF-κB pathway has been widely studied for its involvement in inflammation. It is the main receptor of inflammation and activated in stimuli-challenged macrophages (such as LPS) which further leads to immune cells to release inflammatory cytokines, such as TNF-ɑ, IL-1β, and IL-6, inducing a systemic inflammatory response, followed by the activation of NF-κB.73,117 Zhang et al151 disclosed that Juglanin, a natural compound derived from the crude Polygonum aviculare, could ameliorate LPS induced neuroinflammatory injury through impeding activation of TLR4/NF-κB-mediated inflammatory response, and Neat1 was involved in the TLR4-mediated inflammatory process as identified by Zhang et al.103 Consecutively, Nong et al117 illustrated that knockdown of Neat1 promoted cell viability and ameliorated inflammatory response; besides, it exerted its role through regulating TLR4/NF-κB signaling pathway. TNF receptor-associated factor 6 (TRAF6), an E3 ubiquitin ligase, is a signal transduction molecule shared by the IL-1 receptor/TLR family and the TNFR superfamily.152 To identify that TRAF6 is involved in NF-κB activation, using the TRAF6-deficient mice, Yamamoto et al demonstrated that TRAF6 plays an irreplaceable role in many pathophysiological processes by activating the NF-κB pathway.152 Several previous studies had indicated that down-expression of Neat1 could interact with TRAF6 and decrease its expression level, and significantly inhibited the expression levels of anti-inflammatory cytokines,75,102,104 and Zhang et al104 further hinted that TARF6 could promote the inflammatory injury via activating the NF-κB signaling. Downregulated Neat1 might therefore contribute to suppressing inflammation by regulating TNFR6 and TLR4, which further modulate NF-κB activation. Information regarding this aspect, however, is still scarce. Of note, Akt kinases, also known as protein kinase B, are signaling molecules of cell growth and differentiation and serve as a well-characterized effector of phosphoinositide 3-kinase (PI3K), involved in diverse cellular processes, especially for inflammation.141,153,154 As indicated by Xia et al69 and Wang et al,89 given that Neat1 can modify miRNA/PI3K/AKT axis in cell and animal models exposed to inflammation, it exhibited great potential in inflammatory conditions treatment. Herein, it suggests that pharmacological upregulation of Akt signaling might be a potential target as well for inhibiting inflammation. Besides that, there are, of course, many other pathways that are involved downstream of Neat1 to regulate inflammation as shown in Table 2 and Figure 1.
Conclusion
Inflammation usually leads to an aberrant expression of Neat1 that exerts crucial functions in epigenetic and transcriptional regulation of gene expression. The regulation of Neat1 offers a great opportunity for adjuvant treatment of neurological and peripheral disorders, for which inflammation is an excellent target. There is abundant evidence that Neat1 is directly involved in the regulation of inflammation in both the central nervous system and the peripheral system. Furthermore, inhibiting the activation of microglia and accumulation of lipids has practical implications in regulating inflammation, and a great amount of evidence demonstrates that, for now, the regulation of Neat1 appears to suppress microglial activation, lipid accumulation, and inflammatory responses in clinical and preclinical studies. Among the mechanism of Neat1 in the effect of regulating inflammation, the Neat1-miRNA-mRNA network plays a key role, additionally, it can modify various signaling pathways, such as AKT, TLR4, TRAF6, and NF-κB as well.
Disclosure
The authors declare that they have no competing interests.
References
1. Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454(7203):428–435. doi:10.1038/nature07201
2. Gilroy D, De Maeyer R. New insights into the resolution of inflammation. Semin Immunol. 2015;27(3):161–168. doi:10.1016/j.smim.2015.05.003
3. Broom L, Kogut M. Inflammation: friend or foe for animal production? Poult Sci. 2018;97(2):510–514. doi:10.3382/ps/pex314
