Back to Journals » Drug Design, Development and Therapy » Volume 15
Novel Anti-Tubulin Compounds from Trigonella foenum-graecum Seeds; Insights into In-vitro and Molecular Docking Studies
Authors Alghamdi SS , Suliman RS , Alsaeed AS, Almutairi KK, Aljammaz NA, Altolayyan A, Ali R , Alhallaj A
Received 18 May 2021
Accepted for publication 22 July 2021
Published 5 October 2021 Volume 2021:15 Pages 4195—4211
DOI https://doi.org/10.2147/DDDT.S320793
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianbo Sun
Sahar Saleh Alghamdi,1,2 Rasha Saad Suliman,1,2 Amjad Sulaiman Alsaeed,1 Khlood Khaled Almutairi,1 Norah Abdulaziz Aljammaz,1 Abdulelah Altolayyan,2 Rizwan Ali,2 Alshaimaa Alhallaj2
1College of Pharmacy, King Saud bin Abdulaziz University for Health Sciences, Riyadh, Kingdom of Saudi Arabia; 2Medical Research Core Facility and Platforms, King Abdullah International Medical Research Center (KAIMRC), Ministry of National Guard Health Affairs, Riyadh, Kingdom of Saudi Arabia
Correspondence: Sahar Saleh Alghamdi Tel +966 114299999 ext 99516
Email [email protected]
Background: Fenugreek, also known as Trigonella foenum-graecum L, is a natural plant that belongs to the Fabaceae family and has been known as a promising source of bioactive compounds. It has been widely used as traditional medicine since it has shown to lower blood glucose, manage cholesterol levels and further aid in the prevention and treatment of cancer. Herein, we aim to evaluate the anticancer activity of methanolic fenugreek seed extract against several cancer cell lines.
Methods: We sought to investigate the phytochemical classes present in multiple fenugreek seeds extracts using HPLC-DAD followed by LC/MS, predict and investigate anticancer activity using PASS online webserver, the CellTiter-Glo assay, evaluate ADME properties, and perform molecular docking for all bioactive compounds via Maestro software.
Results: Multiple extracts exhibited distinct phytochemical classes that demonstrated different biological activities. Fenugreek methanolic extract contains flavonoid chemical class, which showed the highest anticancer activity against the HCT8 cell line of colorectal cancer (IC50 of 8.83 μg/mL), followed by KAIMRC1 breast cancer cell line (IC50 of 35.06 μg/mL), HL60 leukemia cell line (37.80 μg/mL), MDA-MB-231 breast cancer cell line (38.51 μg/mL), and lastly, HCT116 colorectal cancer cell line with IC50 of 56.03 μg/mL. In contrast, the chloroform extract was inactive. The molecular docking study for all the bioactive compounds suggested that flavonoids F6 (− 9.713 and − 12.132), F7 (− 10.166 and − 12.411), and F11 (− 10.084 and − 13.516) possess the highest docking scores through SP and XP scores, respectively.
Conclusion: The obtained results confirm that the bioactive compounds present in fenugreek seeds exhibit anticancer activity against several cancer cells that can mediate via tubulin polymerization inhibition. Although our study has evaluated the anticancer potential of Trigonella foenum-graecum as a promising natural source for new anticancer agents, fenugreek biological activity needs further research and investigations on their mechanism of action and toxicity profile.
