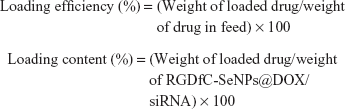Back to Journals » International Journal of Nanomedicine » Volume 13
Novel functionalized nanoparticles for tumor-targeting co-delivery of doxorubicin and siRNA to enhance cancer therapy
Authors Xia Y, Xu T, Wang C, Li Y, Lin Z, Zhao M, Zhu B
Received 11 August 2017
Accepted for publication 13 November 2017
Published 22 December 2017 Volume 2018:13 Pages 143—159
DOI https://doi.org/10.2147/IJN.S148960
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Lei Yang
Yu Xia, Tiantian Xu, Changbing Wang, Yinghua Li, Zhengfang Lin, Mingqi Zhao, Bing Zhu
Central Laboratory, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, People’s Republic of China
Abstract: Human homeobox protein (Nanog) is highly expressed in most cancer cells and has gradually emerged as an excellent target in cancer therapy, owing to its regulation of cancer cell proliferation, metastasis and apoptosis. In this study, we prepared tumor-targeting functionalized selenium nanoparticles (RGDfC-SeNPs) to load chemotherapeutic doxorubicin (DOX) and Nanog siRNA. Herein, RGDfC peptide was used as a tumor-targeting moiety which could specifically bind to αvβ3 integrins overexpressed on various cancer cells. The sizes of RGDfC-SeNPs@DOX nanoparticles (~12 nm) were confirmed by both dynamic light scattering and transmission electron microscopy. The chemical structure of RGDfC-SeNPs@DOX was characterized via Fourier-transform infrared spectroscopy. The RGDfC-SeNPs@DOX was compacted with siRNA (anti-Nanog) by electrostatic interaction to fabricate the RGDfC-SeNPs@DOX/siRNA complex. The RGDfC-SeNPs@DOX/siRNA complex nanoparticles could efficiently enter into HepG2 cells via clathrin-associated endocytosis, and showed high gene transfection efficiency that resulted in enhanced gene silencing. The in vivo biodistribution experiment indicated that RGDfC-SeNPs@DOX/siRNA nanoparticles were capable of specifically accumulating in the tumor site. Furthermore, treatment with RGDfC-SeNPs@DOX/siRNA resulted in a more significant anticancer activity than the free DOX, RGDfC-SeNPs@DOX or RGDfC-SeNPs/siRNA in vitro and in vivo. In summary, this study shows a novel type of DOX and siRNA co-delivery system, thereby providing an alternative route for cancer treatment.
Keywords: nanoparticles, tumor targeting, drug delivery, doxorubicin, Nanog siRNA
Introduction
Hepatocellular carcinoma (HCC) is one of the most common human malignancies and the third leading cause of cancer mortality worldwide.1 Chemotherapy is still a major clinical method for cancer treatments.2 However, the traditional chemotherapeutic drugs are generally nonspecific and have no ability to discriminate between cancer cells and normal cells, which frequently causes systemic side effects.3 To improve these shortcomings, a large number of nanoscale drug carriers, such as polymeric nanomicelles, inorganic nanoparticles and liposomes and so on, have been developed to achieve the tumor-targeted delivery of chemotherapeutic drugs.4–6 Among them, selenium nanoparticles (SeNPs) have received an increasing attention as drug carriers owing to their biocompatibility, low toxicity, simple synthesis process, in vivo degradability and excellent chemopreventive effects.7 Selenium is a mineral trace element of fundamental importance to humans and animals. The role of Se as a potential cancer chemotherapeutic and chemopreventive agent has been supported by many epidemiological, preclinical and clinical studies. Previous studies showed that Se was degradable in vivo and the degraded Se could be used as a nutrient for many kinds of normal cells or as an antiproliferative agent for many kinds of cancer cells.8 In the past years, peptides as a tumor-targeting moiety have been widely used in various kinds of nano-delivery systems due to their high affinity for αvβ3 integrin receptor, which is overexpressed on various cancer cells, including HCC cells.9 A great deal of researches demonstrated that the receptor-mediated active targeting delivery systems significantly improved the antitumor efficacy.10,11 However, increasing evidence indicates that cancers still cannot be cured with single chemotherapeutic drug due to their continuously evolving nature and dysregulated multiplication.12
Recently, the co-delivery of chemotherapeutic drugs and siRNA has gained increasing attention owing to enhanced antitumor efficacy over single administration.13,14 Previous studies showed that simultaneously delivering chemotherapeutic drugs and siRNA via a single targeted vector was more effective in treating some cancers than sequential administration of two separate vectors with one drug in each.15 This finding indicates that simultaneous delivery of chemotherapeutic drugs and suitable amount of siRNA into the same tumor plays an important role in the cancer treatments.16
Emerging evidence has proposed that human homeobox protein (Nanog) is a excellent target in cancer therapy owing to its regulation of cancer cell proliferation, metastasis and apoptosis.17 In addition, Nanog plays a crucial role in maintaining the stemness of cancer cells, such as self-renewal, recurrence and high tumorigenicity in various cancers.18 Previous researches have reported that inhibition of Nanog functions was capable of suppressing cancer cell growth and inducing cancer cell apoptosis.19 Thus, Nanog gene has been developed as a potential cancer target for gene-silencing therapy.
In the past decade, SeNPs have been developed as excellent drug or gene delivery carriers which exhibit many advantages, such as easy preparation, effective cellular uptake, controlled release of drug/gene and absence of hypotoxicity.20 To obtain a combined effect of chemotherapeutic drug and siRNA, the positively charged RGDfC (Arg-Gly-Asp-D-Phe-Cys, tumor-targeting peptide) was coupled with the SeNPs to form peptide-conjugated functionalized selenium nanoparticles (RGDfC-SeNPs) for the targeted co-delivery of doxorubicin (DOX) and siRNA (anti-Nanog) in vitro and in vivo. In vitro studies indicated that the co-delivery system RGDfC-SeNPs@DOX/siRNA exhibited good cellular uptake and high gene transfection efficiency in HepG2 cells. The RGDfC-SeNPs@DOX/siRNA was effectively capable of inhibiting cancer cell proliferation and migration, and inducing cancer cell apoptosis in vitro. The in vivo studies showed that the RGDfC-SeNPs@DOX/siRNA could extensively accumulate in tumor sites, significantly inhibit tumor growth and induce tumor cell apoptosis. Moreover, co-delivery of DOX and siRNA showed a combined effect to inhibit HepG2 tumor growth in vivo, which was more efficient than either DOX- or siRNA-based monotherapy. Such RGDfC peptide-conjugated SeNPs hold great potential applications in HCC therapy for the co-delivery of DOX and siRNA to the HepG2 tumors.