4. Foley J. Focus issue: inflammatory mechanisms. Sci Signal. 2015;8(366):eg2. doi:10.1126/scisignal.aaa9599
5. Chew C, Conos SA, Unal B, Tergaonkar V. Noncoding RNAs: master regulators of inflammatory signaling. Trends Mol Med. 2018;24(1):66–84. doi:10.1016/j.molmed.2017.11.003
6. Zhang L, Wang H. Long non-coding RNA in CNS injuries: a new target for therapeutic intervention. Mol Ther Nucleic Acids. 2019;17:754–766. doi:10.1016/j.omtn.2019.07.013
7. Boloix A, Masanas M, Jiménez C, et al. Long non-coding RNA PVT1 as a prognostic and therapeutic target in pediatric cancer. Front Oncol. 2019;9:1173. doi:10.3389/fonc.2019.01173
8. Lodde V, Murgia G, Simula ER, et al. Long noncoding RNAs and circular RNAs in autoimmune diseases. Biomolecules. 2020;10(7):1044. doi:10.3390/biom10071044
9. Bianchessi V, Badi I, Bertolotti M, et al. The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in endothelial cells. J Mol Cell Cardiol. 2015;81:62–70. doi:10.1016/j.yjmcc.2015.01.012
10. Bao M, Szeto V, Yang BB, et al. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018;9(3):281. doi:10.1038/s41419-018-0282-x
11. Bu F, Wang A, Zhu Y, et al. LncRNA NEAT1: shedding light on mechanisms and opportunities in liver diseases. Liver Int. 2020;40(11):2612–2626. doi:10.1111/liv.14629
12. Zhang P, Cao L, Zhou R, et al. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019;10(1):1495. doi:10.1038/s41467-019-09482-6
13. Nakagawa S, Shimada M, Yanaka K, et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141(23):4618–4627. doi:10.1242/dev.110544
14. Standaert L, Adriaens C, Radaelli E, et al. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014;20(12):1844–1849. doi:10.1261/rna.047332.114
15. Wen S, Wei Y, Zen C, et al. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol Cancer. 2020;19(1):171. doi:10.1186/s12943-020-01293-4
16. Naveed A, Cooper JA, Li R, et al. NEAT1 polyA-modulating antisense oligonucleotides reveal opposing functions for both long non-coding RNA isoforms in neuroblastoma. Cell Mol Life Sci. 2021;78(5):2213–2230. doi:10.1007/s00018-020-03632-6
17. Ghafouri-Fard S, Shirvani-Farsani Z, Hussen BM, et al. The critical roles of lncRNAs in the development of osteosarcoma. Biomed Pharmacother. 2021;135:111217. doi:10.1016/j.biopha.2021.111217
18. Zhao Y, Wang Z, Mao Y, et al. NEAT1 regulates microtubule stabilization via FZD3/GSK3β/P-tau pathway in SH-SY5Y cells and APP/PS1 mice. Aging. 2020;12(22):23233–23250. doi:10.18632/aging.104098
19. Boros F, Vécsei L, Klivényi P. NEAT1 on the field of Parkinson’s disease: offense, defense, or a player on the bench? J Parkinson’s Dis. 2021;11(1):123–138. doi:10.3233/JPD-202374
20. Sunwoo J, Lee S-T, Im W, et al. Altered expression of the long noncoding RNA NEAT1 in Huntington’s disease. Mol Neurobiol. 2017;54(2):1577–1586. doi:10.1007/s12035-016-9928-9
21. Li K, Yao T, Zhang Y, et al. NEAT1 as a competing endogenous RNA in tumorigenesis of various cancers: role, mechanism and therapeutic potential. Int J Biol Sci. 2021;17(13):3428–3440. doi:10.7150/ijbs.62728
22. Yu X, Li Z, Zheng H, et al. NEAT1: a novel cancer-related long non-coding RNA. Cell Prolif. 2017;50(2):e12329.
23. Wang Y, Chen L, Wu Z. Organization and function of paraspeckles. Essays Biochem. 2020;64(6):875–882. doi:10.1042/EBC20200010
24. Zhang M, Weng W, Zhang Q, et al. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018;11(1):113. doi:10.1186/s13045-018-0656-7
25. Zhou K, Zhang C, Yao H, et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17(1):105. doi:10.1186/s12943-018-0849-2
26. Prinz F, Kapeller A, Pichler M, et al. NEAT1The implications of the long non-coding RNA in non-cancerous diseases. Int J Mol Sci. 2019;20(3):627. doi:10.3390/ijms20030627
27. Liu Y, Lu Z. Long non-coding RNA NEAT1 mediates the toxic of Parkinson’s disease induced by MPTP/MPP+ via regulation of gene expression. Clin Exp Pharmacol Physiol. 2018;45(8):841–848. doi:10.1111/1440-1681.12932
28. Paul O, Arora P, Mayer M, et al. Inflammation in periodontal disease: possible link to vascular disease. Front Physiol. 2021;11:609614. doi:10.3389/fphys.2020.609614