Keywords: fenugreek Trigonella foenum-graecum, tubulin inhibitors, computational pharmacokinetics ADME, QTOF-LC/Ms metabolites
Graphical Abstract:
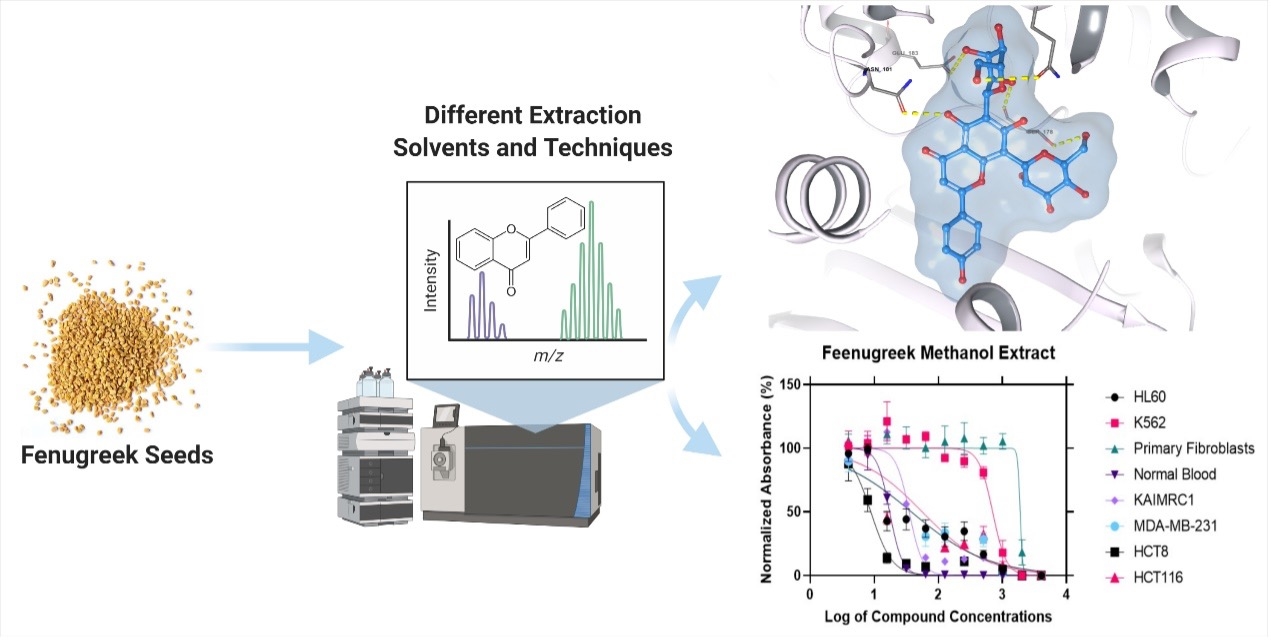
Introduction
Cancer is recognized as a significant public health concern and is expected to rise as a result of changes in human lifestyles.1 Despite the variety of anticancer agents available nowadays, these agents only provide limited therapeutic benefits due to undesirable side effects and the development of chemoresistance.2 In the last few years, researchers have emphasized the need to discover novel chemotherapeutic drugs from natural resources, as the usage of herbs has provided a new therapeutic approach in a variety of diseases. Moreover, it has been reported that the general community is using herbal products to complement or replace modern therapeutic medicine due to their potential safety profile and promising activity.3–6
Fenugreek, known as Trigonella foenum-graecum, or Hilbeh in the Middle East, belongs to the Fabaceae family.7 The extract of fenugreek contains numerous phytochemical classes, including flavonoids, which are recognized as antioxidants that decrease oxidative stress.8 In addition, phytosterol diosgenin, a steroidal saponin with potential biological activities such as glucose preservation and lipid homeostasis, has a cytotoxic effect on lung cancer cells and an inhibitory effect of the hTERT gene. It also exhibits a potential anticarcinogenic activity toward breast cancer tissues due to its strong antioxidant effect.9–12 Moreover, the extract includes the soluble fiber galactomannan, which enhances blood glucose and lowers serum lipid in diabetic animal models, besides 4-Hydroxyisoleucine that stimulates insulin secretion and that has insulin-mimetic properties.13,14
Hence, a wide variety of fenugreek seeds medical applications are well known, including dysmenorrhea, alleviate painful uterine contractions during menstruation, diabetes, wound healing, and stimulation of milk production.15 Furthermore, several in-vivo and in-vitro studies have reported the promising anticancer activity of fenugreek extracts with the advantage of selective toxicity to cancer cells.16
Chatterjee et al studied the Trigonella foenum-graecum seeds extract on skin papilloma tumors. The authors found a remarkable reduction in tumor incidence compared to a control group by inhibiting the tumour necrosis factor (TNF) gene expression.17 Likewise, Trigonella foenum-graecum crude methanol extracts caused apoptosis induction in hepatocellular carcinoma (HepG2) using MTT assay.18 Another study examined the anticancer activities of fenugreek seed extract on MCF-7 breast cancer, HCAM liver cancer, and non-cancerous Vero cell lines and revealed that these seeds diminished cancer cell proliferation within the MCF-7 cell line. Unfortunately, no apoptosis or necrosis was observed.19
In a recent study, T. foenum-graecum extracts were tested on cancer cell lines, including Hep2, MCF7, and normal cell line HCEC. The methanolic extract showed considerable cytotoxicity to Hep2 and MCF7 cell lines, with IC50 values ranging from 2.85 to 3.14 g/L, which was higher than the cytotoxicity of reference drugs Paclitaxel and Tamoxifen. Importantly, these extracts demonstrated no cytotoxicity against normal cells, indicating that they were selectively cytotoxic to cancer cells.20