Experimental
Materials
Ascorbic acid (Vitamin C), doxorubicin hydrochloride (DOX·HCl) and sodium selenite were purchased from Sinopharm (Shanghai, People’s Republic of China). 4′,6-Diamidino-2-phenyindole (DAPI), Annexin V-FITC Apoptosis Kit and cyclic peptide (Arg-Gly-Asp-D-Phe-Cys, RGDfC) were purchased from ChinaPeptides Co. Ltd. TUNEL assay kit and cy5.5 were purchased from Beyotime (Shanghai, People’s Republic of China). All antibodies were purchased from CST. The Nanog siRNAs were purchased from GenePharma (Shanghai, People’s Republic of China), and the sequence was as follows: 5′-CCUCCAUGGAUCUGCUUAUUU-3′.
Synthesis of RGDfC-SeNPs@DOX
The RGDfC-SeNPs were synthesized as previously described.21 Briefly, 1 mg RGDfC powder dissolved in 2 mL water was added to the ascorbic acid and sodium selenite mixed solution under magnetic stirring for 6 h. The RGDfC-SeNPs were synthesized by coating RGDfC onto the surface of SeNPs. Then, the mixed solutions were dialyzed (3.5 kDa) for 12 h to eliminate the redundant RGDfC, ascorbic acid and sodium selenite. To load DOX onto the surfaces of RGDfC-SeNPs, 1 mg DOX·HCl was dissolved in 1 mL dimethyl sulfoxide (DMSO) and dropped to RGDfC-SeNPs solution for 8 h. The reaction solutions were dialyzed using deionized water for 12 h to obtain high-purity RGDfC-SeNPs@DOX. The chemical structure of RGDfC-SeNPs@DOX nanoparticles was confirmed using Fourier-transform infrared spectroscopy (FTIR) analysis. The nanoparticles were characterized by transmission electron microscopy (TEM) and FTIR. Energy-dispersive X-ray spectroscopy analysis was conducted to detect the elemental composition of nanoparticles. The average sizes and zeta potentials of the nanoparticles were tested using Zetasizer particle analyzer (Malvern Instruments Ltd., Malvern, UK).
Synthesis of the RGDfC-SeNPs@DOX/siRNA complexes
The RGDfC-SeNPs@DOX/siRNA complexes were obtained as previously reported.22 Briefly, the RGDfC-SeNPs@DOX was dispersed in DNase/RNase-free water, and then vortexed with an siRNA solution for 1 h to form RGDfC-SeNPs@DOX/siRNA complexes. To examine the DOX- and siRNA-loading capability, we established a calibration curve against the DOX and FAM-siRNA, respectively, and obtained the relative fluorescence intensity of DOX and FAM-siRNA in the nanoparticles, respectively. The test was performed in a multimode plate reader (EnSpire; PerkinElmer, Waltham, MA, USA) (for DOX: Ex 535 nm, Em 590 nm; for FAM-siRNA: Ex 465 nm, Em 520 nm). The loading efficiency and loading content of DOX and siRNA were calculated using the following formulas:
|
The loading efficiency of DOX into the RGDfC-SeNPs@DOX was found to be about 75%.
Agarose gel retardation assay
An agarose gel electrophoresis was carried out to evaluate the ability of RGDfC-SeNPs@DOX to bind siRNA. RGDfC-SeNPs@DOX/siRNA complexes at different N:P ratios from 1:1 to 16:1 were prepared, and the solutions were measured via electrophoresis for 45 min at 85 V. An agarose gel electrophoretic assay was also conducted to determine whether RGDfC-SeNPs@DOX protects siRNA in the presence of serum. Briefly, RGDfC-SeNPs@DOX:siRNA at a ratio of 16:1 were incubated with 50% fetal bovine serum (FBS) at 37°C for 60 min. Then, each sample was collected and subjected to gel electrophoresis in 1.2% agarose gel for 45 min at 85 V. The gel was stained with EB and then imaged by Gel Doc XR+ (Bio-Rad, Hercules, CA, USA).
In vitro release of DOX
The release profiles of DOX from RGDfC-SeNPs@DOX/siRNA were evaluated using the dialysis method.23 Briefly, 1 mg of the RGDfC-SeNPs@DOX/siRNA nanoparticles was suspended in 1 mL of phosphate-buffered saline (PBS) in DEPC and transferred into a dialysis tube. Afterwards, the tubes were immersed in 49 mL of buffer solution at pH 5.4 or pH 7.4 under horizontal shaking (100 rpm). The released DOX was monitored for 30 h by measuring the fluorescence intensity of DOX at excitation/emission wavelengths of 535/590 nm. At predetermined time points, aliquots of 1 mL outside the dialysis tubes were withdrawn for fluorescence test and replenished with the equivalent volume of fresh PBS. The amount of payload released was determined by comparison with an experimentally determined standard curve. All the payload release assays were carried out in triplicate, and the results were expressed as the average data with standard deviation.
Cell lines and cell culture
HCC (HepG2) and human normal liver cell (Lo2) were purchased from ATCC (Manassas, VA, USA). All the cells were incubated in complete Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FBS at 37°C.
Cellular uptake of nanoparticles
For cellular uptake studies using the fluorescence microscope, HepG2 cells (5×104 cells/well) were incubated in 12-well cell culture plate overnight. Then, the previous medium was exchanged with fresh one containing RGDfC-SeNPs@DOX/siRNA (20 μg/mL), and the cells were incubated for different times (1, 2 and 4 h). Following incubation, the cells were gently washed with cold PBS, and the cell nucleus was stained with DAPI. The cellular uptake of nanoparticles in HepG2 cells was visualized by DOX and photographed by a Leica DMi8 fluorescence microscope.
To confirm whether the nanoparticles were located inside HepG2 cells, the cellular uptake was further investigated by TEM. Briefly, HepG2 cells (2×105 cells/mL) were seeded in complete DMEM at 37°C. After culturing with 20 μg/mL RGDfC-SeNPs or RGDfC-SeNPs@DOX/siRNA for 8 h, the cells were washed with cold PBS to eliminate RGDfC-SeNPs or RGDfC-SeNPs@DOX/siRNA that were not internalized into the cells. Then, the cells were harvested and fixed in the 2.5% glutaraldehyde solution. TEM samples were photographed at a 120 kV Tecnai TEM.