29. Sun S. The non-canonical NF-κB pathway in immunity and inflammation. Nat Rev Immunol. 2017;17(9):545–558. doi:10.1038/nri.2017.52
30. Furman D, Campisi J, Verdin E, et al. Chronic inflammation in the etiology of disease across the life span. Nat Med. 2019;25(12):1822–1832. doi:10.1038/s41591-019-0675-0
31. Zhao H, Wu L, Yan G, et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct Target Ther. 2021;6(1):263. doi:10.1038/s41392-021-00658-5
32. Wright H, Moots RJ, Bucknall RC, et al. Neutrophil function in inflammation and inflammatory diseases. Rheumatology. 2010;49(9):1618–1631. doi:10.1093/rheumatology/keq045
33. Schmid T, Brüne B. Prostanoids and resolution of inflammation - beyond the lipid-mediator class switch. Front Immunol. 2021;12:714042. doi:10.3389/fimmu.2021.714042
34. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi:10.1038/nature11233
35. Li J, Wei M, Liu X, et al. The progress, prospects, and challenges of the use of non-coding RNA for diabetic wounds. Mol Ther Nucleic Acids. 2021;24:554–578. doi:10.1016/j.omtn.2021.03.015
36. Beltrami C, Angelini T, Emanueli C. Noncoding RNAs in diabetes vascular complications. J Mol Cell Cardiol. 2015;89:42–50. doi:10.1016/j.yjmcc.2014.12.014
37. Hutchinson J, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi:10.1186/1471-2164-8-39
38. Taiana E, Ronchetti D, Todoerti K, et al. LncRNA NEAT1 in paraspeckles: a structural scaffold for cellular DNA damage response systems? Non-Coding RNA. 2020;6(3):26. doi:10.3390/ncrna6030026
39. An H, Williams N, Shelkovnikova T. NEAT1 and paraspeckles in neurodegenerative diseases: a missing lnc found? Non-Coding RNA Res. 2018;3(4):243–252. doi:10.1016/j.ncrna.2018.11.003
40. Clemson C, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi:10.1016/j.molcel.2009.01.026
41. West J, Davis C, Sunwoo H, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55(5):791–802. doi:10.1016/j.molcel.2014.07.012
42. West J, Mito M, Kurosaka S, et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol. 2016;214(7):817–830. doi:10.1083/jcb.201601071
43. Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol. 2019;13(1):46–60. doi:10.1002/1878-0261.12404
44. Guru S, Agarwal SK, Manickam P, et al. A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res. 1997;7(7):725–735. doi:10.1101/gr.7.7.725
45. Sasaki Y, Ideue T, Sano M, et al. MENε/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106(8):2525–2530. doi:10.1073/pnas.0807899106
46. Isobe M, Toya H, Mito M, et al. Forced isoform switching of Neat1_1 to Neat1_2 leads to the loss of Neat1_1 and the hyperformation of paraspeckles but does not affect the development and growth of mice. RNA. 2020;26(3):251–264. doi:10.1261/rna.072587.119
47. Leng F, Edison P. Neuroinflammation and microglial activation in Alzheimer's disease: where do we go from here? Nat Rev Neurol. 2021;17(3):157–172. doi:10.1038/s41582-020-00435-y
48. Yang Q, Zhou J. Neuroinflammation in the central nervous system: symphony of glial cells. Glia. 2019;67(6):1017–1035. doi:10.1002/glia.23571
49. Dong P, Xiong Y, Yue J, et al. Long non-coding RNA NEAT1: a novel target for diagnosis and therapy in human tumors. Front Genet. 2018;9:471. doi:10.3389/fgene.2018.00471
50. Han D, Zhou Y. YY1-induced upregulation of lncRNA NEAT1 contributes to OGD/R injury-induced inflammatory response in cerebral microglial cells via Wnt/β-catenin signaling pathway. In Vitro Cell Dev Biol Anim. 2019;55(7):501–511. doi:10.1007/s11626-019-00375-y
51. Liu R, Li F, Zhao W. Long noncoding RNA NEAT1 knockdown inhibits MPP-induced apoptosis, inflammation and cytotoxicity in SK-N-SH cells by regulating miR-212-5p/RAB3IP axis. Neurosci Lett. 2020;731:135060. doi:10.1016/j.neulet.2020.135060
52. Chen M, Fan K, Zhao L-J, et al. Long non-coding RNA nuclear enriched abundant transcript 1 (NEAT1) sponges microRNA-124-3p to up-regulate phosphodiesterase 4B (PDE4B) to accelerate the progression of Parkinson’s disease. Bioengineered. 2021;12(1):708–719. doi:10.1080/21655979.2021.1883279
53. Sun Q, Zhang Y, Wang S, et al. neat1 decreasing suppresses Parkinson’s disease progression via acting as miR-1301-3p sponge. J Mol Neurosci. 2021;71(2):369–378. doi:10.1007/s12031-020-01660-2