Microtubule, a protein involved in the critical functions of several cells in the human body, has been proven to be a target in the development of novel anticancer agents.21 Microtubules are formed when α- and β-tubulin heterodimers polymerize to build a tube-shaped structure, which is then depolymerized in a dynamically balanced cycle.22 Since various isotypes of tubulin are expressed in cancerous cells, inhibiting these microtubules can interfere with neoplastic cell cycles and induce apoptosis. Indeed, multiple anti-tubulin agents are naturally derived from plants, such as Taxanes and Vinca Alkaloids. These agents impair cell division by disrupting the equilibrium state of microtubules via inhibiting polymerization or the depolymerization process.23 Similarly, Nagireddy et al found that Noscapine analogs, derived from the Papaver somniferum, can inhibit tubulin protein and were considered a potential chemotherapeutic candidate for the treatment of pancreatic cancer. Furthermore, the presence of bromine in compounds can suppress cell growth at the G2/M phase of MCF-7 cells and induce apoptosis.24
Thus, the main aim of this study is to obtain an appropriate extraction method of fenugreek seeds and to evaluate the anticancer activity against various cancer cell lines. We also predict the pharmacokinetic properties and potential targets for the bioactive constituents that mediate the anticancer activity of fenugreek seed extract.
Materials and Methods
Fenugreek Seed Extraction Protocols
Seeds Collection
Seeds were purchased from a certified company called Mountain Rose Herbs. The purchased seeds are carefully selected to be organic, without additives, and free of preservatives. Moreover, the seeds undergo more than 10 quality control tests. The seeds Lot number is 24330-R. The seeds have an authentication certificate and certificate of analysis. The local herbs shops were avoided because of the high chance of adulterants.
Seeds Extraction
First, we followed the standard method of fenugreek extraction using the Soxhlet apparatus. 250 g of the seeds was grounded by a blender to semi-grounded seeds. An Erlenmeyer of 500 mL was used to weigh 75 g of the grounded seeds. The seeds were placed in 3-filter tube each contains 75 g of grounded seeds to utilize it in the Soxhlet apparatus. Methanol was used as an extraction solvent, the volume was used as recommended by the manufacturer, and a 50-mL volume of solvent was introduced into the extractor thimble from the top to facilitate flushing of the extractor chamber. After the extraction process has been done, the extraction was dried by rotary evaporator apparatus to get a powder form of the extraction. Further extraction has been done using other extract solvents by the maceration method. A 100 g of powder fenugreek was soaked with 1 liter of each solvent as follows: chloroform, water, diethyl ether, and methanol. The mixture was kept soaked under a magnetic stirrer hot plate at 40°C for 48 h. Then, the extract was collected and filtered using filter paper and bacterial filter. A rotary evaporator was used for the drying process, and all the extracts were stored in a 4°C refrigerator to be further used.
Chemical Identification
The extract obtained above was subjected to HPLC-DAD followed by LC/MS using the following protocol.
Chemicals
Formic acid and HPLC grade methanol were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Honeywell (France), respectively.
RP-HPLC Method
All extracts were first injected into the Agilent 1260 Infinity HPLC system (Agilent, Germany) with Diode-Array Detection (DAD) detector. The separation carried out in a reversed-phase mode using Phenomenex Kinetex-C18 column (4.6 mm × 250 mm, 5 μm) with the following elution gradient methanol to ultrapure water; 0–1 min, 5% B; 1–11 min, 5–100% B; 11–13 min, 95% B; 13–15 min, 5% B; 15–16 min, 5% B using mobile phase A (0.1% HCOOH in water) and mobile phase B (0.1% HCOOH in Methanol). Samples were injected with 10 µL, with 16 min run time, and the flow rate was set as 1 mL/min. The DAD collected UV spectrum at 200, 225, 250, 275, 300, 325, and 350. The data were processed using ChemStation software.