Cellular uptake mechanism of RGDfC-SeNPs@DOX/siRNA
To investigate the cellular uptake mechanism, HepG2 cells were incubated with various endocytosis inhibitors. Then, the cells were incubated with 20 μg/mL RGDfC-SeNPs@DOX/siRNA for 4 h at 4°C in the absence of inhibitors, or at 37°C in the presence of inhibitors. Following incubation, the RGDfC-SeNPs@DOX/siRNA in cells was measured by ICP-MS as previously reported.24
Nanog-siRNA transfection
HepG2 cells were cultured on a 12-well cell culture plate and allowed to reach 60% confluence. Then, the cells were incubated with PBS as control group, 100 nM naked FAM-siRNA, Lipofectamine 2000 containing 100 nM of Nanog-siRNA as the positive control and RGDfC-SeNPs@DOX/siRNA containing 100 nM Nanog-siRNA in DMEM for 24 h, respectively. The transfection efficiency was evaluated by a flow cytometer. The mRNA and protein expression levels of Nanog were evaluated via quantitative real-time PCR (qPCR) and Western blotting after 24 h treatment, respectively.
Quantitative real-time PCR
The total RNA of transfected cells was isolated according to the protocol of Total RNA Extraction Reagent (Takara, Beijing, People’s Republic of China). Analysis was carried out by a StepOne™ PCR System. The sequences of primers were as follows: forward 5′-CCGACTGTAAAGAATCTTCACCTATG-3′ and reverse 5′-TCA GGGCTGTCCTGAATAAGC-3′ for Nanog, and forward 5′-GAGTCCACTGGCGTCTTCA-3′ and reverse 5′-GGGGTGCTAAGCAGTTGGT-3′ for GAPDH (as the reference gene). Nanog gene expression level was analyzed using the formula of 2−(ΔΔCT) method.
Cell viability assay
The cytotoxicity of RGDfC-SeNPs/siNC (siNC represents negative control siRNA), naked siRNA, RGDfC-SeNPs/siRNA, Lipofectamine 2000/siRNA, free DOX, RGDfC-SeNPs@DOX and RGDfC-SeNPs@DOX/siRNA on cells was tested using MTT method as previously reported.25 Briefly, the cells were cultured in 96-well plates till they reached 50% confluence, and then treated with RGDfC-SeNPs/siNC, Lipofectamine 2000/siRNA and RGDfC-SeNPs/siRNA at different siRNA dose, or with free DOX, RGDfC-SeNPs@DOX and RGDfC-SeNPs@DOX/siRNA at different equivalent DOX or siRNA concentrations for 48 h. Then, 25 μL MTT (5 mg/mL) solutions were added to each well. After incubation for another 4 h, formazan crystals in each well were dissolved with 150 μL DMSO. The absorbing values were tested at 570 nm using a microplate reader (Bio-Rad).
Cell wound healing assay
HepG2 cells were seeded in 24-well plates and allowed to reach nearly 100% confluence. Linear wounds were made via scratching with a pipette tip, and the exfoliated cells were washed with PBS. Then, the previous medium was exchanged with fresh DMEM containing 2% FBS. After that, the cells were exposed to free DOX, RGDfC-SeNPs@DOX, RGDfC-SeNPs/siRNA and RGDfC-SeNPs@DOX/siRNA (at a concentration equivalent to 2 μg/mL of DOX and/or a concentration equivalent to 37 nM of siRNA) at 37°C. Cell movement into the wound area was monitored and photographed at 0 and 24 h using a light microscope. The average migration rates were calculated as following: cell motility (%) = [1 − (the wound width at 24 h/the wound width at 0 h)] ×100.
Cell invasion assay
The transwell chambers were used to assess cell invasion as reported previously.26 Briefly, cells were seeded in transwell chambers to reach 80% confluence. Then, the cells were exposed to free DOX, RGDfC-SeNPs@DOX, RGDfC-SeNPs/siRNA and RGDfC-SeNPs@DOX/siRNA (at a concentration equivalent to 2 μg/mL of DOX and/or a concentration equivalent to 37 nM of siRNA) for 24 h. Following exposure, the cells across the membrane were counted using a light microscope. Inhibition rates of cell migration were calculated according to the following equation: Inhibition of migration (%) = (Migctrl − Migt)/Migctrl × 100, in which Migctrl and Migt represent the control and treated cells from the lower surface of the membrane, respectively.
Cell apoptosis analysis
Cellular apoptosis in HepG2 cells was analyzed by flow cytometry through staining with PI or co-staining with the Annexin V-FITC/PI as following: Briefly, HepG2 cells were seeded in 12-well plates and allowed to reach 50% confluence, and then treated with the free DOX, RGDfC-SeNPs@DOX, RGDfC-SeNPs/siRNA and RGDfC-SeNPs@DOX/siRNA (at a concentration equivalent to 2 μg/mL of DOX and/or 37 nM of siRNA). The cells were harvested after 24 h of treatment, and stained with PI or Annexin V-FITC/PI in the dark for 30 min. The stained cells were immediately tested using an FACS flow cytometer (BD Biosciences, San Jose, CA, USA). The data were analyzed by a FlowJo software (Treestar, Ashland, OR, USA).
Western blotting assay
Western blotting experiments were performed as previously reported.22 The cells were treated with RGDfC-SeNPs@DOX/siRNA (at an equivalent DOX concentration of 1, 2 and 4 μg/mL) for 24 h, and then they were collected and lysed in 150 μL RIRP with 1% PMSF for 15 min, and centrifuged at 4,000 rpm for 15 min. The proteins were harvested, and the concentrations of proteins were measured using the BCA protein kit.
In vivo biodistribution of RGDfC-SeNPs@DOX/siRNA
HepG2 cells (1×107 cells) were subcutaneously injected into the abdomen of female BALB/c nude mice (5 weeks old). When the tumor grew to about 300 mm3, the cy5.5-loaded RGDfC-SeNPs@DOX/siRNA at a concentration equivalent to 2 mg/kg DOX and 0.25 mg/kg siRNA was injected into the mice via tail vein. Fluorescence imaging assay was carried out at 4 h post-injection, and the fluorescence signal was visualized using IVIS imaging system (Xenogen, Alameda, CA, USA). The exposure time was set as 10 s. After live imaging, mice treated with cy5.5-loaded RGDfC-SeNPs@DOX/siRNA were sacrificed. Then, tumor tissues and main organs were harvested to perform further fluorescence imaging.