54. Xie S, Zhou F, Li J, et al. NEAT1 regulates MPP-induced neuronal injury by targeting miR-124 in neuroblastoma cells. Neurosci Lett. 2019;708:134340. doi:10.1016/j.neulet.2019.134340
55. Zhou S, Zhang D, Guo J, et al. Deficiency of NEAT1 prevented MPP-induced inflammatory response, oxidative stress and apoptosis in dopaminergic SK-N-SH neuroblastoma cells via miR-1277-5p/ARHGAP26 axis. Brain Res. 2021;1750:147156. doi:10.1016/j.brainres.2020.147156
56. Liu T, Zhang Y, Liu W, et al. LncRNA NEAT1 regulates the development of Parkinson’s disease by targeting AXIN1 via sponging miR-212-3p. Neurochem Res. 2021;46(2):230–240. doi:10.1007/s11064-020-03157-1
57. Ban Y, Cui C. Silencing of long non-coding RNA (lncRNA) nuclear paraspeckle assembly transcript 1 (NEAT1) Protects PC-12 Cells from LPS-Induced Injury via Targeting miR-29a. Med Sci Mon. 2020;26:e923914. doi:10.12659/MSM.923914
58. An Q, Lu Z, Xie Y, et al. Knockdown of long non-coding RNA NEAT1 relieves the inflammatory response of spinal cord injury through targeting miR-211-5p/MAPK1 axis. Bioengineered. 2021;12(1):2702–2712. doi:10.1080/21655979.2021.1930925
59. Zhang H, Ouyang B, Ji X-Y, et al. Gastrodin alleviates cerebral ischaemia/reperfusion injury by inhibiting pyroptosis by regulating the lncRNA NEAT1/miR-22-3p axis. Neurochem Res. 2021;46(7):1747–1758. doi:10.1007/s11064-021-03285-2
60. Jin F, Ou W, Wei B, et al. Neat1Transcriptome-wide analysis to identify the inflammatory role of lncRNA in experimental ischemic stroke. J Inflamm Res. 2021;14:2667–2680. doi:10.2147/JIR.S315281
61. Li P, Duan S, Fu A. Long noncoding RNA NEAT1 correlates with higher disease risk, worse disease condition, decreased miR-124 and miR-125a and predicts poor recurrence-free survival of acute ischemic stroke. J Clin Lab Anal. 2020;34(2):e23056. doi:10.1002/jcla.23056
62. Ni X, Su Q, Xia W, et al. Knockdown lncRNA NEAT1 regulates the activation of microglia and reduces AKT signaling and neuronal apoptosis after cerebral ischemic reperfusion. Sci Rep. 2020;10(1):19658. doi:10.1038/s41598-020-71411-1
63. Zhong J, Jiang L, Huang Z, et al. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav Immun. 2017;65:183–194. doi:10.1016/j.bbi.2017.05.001
64. Wan Y, Yang Z. LncRNA NEAT1 affects inflammatory response by targeting miR-129-5p and regulating Notch signaling pathway in epilepsy. Cell Cycle. 2020;19(4):419–431. doi:10.1080/15384101.2020.1711578
65. Murta V, Ferrari C. Peripheral inflammation and demyelinating diseases. Adv Exp Med Biol. 2016;949:263–285.
66. Sato S, Mukai Y. Modulation of chronic inflammation by quercetin: the beneficial effects on obesity. J Inflamm Res. 2020;13:421–431. doi:10.2147/JIR.S228361
67. Huang X, Hussain B, Chang J. Peripheral inflammation and blood-brain barrier disruption: effects and mechanisms. CNS Neurosci Ther. 2021;27(1):36–47. doi:10.1111/cns.13569
68. Süβ P, Lana A, Schlachetzki J. Chronic peripheral inflammation: a possible contributor to neurodegenerative diseases. Neural Regen Res. 2021;16(9):1711–1714. doi:10.4103/1673-5374.306060
69. Xia D, Yao R, Zhou P, et al. LncRNA NEAT1 reversed the hindering effects of miR-495-3p/STAT3 axis and miR-211/PI3K/AKT axis on sepsis-relevant inflammation. Mol Immunol. 2020;117:168–179. doi:10.1016/j.molimm.2019.10.009
70. Huang Q, Huang C, Luo Y, et al. Circulating lncRNA NEAT1 correlates with increased risk, elevated severity and unfavorable prognosis in sepsis patients. Am J Emerg Med. 2018;36(9):1659–1663. doi:10.1016/j.ajem.2018.06.008
71. He F, Zhang C, Huang Q. Long noncoding RNA nuclear enriched abundant transcript 1/miRNA-124 axis correlates with increased disease risk, elevated inflammation, deteriorative disease condition, and predicts decreased survival of sepsis. Medicine. 2019;98(32):e16470. doi:10.1097/MD.0000000000016470
72. Chen J, Xu X, Zhang S. Silence of long noncoding RNA NEAT1 exerts suppressive effects on immunity during sepsis by promoting microRNA-125-dependent MCEMP1 downregulation. IUBMB Life. 2019;71(7):956–968.