LC-QTOF-MS Method
The analysis of all extracts was performed on Agilent 1260 Infinity HPLC system (Agilent, Germany) coupled to Agilent 6530 Quadrupole Time of Flight (Agilent, Singapore). Separation was performed using Agilent Extend-C18 column (2.1 mm × 50 mm, 1.8 μm) with the following elution gradient; 0–1 min, 5% B; 1–11 min, 5–100% B; 11–13 min, 95% B; 13–15 min, 5% B; 15–16 min, 5% B using mobile phase A (0.1% HCOOH in water) and mobile phase B (0.1% HCOOH in Methanol). The injection volume was 10 µL, and the flow rate was set as 300 µL/min. The MS1acquisition method was achieved in positive mode with mass ranged from 100 to 600 m/z. The mass spectrometer parameters were set as follows: Gas Temperature = 300°C; Gas flow = 8 I/min; Nebulizer = 35 psig; SheathGas Temperature = 350 and SheathGas flow was 11. MS1 data was generated using the Agilent MassHunter qualitative analysis software.
In-vitro Anticancer Activity
The anticancer biological experiment was conducted at King Abdullah International Medical Research Center (KAIMRC), Riyadh, Saudi Arabia. To determine the effect of the fenugreek extracts on cell proliferation, MCF-7, MDA MB-231, KAIMRC1 (breast cancer cell lines), HCT8, and HCT116 (colorectal cancer cell lines) were used. All the cell lines were purchased from ATCC, USA, except KAIMRC1, which was isolated and established in the core laboratory facility KAIMRC, Riyadh KSA.25 All the cell lines were maintained in advanced DMEM containing 10% FBS, 1% L-glutamine, and 1% antibiotics (Pen-Strep). Cells were plated on flat-bottom white 96-well plates at a density of 5×103 cells/well in a 100 μL growth medium. Serial dilutions of the extracts, ranging from 100 to 0.01 μg/100µL cell culture media were made in triplicates and transferred to the cell culture plates containing the cells. Additional rows with only cells were added to account for the compounds and cells’ effect. Cells were incubated for 48 h at 37°C with 5% CO2. Cell viability was determined using the CellTiter-Glo assay (Promega) according to the manufacturer’s recommendations. Luminescence was measured using the EnVision plate reader (Perkin Elmer), and readings were normalized to averaged DMSO controls and expressed as a relative percentage. Mitoxantrone was used as a positive control, data were analyzed with GraphPad Prism 8.1 software, and the half-maximal inhibitory concentration (IC50) was determined.
ADME Using SwissADME Webserver
The calculations of the absorption, distribution, metabolism, and elimination (ADME) properties of the fenugreek active compounds were performed using the SwissADME web server.26 The SMILES of each active compound were used as input to generate the ADME data, and the results were then compared and analyzed according to the reported literature values.
Prediction of Biological Activity Using PASS Online
The flavonoid-derived compounds from fenugreek methanolic extract were analyzed using the online service Prediction of Activity Spectra for Substances (PASS) to determine the biological probability of a substance being active (Pa) or inactive (Pi).27
Molecular Docking Study on Tubulin
Protein and Ligands Preparation
The 3D crystal structure of Tubulin (PDB: 4O2B) was obtained from the RCSB protein data bank, and the structure was prepared using the Protein Preparation Wizard tool (PrepWizard, Schrödinger Release 2021-1). The 3D Tubulin structure was refined, minimized, and optimized with the OPLS4 force field. Unnecessary water molecules, substrates, ions, and other subunits were removed and the β-Tubulin subunit that contains the Colchicine binding site was maintained for the docking study. The 2D structures for ligands (F1-11, S1-3, D1-4, and Colchicine) were prepared using Schrödinger’s LigPrep tool, and several conformations were generated, optimized, and minimized for their lowest energy conformation.
Ligand-Protein Molecular Docking
A one-step docking tool was utilized to validate the docking modes, and the root-mean-square deviation (RMSD) values were below 0.366 Å for both standard precision (SP) and extra precision (XP) scoring functions of GLIDE. All the compounds were docking using SP and XP scoring functions, and post-docking analysis for the docked poses was performed. Moreover, the energies or the relative binding affinity for the best docking (complex) structures were carried out using Prime MM-GBSA in which VSGB was used as a solvation model and OPLS4 force field.
QikProp for Absorption, Distribution, Metabolism, and Excretion (ADME) Predictions
For all the ligands, the ADME parameters were calculated and computed using the QikProp tool, Schrödinger, LLC, New York, NY, 2021. More than twenty physical descriptors were generated, and the most important parameters for drug development were chosen for further analysis.