Xenograft mouse model
The tumor model was established via subcutaneously injecting HepG2 cells (1×107 cells for each mouse) into the abdomen of mice. After the tumor grew to 100 mm3, the mice were subcutaneously injected with saline (as control group), free DOX, RGDfC-SeNPs@DOX, RGDfC-SeNPs/siRNA and RGDfC-SeNPs@DOX/siRNA (final concentration equivalent to 2 mg/kg DOX and/or 0.25 mg/kg siRNA) once every other day, respectively. The estimated tumor volume was calculated via the following formula:
|
Histology and immunohistochemistry
Tissues of tumor and main organs (heart, liver, spleen, lung and kidney) were fixed with 3.7% paraformaldehyde for 24 h at room temperature, processed into paraffin and sectioned at a thickness of 5 μm. After deparaffinization, the tissues were stained with hematoxylin and eosin (H&E) for histological analysis. The expression level of Ki67 protein associated with tumor growth was examined by immunohistochemical methods. For cancer cell apoptosis analysis, TUNEL apoptosis assay kit was used to detect the apoptosis of tumor cells. The apoptosis-related proteins in tumor tissues, including caspase-3 and pp53, were further measured by immunohistochemistry as previously reported. The main organs from each group were harvested, and then processed as described above. The tissue sections were examined by a digital microscope (Leica DMi8). All animal experiments were approved by the Ethics Committee of Guangzhou Medical University and performed according to the protocols and guidelines of the Experimental Animal Center of Guangzhou Medical University.
Statistical analysis
All experiments were carried out at least three times, and the data were expressed as mean ± SD. The statistical significance of differences was determined using SPSS statistical software package (SPSS Inc., Chicago, IL, USA). Statistical significance of data was analyzed using *p<0.05 or **p<0.01. The differences among different groups were analyzed using one-way ANOVA (multiple comparisons).
Results and discussion
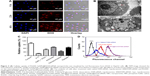
In the current study, a novel tumor-targeting co-delivery system RGDfC-SeNPs@DOX/siRNA was designed and synthesized, as illustrated in Scheme 1. In this co-delivery system, selenite was reduced to Se atoms by redox reaction after adding ascorbic acid and the accumulated Se atoms led to the formation of SeNPs. TEM images demonstrated that RGDfC-SeNPs@DOX nanoparticles presented spherical morphology with a size of about 12 nm, and the spherical SeNPs were spontaneously arranged in dendritic manner due to the interconnection between SeNPs and RGDfC via electrostatic interaction (Figure 1A and B). Nano ZS particle analyzer further confirmed the average size of RGDfC-SeNPs@DOX (Figure 1C). As shown in Figure 1D, the marker signals of the S atom (from the RGDfC molecule) and the Cl atom (from the DOX·HCl molecule) were observed during the elemental composition analysis of RGDfC-SeNPs@DOX, indicating RGDfC and DOX were successfully conjugated to the surface of SeNPs. The FTIR spectrums of SeNPs, RGDfC, DOX and RGDfC-SeNPs@DOX are shown in Figure 1E. The characteristic peaks of SeNPs were also observed in the spectrum of RGDfC-SeNPs@DOX. The peaks at 1,670 and 1,545 cm−1 were assigned to amide bands from RGDfC. After conjugating RGDfC with SeNPs, the obvious characteristic amide bands with a slight blue shift (1,655 and 1,543 cm−1) were also observed, indicating the successful conjugation of SeNPs with RGDfC by the amide bond. The peak between 3,500 and 3,250 cm−1 (blue pane) attributed to the large number of hydroxyl groups from DOX occurred in the spectrum of RGDfC-SeNPs@DOX, which sufficiently verified the successful loading of DOX onto the surface of RGDfC-SeNPs by the hydroxyl bond. The potentials of nanoparticles and siRNA are shown in Figure S1; the zeta potentials of SeNPs were changed from negative to positive after loading with RGDfC and DOX, suggesting that the electrostatic interaction might also contribute to the conjugation between the RGDfC or DOX and SeNPs. The positively charged nanoparticles would be beneficial to load the negatively charged siRNA by electrostatic interaction.
  | Scheme 1 Schematic illustration of the formation of RGDfC-SeNPs@DOX/siRNA. |
siRNA loading capability of nanoparticles and serum stability study
The agarose gel electrophoresis assay was carried out to research the loading capability of RGDfC-SeNPs@DOX as siRNA carrier. The migrations of RGDfC-SeNPs@DOX-bound siRNA were thoroughly blocked at the ratio of 8:1, compared to naked siRNA (Figure 2A), which indicated that RGDfC-SeNPs@DOX could strongly bind siRNA to resist dissociation during electrophoresis.
The RNAse in serum easily leads to complete degradation of siRNA, and thus, the increased stability of siRNA in the presence of serum is essential for improving the in vitro and in vivo transfection efficiency. In order to assess the ability of RGDfC-SeNPs@DOX to protect siRNA from serum-mediated degradation, the RGDfC-SeNPs@DOX/siRNA complexes were incubated in a medium containing 50% serum and assessed by a gel retardation assay. As shown in Figure S2, the naked siRNA was partially degraded after 1 h of incubation in 50% serum and almost totally degraded after 2 h of incubation. In contrast, the siRNA loaded onto the RGDfC-SeNPs@DOX was efficiently protected when incubated in the presence of 50% FBS during 2 h. This finding strongly suggests that the RGDfC-SeNPs@DOX is able to condense and protect the siRNA and delays the degradation by serum nucleases.
In vitro release of DOX
To evaluate the in vitro release behavior of RGDfC-SeNPs@DOX/siRNA, we investigated the release profiles in two kinds of buffer solutions (pH 5.4 or 7.4) at 37°C using the dialysis method. The two different pH values were used to simulate the normal physiological environment and the endosomal/lysosomal microenvironments of cancer cells which are more acidic (pH 4–6). The typical release profiles of DOX from the RGDfC-SeNPs@DOX/siRNA are shown in Figure 2B, and the data revealed that its accumulative release behavior was acid-dependent and that there was significant initial burst payload release during the initial 2 h. The release rates of DOX significantly increased when the pH values changed from 7.4 to 5.4. As indicated in Figure 2B, at pH 5.4, nearly 81.1% of DOX was released in a sustained manner during 30 h. However, at physiological pH (pH 7.4), DOX was released at a slower rate (51.8%). The faster release in acidic environment may be due to the decrease of the surface negative charge of the SeNPs in acidic pH, which weakens the electrostatic attraction of DOX and facilitates the release of DOX from the SeNPs.27 This pH-dependent DOX release property of the RGDfC-SeNPs@DOX/siRNA is quite beneficial for drug delivery systems in cancer therapy, because most tumors present a more acidic condition than normal tissues. Thus, the RGDfC-SeNPs have great potential in carrying and releasing therapeutics for treating cancers.