73. Li Y, Guo W, Cai Y. NEAT1 promotes LPS-induced inflammatory injury in macrophages by regulating MiR-17-5p/TLR4. Open Med. 2020;15:38–49. doi:10.1515/med-2020-0007
74. Liu L, Liu F, Sun Z, et al. LncRNA NEAT1 promotes apoptosis and inflammation in LPS-induced sepsis models by targeting miR-590-3p. Exp Ther Med. 2020;20(4):3290–3300. doi:10.3892/etm.2020.9079
75. Wang W, Guo Z. Downregulation of lncRNA NEAT1 ameliorates LPS-induced inflammatory responses by promoting macrophage M2 polarization via miR-125a-5p/TRAF6/TAK1 axis. Inflammation. 2020;43(4):1548–1560. doi:10.1007/s10753-020-01231-y
76. Wu X, Fang Y, Zheng F-X, et al. LncRNA NEAT1 facilitates the progression of sepsis through up-regulating TSP-1 via sponging miR-370-3p. Eur Rev Med Pharmacol Sci. 2020;24(1):333–344. doi:10.26355/eurrev_202001_19931
77. Xiao T, Sun C, Xiao Y, et al. lncRNA NEAT1 mediates sepsis progression by regulating Irak2 via sponging miR-370-3p. Biol Open. 2020;9(6). doi:10.1242/bio.049353
78. Yang Y, Xue J, Qin L, et al. LncRNA NEAT1 promotes inflammatory response in sepsis via the miR-31-5p/POU2F1 axis. Inflammation. 2021;44(4):1518–1528. doi:10.1007/s10753-021-01436-9
79. Gast M, Rauch BH, Haghikia A, et al. Long noncoding RNA NEAT1 modulates immune cell functions and is suppressed in early onset myocardial infarction patients. Cardiovasc Res. 2019;115(13):1886–1906. doi:10.1093/cvr/cvz085
80. Chen X, Jiang Y-J, Zeng T, et al. Overexpression of the long noncoding RNA NEAT1 protects against As2O3-induced injury of cardiomyocyte by inhibiting the miR-124/NF-κB signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(3):1378–1390. doi:10.26355/eurrev_202002_20195
81. Wang S, Liu G-Q, Xian H-B, et al. LncRNA NEAT1 alleviates sepsis-induced myocardial injury by regulating the TLR2/NF-κB signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(11):4898–4907. doi:10.26355/eurrev_201906_18078
82. Wei J, Wu C-J, Chen J-J, et al. LncRNA NEAT1 promotes the progression of sepsis-induced myocardial cell injury by sponging miR-144-3p. Eur Rev Med Pharmacol Sci. 2020;24(2):851–861. doi:10.26355/eurrev_202001_20069
83. Huang-Fu N, Cheng J-S, Wang Y, et al. Neat1 regulates oxidized low-density lipoprotein-induced inflammation and lipid uptake in macrophages via paraspeckle formation. Mol Med Rep. 2018;17(2):3092–3098. doi:10.3892/mmr.2017.8211
84. Chen D, Hui LL, Zhang XC, et al. NEAT1 contributes to ox-LDL-induced inflammation and oxidative stress in macrophages through inhibiting miR-128. J Cell Biochem. 2019;120:2493–2501.
85. Wang L, Xia J-W, Ke Z-P, et al. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342-3p in human macrophages THP-1 cells. J Cell Physiol. 2019;234(4):5319–5326. doi:10.1002/jcp.27340
86. Zhou H, Wang X, Zhang B. Depression of lncRNA NEAT1 antagonizes LPS-evoked acute injury and inflammatory response in alveolar epithelial cells via HMGB1-RAGE signaling. Mediators Inflamm. 2020;2020:8019467. doi:10.1155/2020/8019467
87. Li X, Ye S, Lu Y. Long non-coding RNA NEAT1 overexpression associates with increased exacerbation risk, severity, and inflammation, as well as decreased lung function through the interaction with microRNA-124 in asthma. J Clin Lab Anal. 2020;34(1):e23023. doi:10.1002/jcla.23023
88. Wang X, Xu R, Chi D, et al. Role of NEAT1/MiR-9-5p/SLC26A2 pathway on human airway smooth muscle cell. Yonsei Med J. 2021;62(9):858–867. doi:10.3349/ymj.2021.62.9.858
89. Wang K, Zhang Z, Liu K, et al. Neat1-miRNA204-5p-PI3K-AKT axis as a potential mechanism for photodynamic therapy treated colitis in mice. Photodiagnosis Photodyn Ther. 2018;24:349–357. doi:10.1016/j.pdpdt.2018.10.020
90. Liu R, Tang A, Wang X, et al. Inhibition of lncRNA NEAT1 suppresses the inflammatory response in IBD by modulating the intestinal epithelial barrier and by exosome-mediated polarization of macrophages. Int J Mol Med. 2018;42(5):2903–2913. doi:10.3892/ijmm.2018.3829
91. Guo J, Wang L, Yu H. Knockdown of NEAT1 mitigates ox-LDL-induced injury in human umbilical vein endothelial cells via miR-30c-5p/TCF7 axis. Eur Rev Med Pharmacol Sci. 2020;24(18):9633–9644. doi:10.26355/eurrev_202009_23052
92. Shao K, Xi L, Cang Z, et al. Knockdown of NEAT1 exerts suppressive effects on diabetic retinopathy progression via inactivating TGF-β1 and VEGF signaling pathways. J Cell Physiol. 2020;235(12):9361–9369. doi:10.1002/jcp.29740
93. Feng Y, Liu J, Wu R, et al. NEAT1 aggravates sepsis-induced acute kidney injury by sponging miR-22-3p. Open Med. 2020;15(1):333–342. doi:10.1515/med-2020-0401
94. Wang J, Chen Y, Tang Z, et al. LncRNA NEAT1 regulated inflammation and apoptosis in a rat model of sepsis-induced acute kidney injury via MiR-27a-3p/TAB3 axis. Biosci Biotechnol Biochem. 2020;84(11):2215–2227. doi:10.1080/09168451.2020.1792760