Results
Chemical Analysis and Compound Identifications in the LC/MS Based on the Solvent Used
Soxhlet Methanolic Extract
The extracted yield obtained was as follows: Soxhlet methanolic extract 68% of the started weight, chloroform extract 45%, water extract 73%, diethyl ether extract 67%, and methanol extract 84%. The obtained extracts were injected into LC/Ms, and the peaks total ion chromatogram, which shows high abundancy, was examined as shown in Figure 1. QTOF-LCMS is one of the recent methods that could be used to identify metabolites, since we are handling the same seed that was analysed previously and identified, and their chemical structure was illustrated using NMR and chromatography techniques. It is valid to tentatively map the specific mass value with the previously reported compounds isolated from the same seed.
 |
Figure 1 The LC/MS spectrum for soxhlet methanolic extract. |
Chloroform Extract
The obtained chloroform extract was injected into LC/Ms, and the peaks total ion chromatogram, which shows high abundancy, was examined as shown in Figure 2.
 |
Figure 2 The LC/MS spectrum for chloroform extract. |
Water Extract
The obtained water extract was injected into LC/Ms, and the peaks total ion chromatogram, which shows high abundancy, was examined as shown in Figure 3.
 |
Figure 3 The LC/MS spectrum for water extract. |
Diethyl Ether Extract
The obtained water extract was injected into LC/Ms, and the peaks total ion chromatogram, which shows high abundancy, was examined as shown in (Figure 4).
 |
Figure 4 The LC/MS spectrum for diethyl ether extract. |
Predicted Biological Activity of Bioactive Compounds Using PASS Online
The metabolic profiling results showed that the fenugreek seed extracts exhibited different phytochemical classes depending on the solvent used. Our results showed that methanol extract was rich in flavonoids, while the chloroform extract was rich with Phytosterol Diosgenin (Spirostan) derivatives. The Prediction of Activity Spectra for Substances (PASS) online service was used to assess the potential pharmacological activities of all the three different phytochemical classes that are present in different extracts.27 As a result, compounds F6, F7, and F11 exhibited the highest probability of being active as anticarcinogenic and antineoplastic relative to the other compounds present in fenugreek extracts, as summarized in Table 1.
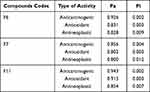 |
Table 1 Biological Activity Predictions Using PASS Online Webserver |
In-vitro AntiCancer Activity of Fenugreek Extracts Against Several Cancer Cell Lines
To confirm the predicted anticancer activity of fenugreek phytochemical compounds, the methanol and chloroform fenugreek extracts were examined against Leukemia cancer cell lines (HL60 and K562), breast cancer cell lines (KAIMRC1 and MDA-MB-231), colorectal cancer cell lines (HCT8 and HCT116), and also against normal primary fibroblasts P1 and normal blood cells N1 to determine the selectivity of these chemical classes. Methanol extract exhibited a promising anticancer activity on all the cell lines except the K562 Leukemia cell, as plotted in Figure 5. The highest inhibitory activity of the methanol extract was against the HCT8 cell line of colorectal cancer with an IC50 of 8.83 μg/mL, followed by KAIMRC1 breast cancer cell line, HL60 leukemia cell line, MDA-MB-231 breast cancer cell line, and lastly HCT116 colorectal cancer cell line, with IC50 of 35.06, 37.80, 38.51, and 56.03 μg/mL, respectively (presented in Table 2). On the contrary, the chloroform extract was inactive on all cell lines, as seen in Figure 6.
 |
Table 2 The IC50 Values of Fenugreek Extracts Against Six Cancer Cell Lines |
 |
Figure 5 The inhibitory activity of fenugreek methanol extract against six cancer cell lines. |
 |
Figure 6 The inhibitory activity of fenugreek chloroform extract against six cancer cell lines. |
ADME Properties of Fenugreek Extracts Bioactive Compounds
The selection of the ADME properties that were assessed in this study was based on Lipinski’s rule of five (ROF) that estimates the drug-likeness properties of tested compounds.28,29 The pharmacokinetic properties of flavonoid derivatives (F1-F11) were within the acceptable range <500 Daltons in the molecular weight, except for F9 and F10 that were out of range, suggesting the bulkiness of these two compounds (Table 3, Figure 7).28 Flavonoid derivatives F6 to F11 showed greater than 5 in the number of H-bond donors, whereas F9 to F11 were out of range in the number of H-bond acceptors, which consequently would affect the blood-brain barrier (BBB) permeability and GI absorption. Moreover, all the flavonoid derivatives (F1-F11) showed low lipophilicity and high solubility profile that affected the BBB penetration and oral activity. Furthermore, it was predicted that F1 to F8 could potentially have oral activities; however, F9 to F11 are orally inactive due to violation of two or more of Lipinski’s rules.