Cellular uptake of RGDfC-SeNPs@DOX/siRNA
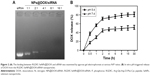
A fluorescence microscope was used to observe the cellular uptake of RGDfC-SeNPs@DOX/siRNA in HepG2 cells. As shown in Figure 3A, after 1 h of incubation with RGDfC-SeNPs@DOX/siRNA, weak red fluorescence was observed in the cytoplasm. The intensity of the fluorescence increased as the incubation time prolonged to 4 h. It indicated that RGDfC-SeNPs@DOX/siRNA could rapidly enter HepG2 cells and mainly stayed in the cytoplasm. TEM is another advisable tool to observe the intracellular distribution of nanoparticles. As shown in Figure 3B, TEM images showed that the cells exposed to the delivery carrier RGDfC-SeNPs or RGDfC-SeNPs@DOX/siRNA incorporated the nanoparticles in perinuclear compartments and vesicular structures (red arrows). Interestingly, we also observed that the in vitro treatment with RGDfC-SeNPs@DOX/siRNA in HepG2 cells resulted in the nuclear condensation, fragmentation and increase of apoptotic index. These TEM images confirmed the efficient cellular uptake and internalization of nanoparticles.
SeNPs have been reported to enter the cells via the energy-dependent endocytic pathways.28 Therefore, we researched whether temperature influenced the cellular entry process. The cellular uptake of cells incubated at 4°C decreased by ~82.7% (Figure 3C), indicating that the energy-dependent cellular uptake played a significant role in the internalization of RGDfC-SeNPs@DOX/siRNA. The cellular uptake significantly decreased after the incubation with NaN3/DOG, a cell energy metabolism inhibitor, confirming that the endocytosis of RGDfC-SeNPs@DOX/siRNA was an active and energy-dependent process. Furthermore, we utilized different endocytosis inhibitors to analyze the effect of RGDfC-SeNPs@DOX/siRNA on the cellular uptake. Chlorpromazine, amiloride and nystatin are known to inhibit clathrin-associated endocytosis, micropinocytosis and caveolae-mediated cellular uptake, respectively. The cellular uptake of nanoparticles pretreated with amiloride and nystatin was reduced by about 31.2% and 33.6%, respectively. However, the pretreatment with chlorpromazine led to 65.2% decrease in cellular uptake of nanoparticles, indicating that clathrin-associated endocytosis mainly contributed to the internalization of RGDfC-SeNPs@DOX/siRNA.
To precisely examine the transfection efficiency of siRNA in HepG2 cells, we incubated HepG2 cells with naked FAM-siRNA, RGDfC-SeNPs@DOX/FAM-siRNA and Lipofectamine 2000/FAM-siRNA for 24 h, and then analyzed the cells by FACS. As shown in Figure 3D, the cells incubated with naked FAM-siRNA showed minimal fluorescence intensity; however, strong fluorescence was observed in the cells cultured with RGDfC-SeNPs@DOX/FAM-siRNA. The fluorescence of cells treated with Lipofectamine 2000/FAM-siRNA was weaker than those treated with RGDfC-SeNPs@DOX/FAM-siRNA, suggesting that RGDfC-SeNPs@DOX/FAM-siRNA exhibited higher gene transfection efficiency, compared to Lipofectamine 2000/FAM-siRNA.
Downregulation of gene expression
Herein, we used the tumor-targeting functionalized SeNPs to deliver Nanog-siRNA into HepG2 cells and investigated the effect caused by Nanog knockdown on the HepG2 cells. After the cells were transfected with RGDfC-SeNPs@DOX/siRNA for 24 h, the level of Nanog mRNA was analyzed by qPCR. As shown in Figure 4A, RGDfC-SeNPs@DOX/siNC used as negative control group failed to silence Nanog expression, and the naked siRNA did not obviously affect the mRNA expression of Nanog, owing to its barrier to get across cell membranes and labile degradation by endogenous nucleases, whereas RGDfC-SeNPs@DOX/siRNA remarkably decreased the Nanog mRNA expression in HepG2 cells. RGDfC-SeNPs@DOX/siRNA resulted in 57.2% reduction in the level of Nanog mRNA, exhibiting stronger ability to silence the Nanog expression than that of Lipofectamine 2000/siRNA (31.2%).
Western blotting assay was used to determine the Nanog protein level in HepG2 cells. As shown in Figure 4B, the delivery of Nanog-siRNA via RGDfC-SeNPs@DOX significantly reduced the protein expression of Nanog, whereas RGDfC-SeNPs@DOX/siNC did not evidently influence the expression of Nanog, indicating no nonspecific gene silencing by this gene transfection system. The naked siRNA hardly affected the protein expression of Nanog. Meanwhile, the Lipofectamine 2000/siRNA as positive control also significantly inhibited the protein expression of Nanog. Both qPCR and Western blotting proved that RGDfC-SeNPs@DOX/siRNA was capable of silencing Nanog expression, which might control the cell fate via regulating the downstream effectors. The expression level of Nanog in normal cells and cancer cells was also examined by Western blotting assay. Figure S3 shows that the protein expression level of Nanog in HepG2 cells was remarkably higher than that in Lo2 cells, indicating Nanog-guided selectivity between Lo2 and HepG2 cells.
In vitro cytotoxicity study
MTT assay was employed to investigate in vitro anticancer activity of the nanoparticles. As shown in Figure S4, compared with untreated cells, viability of cells treated with drug carrier RGDfC-SeNPs up to a concentration of 8 μg/mL remained nearly 90% after 48 h incubation, indicating the good biocompatibility of RGDfC-SeNPs in HepG2 cells. The viability of cells treated with RGDfC-SeNPs/siNC remained about 94%, compared with untreated cells (Figure 5A), indicating no nonspecific cytotoxicity against HepG2 cells by this gene transfection system. The naked siRNA hardly exhibited cytotoxicity against HepG2 cells at the tested concentrations. RGDfC-SeNPs/siRNA exhibited a stronger tendency to dose-dependently inhibit the proliferation of HepG2 cells than Lipofectamine 2000/siRNA (Figure 5A), but was nontoxic on normal human liver Lo-2 cells (Figure S5). The results indicated that siRNA could be effectively delivered to HepG2 cells by tumor-targeting SeNPs and showed high cell proliferation inhibition activity against the HepG2 cells.