95. Zhang C, Niu F. LncRNA NEAT1 promotes inflammatory response in sepsis-induced liver injury via the Let-7a/TLR4 axis. Int Immunopharmacol. 2019;75:105731. doi:10.1016/j.intimp.2019.105731
96. Jin S, Lin X-F, Zheng J-Z, et al. lncRNA NEAT1 regulates fibrosis and inflammatory response induced by nonalcoholic fatty liver by regulating miR-506/GLI3. Eur Cytokine Netw. 2019;30(3):98–106. doi:10.1684/ecn.2019.0432
97. Ye J, Lin Y, Yu Y, et al. LncRNA NEAT1/microRNA-129-5p/SOCS2 axis regulates liver fibrosis in alcoholic steatohepatitis. J Transl Med. 2020;18(1):445. doi:10.1186/s12967-020-02577-5
98. Wang Q, Liu S, Wang H, et al. Silencing long noncoding RNA NEAT1 alleviates acute liver failure via the EZH2-mediated microRNA-139/PUMA axis. Aging. 2021;13(9):12537–12551. doi:10.18632/aging.202927
99. Xu Y, Cao Z, Ding Y, et al. Long non-coding RNA NEAT1 alleviates acute-on-chronic liver failure through blocking TRAF6 mediated inflammatory response. Front Physiol. 2019;10:1503. doi:10.3389/fphys.2019.01503
100. Zhang Z, Wen H, Peng B, et al. Downregulated microRNA-129-5p by long non-coding RNA NEAT1 upregulates PEG3 expression to aggravate non-alcoholic steatohepatitis. Front Genet. 2020;11:563265. doi:10.3389/fgene.2020.563265
101. Chen C, Zhang H, Ge M, et al. LncRNA NEAT1 acts as a key regulator of cell apoptosis and inflammatory response by the miR-944/TRIM37 axis in acute lung injury. J Pharmacol Sci. 2021;145(2):202–212. doi:10.1016/j.jphs.2020.11.009
102. Gao C, Zou X, Chen H, et al. Long non-coding RNA nuclear paraspeckle assembly transcript 1 (NEAT1) relieves sepsis-induced kidney injury and lipopolysaccharide (LPS)-induced inflammation in HK-2 cells. Med Sci Mon. 2020;26:e921906. doi:10.12659/MSM.921906
103. Zhang F, Wu L, Qian J, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. 2016;75:96–104. doi:10.1016/j.jaut.2016.07.012
104. Zhang L, Xiao B, Zhong M, et al. LncRNA NEAT1 accelerates renal mesangial cell injury via modulating the miR-146b/TRAF6/NF-κB axis in lupus nephritis. Cell Tissue Res. 2020;382(3):627–638. doi:10.1007/s00441-020-03248-z
105. Liu F, Liu X, Yang Y, et al. NEAT1/miR-193a-3p/SOX5 axis regulates cartilage matrix degradation in human osteoarthritis. Cell Biol Int. 2020;44(4):947–957. doi:10.1002/cbin.11291
106. Wang Z, Hao J, Chen D. Long noncoding RNA nuclear enriched abundant transcript 1 (NEAT1) regulates proliferation, apoptosis, and inflammation of chondrocytes via the miR-181a/glycerol-3-phosphate dehydrogenase 1-like (GPD1L) axis. Med Sci Mon. 2019;25:8084–8094. doi:10.12659/MSM.918416
107. Tu Y, Ma T, Wen T, et al. MicroRNA-377-3p alleviates IL-1β-caused chondrocyte apoptosis and cartilage degradation in osteoarthritis in part by downregulating ITGA6. Biochem Biophys Res Commun. 2020;523(1):46–53. doi:10.1016/j.bbrc.2019.11.186
108. Sheng B, Zhao L, Zang X, et al. Quercetin inhibits caerulein-induced acute pancreatitis through regulating miR-216b by targeting MAP2K6 and NEAT1. Inflammopharmacology. 2021;29(2):549–559. doi:10.1007/s10787-020-00767-7
109. Zhu Q, Zhao C, Wang Y, et al. LncRNA NEAT1 promote inflammatory responses in coronary slow flow through regulating miR-148b-3p/ICAM-1 axis. J Inflamm Res. 2021;14:2445–2463. doi:10.2147/JIR.S312583
110. Wang Q, Wang W, Zhang F, et al. NEAT1/miR-181c regulates osteopontin (OPN)-mediated synoviocyte proliferation in osteoarthritis. J Cell Biochem. 2017;118(11):3775–3784. doi:10.1002/jcb.26025
111. Li C, Liu Y-F, Huang C, et al. Long noncoding RNA NEAT1 sponges miR-129 to modulate renal fibrosis by regulation of collagen type I. Am J Physiol Renal Physiol. 2020;319(1):F93–F105. doi:10.1152/ajprenal.00552.2019
112. Pan S, Liu R, Wu X, et al. LncRNA NEAT1 mediates intestinal inflammation by regulating TNFRSF1B. Ann Transl Med. 2021;9(9):773. doi:10.21037/atm-21-34
113. Wang S, Zhang Q, Wang Q, et al. NEAT1 paraspeckle promotes human hepatocellular carcinoma progression by strengthening IL-6/STAT3 signaling. Oncoimmunology. 2018;7(11):e1503913. doi:10.1080/2162402X.2018.1503913
114. Sun Q, Shen X, Ma J, et al. LncRNA NEAT1 participates in inflammatory response in macrophages infected by mycobacterium tuberculosis through targeted regulation of miR-377-3p. Microb Pathog. 2021;150:104674. doi:10.1016/j.micpath.2020.104674
115. Wang L, Qu P, Yin W, et al. Lnc-NEAT1 induces cell apoptosis and inflammation but inhibits proliferation in a cellular model of hepatic ischemia/reperfusion injury. J Int Med Res. 2021;49(3):300060519887251. doi:10.1177/0300060519887251
116. Wang R, et al. The correlation of long non-coding RNA NEAT1 and its targets microRNA (miR)-21, miR-124, and miR-125a with disease risk, severity and inflammation of allergic rhinitis. Medicine. 2021;100(4):e22946.