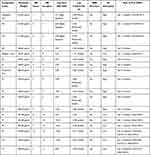 |
Table 3 The Pharmacokinetic ADME Properties of Fenugreek Bioactive Constituents |
 |
Figure 7 Log Po/w and LogS of fenugreek phytochemical classes. |
The pharmacokinetic properties of diosgenin derivatives were computed (Table 3, Figure 7), and all the four derivatives were within the acceptable range <500 Daltons in the molecular weight and hydrogen bond donors and acceptors requirements. The computed lipophilicity (Log Po/w) and solubility (LogS) parameters of diosgenin derivatives were higher compared to the optimal range except for D4, suggesting these compounds D1 to D3 are highly lipophilic and thus poorly soluble.30,31 Additionally, most of the compounds were permeable to the BBB except for D2, and all the four derivatives demonstrated high GI absorption suggesting these diosgenin derivatives could be orally active, with no violation of two or more of Lipinski’s rules.
Moreover, the ADME predictions (Table 3, Figure 7) of polyphenol stilbenes derivatives were within the acceptable range <500 Daltons in the molecular weight and hydrogen bond donors and acceptors except for S1 that demonstrated H-bond donors of more than 5. All three derivatives showed good lipophilicity (LogPo/w) and solubility (LogS) values that were within the recommended range. Also, ADME results showed that S3 could penetrate the BBB, and this might be due to the balance between the solubility and lipophilicity values; however, the other two stilbenes S1 and S2 derivatives could not penetrate BBB due to their low lipophilicity and high solubility.32 Additionally, S2 and S3 showed high GI absorption, whereas S1 showed low GI absorption that correlates with H-bond donor ability that affects the absorption. Overall, all the Stilbenes derivatives were predicted to be orally active due to no violation of two or more of Lipinski’s rules.
Docking of Fenugreek Bioactive Chemical Classes into Tubulin Crystal Structure
In this study, we sought to determine the potential mechanism that mediates the anticancer activity of the fenugreek bioactive constituents. The colchicine (reference ligand) docking score with SP (−11.572) method was better compared to the XP scoring (−7.707) function and a similar observation was found with MM/GBSA binding energy in which SP (−88.10 Kcal mol−1) was significantly higher compared to XP (−20.81 Kcal mol−1) docking (Table 4). From the docked compounds, the best ligand was F7 that demonstrated a value of −10.166 and −12.411 through SP and XP docking methods, respectively. It is worth mentioning that ligands demonstrated higher docking scores with XP scoring function; however, this was not the case with the reference ligand (colchicine). Moreover, ligand F11 exhibited the third-best docking scores with values of −10.084 for SP and −13.516 for XP docking methods. Ligand F6 exhibited the fourth-best docking values with −9.713 and −12.132, respectively, through SP and XP scoring functions. In the case of MM/GBSA binding energy, the order of the ligands is different, as ligand F6 exhibits the highest binding energy, while F11 demonstrates the lowest with both SP and XP scoring methods. Of note, all the ligands were docked into β-Tubulin using SP and XP; however, our results showed that the flavonoid chemical class exhibit the highest docking scores with tubulin confirming the above-discussed results.
 |
Table 4 SP and XP Glide Scores for the Best Docked Compounds/Protein Complexes with MM/GBSA Binding Energy Calculations |
The post-docking analysis of the ligands with SP docking function exhibited several valuable molecular interactions. For instance, colchicine exhibits multiple hydrogen bond (H-bond) interactions with residues Cys241, Val238, Val181, and Thr353 (Figure 8). While XP docking interaction demonstrates less interaction while maintaining only one H-bond with Cys241 (Figure 9) that could explain the lower docking score compared to SP, and a similar trend was observed with the docked ligands. Moreover, multiple H-bonds interactions were demonstrated with the SP best docking pose of ligand F6 with Asn249, Gln11, Glu183, Asn101, and Thr179. Using XP docking, ligand F6 demonstrated additional two H-bonds interactions with Gln247 and Tyr224 residues. The highest SP docking score was attained by ligand F7 that showed H-bonds interactions with Asn101, Ser178, Lys254, Gln11, Gln247, and Val315. Using the XP scoring function, ligand F7 exhibited additional H-bonds with residues Cys241, Val238, Asn101, Asn249, Gln11, Gln247, and Glu183. Additionally, the H-bonds interactions exhibited by ligand F11 within the binding site involve Cys241, Asn101, Ser178, Val181, and Asn350 through the SP docking method, while XP docking exhibited six H-bonds interactions with Cys241, Asn101, Lys254, Ser178, Val181, and Val315.