To evaluate the active targeting ability of the delivery system, the cytotoxicity of the three formulations of DOX against HepG2 cells was tested by MTT assay. As shown in Figure 5B, the cell viability of HepG2 cells treated with free DOX, RGDfC-SeNPs@DOX and RGDfC-SeNPs@DOX/siRNA gradually decreased with the increasing equivalent DOX concentrations. The cell-killing efficiency of the three formulations was ranked (from the most to least) as following: RGDfC-SeNPs@DOX/siRNA > RGDfC-SeNPs@DOX > free DOX. For example, the three formulations at an equivalent DOX concentration of 8 μg/mL significantly inhibited cancer cell proliferation, and the corresponding cell viabilities decreased to 20.5%, 38.6% and 53.3%, respectively. The RGDfC-SeNPs@DOX resulted in lower cell viability than free DOX, which was likely due to the enhanced cellular uptake of RGDfC-SeNPs@DOX nanoparticles. These results showed that the co-delivery of DOX and Nanog siRNA could efficaciously improve the chemotherapeutic efficacy of DOX, compared with delivery of DOX or siRNA alone.
Wound healing and transwell studies
The effects of different formulations of DOX on HepG2 cell migration and invasion were evaluated via wound healing and transwell assays, respectively. As shown in Figure 6A, the wound healing results revealed that RGDfC-SeNPs@DOX/siRNA significantly reduced the migration of HepG2 cells during 24 h interval (migration rate of control group, 26.8%; Figure 6B). Also, RGDfC-SeNPs@DOX, RGDfC-SeNPs/siRNA and free DOX reduced the rate of migration of the HepG2 cells compared to control cells. Similarly, HepG2 cells invasion was strongly reduced by RGDfC-SeNPs@DOX/siRNA (Figure 6C and D). Collectively, we conclude that RGDfC-SeNPs@DOX/siRNA has greater ability to suppress HepG2 cell migration and invasion through active targeting co-delivery of DOX and siRNA than delivery of DOX or siRNA alone.
Flow cytometry studies
Flow cytometry was utilized to analyze apoptotic cells with DNA fragmentation reflected as the sub-G1 peak. As shown in Figure 7A, cells treated with RGDfC-SeNPs@DOX/siRNA exhibited an obvious sub-G1 peak (65.06%), compared to the control group (4.84%). The sub-G1 apoptotic peak was stronger in the RGDfC-SeNPs@DOX/siRNA group than free DOX, RGDfC-SeNPs@DOX or RGDfC-SeNPs/siRNA at the equivalent DOX or siRNA dose, suggesting that RGDfC-SeNPs@DOX/siRNA exhibited greater ability to induce HepG2 cells apoptosis, while no significant differences was observed in cell cycle distribution.
Annexin-V/PI double staining is a more effective method for examining cell apoptosis.29 As shown in Figure 7B, RGDfC-SeNPs@DOX/siRNA treatment significantly induced HepG2 cells apoptosis, and led to higher cell apoptosis rate (early apoptosis: 27.5% and late apoptosis: 45.7%), compared to cells treated with free DOX (17.3% and 23.4%), RGDfC-SeNPs@DOX (22.4% and 35.8%) or RGDfC-SeNPs/siRNA (22.9% and 36.6%). This result was in agreement with the above one that RGDfC-SeNPs@DOX/siRNA significantly induced HepG2 cells apoptosis in vitro.
Western blotting analysis
In the present study, two proteins related to cell metastasis, MMP2 and MMP9,30 were observed to be decreased at protein levels in the RGDfC-SeNPs@DOX/siRNA treatment group (Figure 8), indicating RGDfC-SeNPs@DOX/siRNA might inhibit the HepG2 cell migration through downregulating the protein expression of MMP2 and MMP9.
Caspase-3 is thought to be the main executioner of cell apoptosis in the apoptosis-related signaling pathways, because it is responsible for the cleavage of many proteins.31 PARP is one of the significant cleavage targets of caspase-3, and is in the downstream of caspase family proteins.32 Herein, the protein expression levels of caspase-3 and PARP were tested by Western blotting to assess their contribution to apoptosis. Figure 8 shows that the treatment with RGDfC-SeNPs@DOX/siRNA resulted in the increased expression levels of caspase-3 and PARP, compared with the control group. The results indicated that RGDfC-SeNPs@DOX/siRNA significantly activated caspase-3 in apoptosis signaling pathway.
In vivo biodistribution of RGDfC-SeNPs@DOX/siRNA
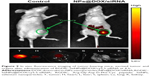
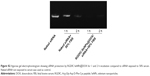
As an effective delivery platform for drugs, the carriers should have the ability to selectively deliver drugs to the tumor site for achieving the tumor-targeting effect. The biodistribution of RGDfC-SeNPs@DOX/siRNA nanoparticles was evaluated by monitoring the fluorescence of cy5.5-loaded RGDfC-SeNPs@DOX/siRNA at 4 h postinjection. As shown in Figure 9, no fluorescence signal was detected in the tumor site of blank group (no injection with cy5.5-loaded RGDfC-SeNPs@DOX/siRNA), indicating that there was no autofluorescence in the mice, and the fluorescence signal of imaging was close to the true reflection of cy5.5-loaded RGDfC-SeNPs@DOX/siRNA. However, the fluorescence signals were clearly accumulated in the tumor site after intravenous administration of cy5.5-loaded RGDfC-SeNPs@DOX/siRNA. Additionally, the tumor tissues and main organs were collected and photographed. As shown in Figure 9, the cy5.5-loaded RGDfC-SeNPs@DOX/siRNA mainly distributed in tumor, but weak fluorescence signal was detected in other organs. It is well known that nanoparticles smaller than 200 nm possess passive targeting ability in solid tumors through the enhanced permeation and retention (EPR) effect.33 In addition, RGDfC peptide, an extremely effective tumor-targeting moiety, can bind specifically to αvβ3 integrin receptors overexpressed on tumor cells.34 Thus, the cy5.5-loaded RGDfC-SeNPs@DOX/siRNA readily accumulated in the tumors owing to the combined contribution of the EPR effect and peptide–integrin specific binding.
In vivo antitumor efficacy
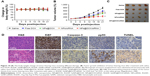
The antitumor efficacy of RGDfC-SeNPs@DOX/siRNA was further researched in the HepG2 tumor xenograft model. The mice were randomly assigned to five groups and then treated with saline, free DOX, RGDfC-SeNPs@DOX, RGDfC-SeNPs/siRNA and RGDfC-SeNPs@DOX/siRNA, respectively, to test the antitumor activity. The body weight and tumor volume were measured every other day up to 21 d. No obvious body weight loss was recorded after the various treatments, indicating the minimal side effect of the drugs at the tested doses (Figure 10A). As shown in Figure 10B, rapid tumor growth was observed in the mice treated with saline. Compared with the groups treated with saline, delivery of separate DOX or siRNA by tumor-targeting delivery carrier led to moderate inhibition of tumor growth. Meanwhile, free DOX showed relatively slight inhibition of tumor growth. Excitingly, the tumor-targeting co-delivery system RGDfC-SeNPs@DOX/siRNA showed the most effective antitumor activity, which attributed to the combined effect in treating cancer through the in vivo co-delivery of DOX and Nanog siRNA. The tumor image (Figure 10C) also indicated that RGDfC-SeNPs@DOX/siRNA showed remarkable antitumor activity.