117. Nong W. Long non-coding RNA NEAT1/miR-193a-3p regulates LPS-induced apoptosis and inflammatory injury in WI-38 cells through TLR4/NF-κB signaling. Am J Transl Res. 2019;11(9):5944–5955.
118. Dai W, Wang M, Wang P, et al. lncRNA NEAT1 ameliorates LPS‑induced inflammation in MG63 cells by activating autophagy and suppressing the NLRP3 inflammasome. Int J Mol Med. 2021;47(2):607–620. doi:10.3892/ijmm.2020.4827
119. Wang Y, Hou L, Yuan X, et al. LncRNA NEAT1 targets fibroblast-like synoviocytes in rheumatoid arthritis via the miR-410-3p/YY1 axis. Front Immunol. 2020;11:1975. doi:10.3389/fimmu.2020.01975
120. Rao Y, Fang Y, Tan W, et al. Delivery of long non-coding RNA NEAT1 by peripheral blood mononuclear cells-derived exosomes promotes the occurrence of rheumatoid arthritis via the microRNA-23a/MDM2/SIRT6 axis. Front Cell Dev Biol. 2020;8:551681. doi:10.3389/fcell.2020.551681
121. Zhan J, Huang H-W, Huang C, et al. Long non-coding RNA NEAT1 regulates pyroptosis in diabetic nephropathy via mediating the miR-34c/NLRP3 axis. Kidney Blood Press Res. 2020;45(4):589–602. doi:10.1159/000508372
122. Fakhoury M. Microglia and astrocytes in Alzheimer’s disease: implications for therapy. Curr Neuropharmacol. 2018;16(5):508–518. doi:10.2174/1570159X15666170720095240
123. Xin W, Wei W, Pan Y-L, et al. Modulating poststroke inflammatory mechanisms: novel aspects of mesenchymal stem cells, extracellular vesicles and microglia. World J Stem Cells. 2021;13(8):1030–1048. doi:10.4252/wjsc.v13.i8.1030
124. Iadecola C, Anrather J. The immunology of stroke: from mechanisms to translation. Nat Med. 2011;17(7):796–808. doi:10.1038/nm.2399
125. Jin R, Yang G, Li G. Inflammatory mechanisms in ischemic stroke: role of inflammatory cells. J Leukoc Biol. 2010;87(5):779–789. doi:10.1189/jlb.1109766
126. Lehrmann E, et al. Microglial and macrophage reactions mark progressive changes and define the penumbra in the rat neocortex and striatum after transient middle cerebral artery occlusion. J Comp Neurol. 1997;386(3):461–476.
127. Kettenmann H, Hanisch U-K, Noda M, et al. Physiology of microglia. Physiol Rev. 2011;91(2):461–553. doi:10.1152/physrev.00011.2010
128. MacMicking J, Xie Q, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi:10.1146/annurev.immunol.15.1.323
129. Yenari M, Kauppinen T, Swanson R. Microglial activation in stroke: therapeutic targets. Neurotherapeutics. 2010;7(4):378–391. doi:10.1016/j.nurt.2010.07.005
130. Varnum M, Ikezu T. The classification of microglial activation phenotypes on neurodegeneration and regeneration in Alzheimer’s disease brain. Arch Immunol Ther Exp. 2012;60(4):251–266. doi:10.1007/s00005-012-0181-2
131. Cherry J, Olschowka J, O’Banion M. Neuroinflammation and M2 microglia: the good, the bad, and the inflamed. J Neuroinflammation. 2014;11:98. doi:10.1186/1742-2094-11-98
132. Ponomarev E, Veremeyko T, Weiner H. MicroRNAs are universal regulators of differentiation, activation, and polarization of microglia and macrophages in normal and diseased CNS. Glia. 2013;61(1):91–103. doi:10.1002/glia.22363
133. Zhang C, Wang K, Yang L, et al. Lipid metabolism in inflammation-related diseases. Analyst. 2018;143(19):4526–4536. doi:10.1039/C8AN01046C
134. Bennett M, Gilroy D, Gordon S. Lipid Mediators in Inflammation. Microbiol Spectr. 2016;4(6). doi:10.1128/microbiolspec.MCHD-0035-2016
135. Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med. 2011;17(11):1410–1422. doi:10.1038/nm.2538
136. Alves-Bezerra M, Cohen D. Triglyceride metabolism in the liver. Compr Physiol. 2017;8(1):1–8. doi:10.1002/cphy.c170012
137. Fan G, Zhang C, Wei X, et al. NEAT1/hsa-miR-372-3p axis participates in rapamycin-induced lipid metabolic disorder. Free Radic Biol Med. 2021;167:1–11. doi:10.1016/j.freeradbiomed.2021.02.033