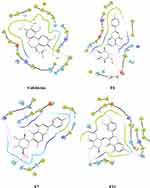 |
Figure 8 Ligand and protein molecular interactions for colchicine, F6, F7, and F11 with the β-Tubulin binding site using glide SP scoring tool. |
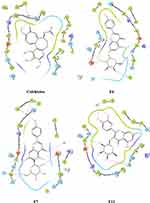 |
Figure 9 Ligand and protein molecular interactions for colchicine, F6, F7, and F11 with the β-Tubulin binding site using glide XP scoring tool. |
Discussion
In Soxhlet methanolic extract, the first spectrum screening at Rt (0.691–1.294 min): The peaks, which appear at Rt range (0.691–1.294 min), are correlated with the compounds Quercitrin (F11) and Vicenin-1 (F9), respectively,33 F11 with [M+H]+ 448.101 and molecular formula of [C21H20O11] +, and compound F9 with [M+H]+ 564.148 and molecular formula of [C26H28O14]+. Second spectrum screening at Rt (4.712–5.110 min): The peaks, which appear at Rt range (4.712–5.110 min), are correlated with the compounds Afroside (F6),34 F6 with [M+H]+ 432.106 and molecular formula of [C21H20O10]+. Third spectrum screening at Rt (10.816–12.825 min): The peaks, which appear at Rt range (10.816–12.825 min), are correlated with the compounds Naringenin (F2) and Vicenin-2 (F10), respectively,34 F2 with [M+H]+ 270.053 and molecular formula of [C15H10O5] +, and compound F10 with [M+H]+ 594.158 and molecular formula of [C27H30O15]+. Fourth spectrum screening at Rt (25.146–25.411 min): The peaks, which appear at Rt range (25.146–25.411 min), are associated with the compounds Quercetin (F3) and Tricin-2 (F4), respectively,34 F2 with [M+H]+ 270.053 and molecular formula of [C15H10O5] +, and compound F10 with [M+H]+ 594.158 and molecular formula of [C27H30O15]+.
In chloroform extract, the first spectrum screening at Rt (10.348–12.645 min): The peaks that appear at Rt range (10.348–12.645 min) are correlated with the compounds Diosgenin-1 (D1),33 D1 with [M+H]+ 414.621 and molecular formula of [C27H42O3]+. Second spectrum screening at Rt (25.835–26.934 min): The peaks that appear at Rt range (25.835–26.934 min) are related with the compounds (25R)-5x-Spirost-2-en-6-one (D3) and (25R)-2a,3az-Dihydroxy-5a-spirostan-6-one (D4), respectively,33 Compound D3 with [M+H]+ 412.298 and a molecular formula of [C27H40O3]+, and compound D4 with [M+H]+ 446.303 and a molecular formula of [C27H42O5]+. Third spectrum screening at Rt (27.805–28.692 min): The peaks that appear at Rt range (27.805–28.692 min) are correlated with the compounds (25R)-3cx,5-Cyclo-5a-spirostan-6-one (D2) Compound D2 with [M+H]+ 432.106 and a molecular formula of [C21H20O10]+.33
In water extract, the first spectrum screening at Rt (0.752–1.266 min): The peaks that appear at Rt range (0.752–1.266 min) are associated with the compounds Rhapontigenin (S3) and Afroside (F6), respectively,34,35 compound S3 with [M+H]+ 258.089 and molecular formula of [C15H14O4]+, and compound F6 with [M+H]+ 432.106 and molecular formula of [C21H20O10]+. Second spectrum screening at Rt (25.295–25.884 min): The peaks that appear at Rt range (25.295–25.884 min) are correlated with the compounds Desoxyrhaponticin (S2) and Vicenin-1 (F9), respectively,34,35 compound S2 with [M+H]+ 402.147 and a molecular formula of [C21H24O8]+, and compound F9 with [M+H]+ 564.148 and a molecular formula of [C26H28O14]+. Third spectrum screening at Rt (28.046–28.510 min): The peak that appears at Rt range (28.046–28.510 min) is correlated with the compound Rhaponticin (S1).35 Compound S1 with [M+H]+ 420.3362 and a molecular formula of [C21H24O9]+.