Cell proliferation and apoptosis in the tumors were investigated by H&E staining, Ki67, caspase-3, pp53 and TUNEL assays after treatment with RGDfC-SeNPs@DOX/siRNA (Figure 10D). The H&E-stained section of tumor tissue in saline group appeared to be hypercellular, and the nuclear polymorphism in tumor tissue was very evident. In contrast, most of the cancer cells in RGDfC-SeNPs@DOX/siRNA treatment group were lysed and destroyed, indicating that the RGDfC-SeNPs@DOX/siRNA treatment had induced significant cancer cell apoptosis and necrosis in vivo. Meanwhile, RGDfC-SeNPs@DOX/siRNA treatment resulted in an obvious decrease of Ki67-positive cancer cells, suggesting that the cancer cell proliferation had been significantly suppressed by RGDfC-SeNPs@DOX/siRNA. As expected, caspase-3, pp53 and TUNEL assays confirmed that the treatment with RGDfC-SeNPs@DOX/siRNA led to enhanced apoptosis and cell death compared with the saline treatment groups, which was in agreement with the results obtained from H&E and Ki67 analysis. The results demonstrated that RGDfC-SeNPs@DOX/siRNA presented a great potential in cancer therapy through inhibiting the cancer cell proliferation and inducing the cancer cell apoptosis.
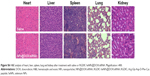
Apart from the antitumor efficacy, the in vivo toxicity assessment is crucial for chemotherapeutics.35 Histological analysis of main organs was performed by H&E staining. As shown in Figure S6, compared with the saline treatment group, no obvious difference was observed in the RGDfC-SeNPs@DOX/siRNA treatment group. The integrated results certified that RGDfC-SeNPs@DOX/siRNA nanoparticles were well tolerated in vivo at the tested dose. The low toxicity of RGDfC-SeNPs@DOX/siRNA may be attributed to the decreased drug distribution in normal tissues due to the improved tumor targeting of nanoparticles in mice. Analyzing these issues, we conclude that the RGDfC-SeNPs@DOX/siRNA nanoparticles have a great potential as a cancer-targeting co-delivery system for effective HCC therapy with a reduced systemic toxicity.
Conclusion
We successfully synthesized a novel type of tumor-targeting functionalized SeNPs RGDfC-SeNPs@DOX/siRNA for effective co-delivery of DOX and anti-Nanog siRNA to treat HCC. The RGDfC-SeNPs@DOX/siRNA showed excellent cellular uptake in HepG2 cells, resulting in significant gene silencing efficacy in vitro and in vivo. The RGDfC-SeNPs@DOX/siRNA could significantly inhibit HepG2 cells proliferation and migration, and induce cells apoptosis in vitro. Most importantly, RGDfC-SeNPs@DOX/siRNA exhibited stronger in vivo antitumor effectiveness compared to free DOX and single DOX- or siRNA-loaded RGDfC-SeNPs. The enhanced antitumor activity of RGDfC-SeNPs@DOX/siRNA might be attributed to the unique functions of RGDfC-SeNPs@DOX/siRNA in the specific simultaneous delivery of two drugs with different physicochemical properties to the same tumor cells. These findings indicate that the tumor-targeting co-delivery system RGDfC-SeNPs@DOX/siRNA holds enormous potential for combined drug/gene treatment of HCC.
Acknowledgments
This work was supported by the China Postdoctoral Science Foundation (No 2017M612632), the Technology Planning Project of Guangdong Province (No 2014A020212697) and the Technology Planning Project of Guangzhou City (No 201607010120). Yu Xia is particularly grateful to Qiumei Yang for providing inspiration in designing this paper.
Disclosure
The authors report no conflicts of interest in this work.
References
Shen JM, Li XX, Fan LL, et al. Heterogeneous dimer peptide-conjugated polylysine dendrimer-Fe3O4 composite as a novel nanoscale molecular probe for early diagnosis and therapy in hepatocellular carcinoma. Int J Nanomedicine. 2017;12(2):1183–1200. | ||
Ramasamy T, Ruttala HB, Gupta B, et al. Smart chemistry-based nanosized drug delivery systems for systemic applications: a comprehensive review. J Control Release. 2017;258(8):226–253. | ||
Lv Y, Li J, Chen H, Bai Y, Zhang L. Glycyrrhetinic acid-functionalized mesoporous silica nanoparticles as hepatocellular carcinoma-targeted drug carrier. Int J Nanomedicine. 2017;12(6):4361–4370. | ||
Sun L, Zheng C, Webster T. Self-assembled peptide nanomaterials for biomedical applications: promises and pitfalls. Int J Nanomedicine. 2016;12(12):73–86. | ||
Lin X, Yang S, Lai K, Yang H, Webster TJ, Yang L. Orthopedic implant biomaterials with both osteogenic and anti-infection capacities and associated in vivo evaluation methods. Nanomedicine. 2017;13(1):123–142. | ||
Kevadiya BD, Bade AN, Woldstad C, et al. Development of europium doped core-shell silica cobalt ferrite functionalized nanoparticles for magnetic resonance imaging. Acta Biomater. 2017;49(2):507–520. | ||
Zhao S, Yu Q, Pan J, et al. Redox-responsive mesoporous selenium delivery of doxorubicin targets MCF-7 cells and synergistically enhances its anti-tumor activity. Acta Biomater. 2017;54(5):294–306. | ||
Xia Y, Lin Z, Li Y, et al. Targeted delivery of siRNA using RGDfC-conjugated functionalized selenium nanoparticles for anticancer therapy. J Mater Chem B. 2017;5(33):6941–6952. | ||
Li Y, Lin R, Wang L, et al. PEG-b-AGE polymer coated magnetic nanoparticle probes with facile functionalization and anti-fouling properties for reducing non-specific uptake and improving biomarker targeting. J Mater Chem B. 2015;3(17):3591–3603. | ||
Tang J, Huang N, Zhang X, et al. Aptamer-conjugated PEGylated quantum dots targeting epidermal growth factor receptor variant III for fluorescence imaging of glioma. Int J Nanomedicine. 2017;12(5):3899–3911. | ||