138. Chen X, Tan X-R, Li S-J, et al. LncRNA NEAT1 promotes hepatic lipid accumulation via regulating miR-146a-5p/ROCK1 in nonalcoholic fatty liver disease. Life Sci. 2019;235:116829. doi:10.1016/j.lfs.2019.116829
139. Chatterjee S, Bhattcharjee D, Misra S, et al. Increase in MEG3, MALAT1, NEAT1 significantly predicts the clinical parameters in patients with rheumatoid arthritis. Per Med. 2020;17(6):445–457. doi:10.2217/pme-2020-0009
140. Xiong W, Qu Y, Chen H, et al. Insight into long noncoding RNA-miRNA-mRNA axes in myocardial ischemia-reperfusion injury: the implications for mechanism and therapy. Epigenomics. 2019;11(15):1733–1748. doi:10.2217/epi-2019-0119
141. Pan Y, Jiao Q, Wei W, et al. Emerging role of LncRNAs in ischemic stroke-novel insights into the regulation of inflammation. J Inflamm Res. 2021;14:4467–4483. doi:10.2147/JIR.S327291
142. He L, Chen Y, Hao S, et al. Uncovering novel landscape of cardiovascular diseases and therapeutic targets for cardioprotection via long noncoding RNA-miRNA-mRNA axes. Epigenomics. 2018;10(5):661–671. doi:10.2217/epi-2017-0176
143. Archer K, Broskova Z, Bayoumi A, et al. Long non-coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci. 2015;16(10):23651–23667. doi:10.3390/ijms161023651
144. Huang S, Ge X, Yu J, et al. Increased miR-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J. 2018;32(1):512–528. doi:10.1096/fj.201700673r
145. Yang Z, Zeng B, Wang C, et al. MicroRNA-124 alleviates chronic skin inflammation in atopic eczema via suppressing innate immune responses in keratinocytes. Cell Immunol. 2017;319:53–60. doi:10.1016/j.cellimm.2017.08.003
146. Zhao Y, Ma T, Chen W, et al. MicroRNA-124 promotes intestinal inflammation by targeting Aryl hydrocarbon receptor in Crohn’s disease. J Crohn’s Colitis. 2016;10(6):703–712. doi:10.1093/ecco-jcc/jjw010
147. Barnabei L, Laplantine E, Mbongo W, et al. NF-κB: at the borders of autoimmunity and inflammation. Front Immunol. 2021;12:716469. doi:10.3389/fimmu.2021.716469
148. Tian J, Liu Y, Wang Z, et al. LncRNA Snhg8 attenuates microglial inflammation response and blood-brain barrier damage in ischemic stroke through regulating miR-425-5p mediated SIRT1/NF-κB signaling. J Biochem Mol Toxicol. 2021;35(5):e22724. doi:10.1002/jbt.22724
149. Jie Z, Ko C-J, Wang H, et al. Microglia promote autoimmune inflammation via the noncanonical NF-κB pathway. Sci Adv. 2021;7(36):eabh0609. doi:10.1126/sciadv.abh0609
150. Xiao J, Wang R, Zhou W, et al. LncRNA NEAT1 regulates the proliferation and production of the inflammatory cytokines in rheumatoid arthritis fibroblast-like synoviocytes by targeting miR-204-5p. Hum Cell. 2021;34(2):372–382. doi:10.1007/s13577-020-00461-4
151. Zhang F, Xu R. Juglanin ameliorates LPS-induced neuroinflammation in animal models of Parkinson’s disease and cell culture via inactivating TLR4/NF-κB pathway. Biomed Pharmacother. 2018;97:1011–1019. doi:10.1016/j.biopha.2017.08.132
152. Yamamoto M, Gohda J, Akiyama T, et al. TNF receptor-associated factor 6 (TRAF6) plays crucial roles in multiple biological systems through polyubiquitination-mediated NF-κB activation. Proc Japan Acad Ser B. 2021;97(4):145–160. doi:10.2183/pjab.97.009
153. Lv H, Li J, Che Y. CXCL8 gene silencing promotes neuroglial cells activation while inhibiting neuroinflammation through the PI3K/Akt/NF-κB-signaling pathway in mice with ischemic stroke. J Cell Physiol. 2019;234(5):7341–7355. doi:10.1002/jcp.27493
154. Abeyrathna P, Su Y. The critical role of Akt in cardiovascular function. Vascul Pharmacol. 2015;74:38–48. doi:10.1016/j.vph.2015.05.008
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.