Nie X, Zhang J, Xu Q, et al. Targeting peptide iRGD-conjugated amphiphilic chitosan-co-PLA/DPPE drug delivery system for enhanced tumor therapy. J Mater Chem B. 2014;2(21):3232–3242. | ||
Zhang H, Li W, Guo X, et al. Specifically increased paclitaxel release in tumor and synergetic therapy by a hyaluronic acid-tocopherol nanomicelle. ACS Appl Mater Interfaces. 2017;9(24):20385–20398. | ||
McMahon KM, Plebanek MP, Thaxton CS. Properties of native high-density lipoproteins inspire synthesis of actively targeted in vivo siRNA delivery vehicles. Adv Funct Mater. 2016;26(43):7824–7835. | ||
Zheng N, Song Z, Yang J, et al. Manipulating the membrane penetration mechanism of helical polypeptides via aromatic modification for efficient gene delivery. Acta Biomater. 2017;58(8):146–157. | ||
Luo X, Peng X, Hou J, Wu S, Shen J, Wang L. Folic acid-functionalized polyethylenimine superparamagnetic iron oxide nanoparticles as theranostic agents for magnetic resonance imaging and PD-L1 siRNA delivery for gastric cancer. Int J Nanomedicine. 2017;12(7):5331–5343. | ||
Ni Q, Teng Z, Dang M, et al. Gold nanorod embedded large-pore mesoporous organosilica nanospheres for gene and photothermal cooperative therapy of triple negative breast cancer. Nanoscale. 2017;9(4):1466–1474. | ||
Sultana R, Kataki AC, Borthakur BB, Basumatary TK, Bose S. Imbalance in leptin-adiponectin levels and leptin receptor expression as chief contributors to triple negative breast cancer progression in Northeast India. Gene. 2017;621:51–58. | ||
Park SW, Do HJ, Choi W, Song H, Chung HJ, Kim JH. NANOG gene expression is regulated by the ETS transcription factor ETV4 in human embryonic carcinoma NCCIT cells. Biochem Biophys Res Commun. 2017;487(3):532–538. | ||
Guo W, Liu S, Cheng Y, et al. ICAM-1-related noncoding RNA in cancer stem cells maintains ICAM-1 expression in hepatocellular carcinoma. Clin Cancer Res. 2016;22(8):2041–2050. | ||
Zheng W, Yin T, Chen Q, et al. Co-delivery of Se nanoparticles and pooled SiRNAs for overcoming drug resistance mediated by P-glycoprotein and class III beta-tubulin in drug-resistant breast cancers. Acta Biomater. 2016;31(2):197–210. | ||
Chen Q, Yu Q, Liu Y, et al. Multifunctional selenium nanoparticles: chiral selectivity of delivering MDR-siRNA for reversal of multidrug resistance and real-time biofluorescence imaging. Nanomedicine. 2015;11(7):1773–1784. | ||
Wang Y, Yang C, Hu R, et al. Assembling Mn:ZnSe quantum dots-siRNA nanoplexes for gene silencing in tumor cells. Biomater Sci. 2015;3(1):192–202. | ||
Jones SK, Sarkar A, Feldmann DP, Hoffmann P, Merkel OM. Revisiting the value of competition assays in folate receptor-mediated drug delivery. Biomaterials. 2017;138(9):35–45. | ||
Xu L, Yeudall WA, Yang H. Folic acid-decorated polyamidoamine dendrimer exhibits high tumor uptake and sustained highly localized retention in solid tumors: its utility for local siRNA delivery. Acta Biomater. 2017;57(7):251–261. | ||
Xia Y, Chen Q, Qin X, Sun D, Zhang J, Liu J. Studies of ruthenium(ii)-2,2′-bisimidazole complexes on binding to G-quadruplex DNA and inducing apoptosis in HeLa cells. New J Chem. 2013;37(11):3706–3715. | ||
Xia Y, You P, Xu F, Liu J, Xing F. Novel functionalized selenium nanoparticles for enhanced anti-hepatocarcinoma activity in vitro. Nanoscale Res Lett. 2015;10(1):349–363. | ||
Qiu L, Hu Q, Cheng L, et al. cRGDyK modified pH responsive nanoparticles for specific intracellular delivery of doxorubicin. Acta Biomater. 2016;30(1):285–298. | ||
Zhou Y, Yu Q, Qin X, et al. Improving the anticancer efficacy of laminin receptor-specific therapeutic ruthenium nanoparticles (RuBB-loaded EGCG-RuNPs) via ROS-dependent apoptosis in SMMC-7721 cells. ACS Appl Mater Interfaces. 2016;8(24):15000–15012. | ||
Li Y, Li X, Zheng W, Fan C, Zhang Y, Chen T. Functionalized selenium nanoparticles with nephroprotective activity, the important roles of ROS-mediated signaling pathways. J Mater Chem B. 2013;1(46):6365–6372. | ||
Rodriguez-Yanez Y, Bahena-Uribe D, Chavez-Munguia B, et al. Commercial single-walled carbon nanotubes effects in fibrinolysis of human umbilical vein endothelial cells. Toxicol In Vitro. 2015;29(5):1201–1214. | ||
Choi JY, Ramasamy T, Tran TH, et al. Systemic delivery of axitinib with nanohybrid liposomal nanoparticles inhibits hypoxic tumor growth. J Mater Chem B. 2015;3(3):408–416. | ||
Zhou Y, Xu M, Liu Y, et al. Green synthesis of Se/Ru alloy nanoparticles using gallic acid and evaluation of theiranti-invasive effects in HeLa cells. Colloid Surf B Biointerfaces. 2016;144(8):118–124. | ||
Huo ZJ, Wang SJ, Wang ZQ, et al. Novel nanosystem to enhance the antitumor activity of lapatinib in breast cancer treatment: therapeutic efficacy evaluation. Cancer Sci. 2015;106(10):1429–1437. | ||
Wang F, Gao L, Meng LY, Xie JM, Xiong JW, Luo Y. A neutralized noncharged polyethylenimine-based system for efficient delivery of siRNA into heart without toxicity. ACS Appl Mater Interfaces. 2016;8(49):33529–33538. | ||
Hong J, Sun Z, Li Y, et al. Folate-modified annonaceous acetogenins nanosuspensions and their improved antitumor efficacy. Int J Nanomedicine. 2017;12(7):5053–5067. |
Supplementary materials
  | Figure S3 The protein expressions of Nanog in HepG2 and Lo-2 cells. |
  | Figure S4 The in vitro cytotoxicity of RGDfC-SeNPs on HepG2 cells. |
  | Figure S5 In vitro cytotoxicity of RGDfC-SeNPs/siRNA on Lo2 cells. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.