In diethyl ether, the first spectrum screening at Rt (25.084–25.766 min): The peaks that appear at Rt range (25.084–25.766 min) are related with the compounds Desoxyrhaponticin (S2) and Vicenin-1 (F9), respectively,34,35 compound S2 with [M+H]+ 402.147 and a molecular formula of [C21H24O8]+, and compound F9 with [M+H]+ 564.148 and a molecular formula of [C26H28O14]+. Second spectrum screening at Rt (25.146–25.411 min): The peaks that appear at Rt range (25.146–25.411 min) are correlated with the compounds Desoxyrhaponticin (S2) and Rhaponticin (S1), respectively,35 compound S2 with [M+H]+ 402.147 and a molecular formula of [C21H24O8]+, and compound S1 with [M+H]+ 420.3362 and a molecular formula of [C21H24O9]+.
Using the PASS online webserver, the flavonoid class of compounds demonstrated the most promising anticancer and anti-neoplastic activities among others, while the polyphenol stilbenes and phytosterol diosgenin derivatives showed lower activity compared to flavonoids, suggesting the flavonoid class of compounds could be responsible for the anticancer activity of the fenugreek plant. Since flavonoids predominate in the methanolic extract, our in-vitro cytotoxicity findings appear to be consistent with previously reported studies that support its anticancer properties. For example, Torres et al found that THDF, a natural flavonoid, is a potent inhibitor of cytotoxic activity on human myeloid leukemia HL-60 and U937 cells.36 Moreover, Centaureidin, a flavone derived from Polymnia fruticose, has been shown to inhibit tubulin polymerization, making it one of the first known flavonoids with an antimitotic effect.37 Furthermore, Liu et al proposed that Casticin, a polymethoxyflavone derived from Vitex rotundifolia, reduced the viability of glioma cells U251, U87, and U373, through a variety of mechanisms, one of which is attenuating the polymerization of tubulin.38 Similarly, Isoliquiritigenin, a flavonoid isolated from liquorice, has been shown to suppress proliferation and promote differentiation in glioma stem cells isolated from SHG44 human glioma cells.39 These studies confirm our results and suggest that fenugreek methanolic extract possesses a promising anticancer activity that can mediate through inhibition of tubulin polymerization as reported in the literature. Meanwhile, chloroform extract demonstrated less anticancer activity, confirming the predicted activities using PASS online webserver.
Furthermore, we assessed and evaluated the absorption, distribution, metabolism, and elimination (ADME) properties of natural active compounds present in the fenugreek seeds extract. Understanding the ADME parameters for the tested compounds allows the design and development of highly effective therapeutic agents with good pharmaceutical properties.40 Therefore, our results show that the most important ADME parameters that possess a crucial role in the drug discovery process are acceptable among the three different classes. Additionally, since several reported studies have suggested β-Tubulin could mediate beneficial flavonoid effects, a molecular docking study with β-Tubulin was carried out, and our docking results showed that flavonoids exhibit the highest docking scores with tubulin crystal structure, suggesting the cytotoxic activity of flavonoids could be mediated through inhibition of tubulin polymerization, which warrants further investigation.41–44
Conclusion
This study demonstrates that the use of solvents has a significant impact on the obtained phytochemical class present in the fenugreek seeds. Among the four various solvents used, the phytochemical class obtained with maceration using chloroform is steroidal skeleton diosgenin, while methanol solvent contained the flavonoids. Furthermore, the computational activity predictions and in-vitro anticancer results confirmed the promising anticancer activity of flavonoids against several cancer cell lines that can potentially mediate through inhibition of tubulin polymerization. Thus, fenugreek methanolic extract obtained from Trigonella foenum-graecum seeds exhibits anti-tubulin activity, and further research is needed to identify new chemotherapeutic agents with less harmful side effects.
Data Sharing Statement
The data associated with the paper are available, and under what conditions the data can be accessed.
Ethical Statement
Written consent was obtained from the donors under project # RC16/096 according to the Institutional Review Board (IRB) of King Abdullah International Medical Research Center (KAIMRC), Riyadh, Saudi Arabia. All the used cancer cell lines were purchased from authorized accredited commercial suppliers. The cell lines are patented and previously reported and published.
Acknowledgments
The authors want to express their sincerest gratitude to the College of Pharmacy (COP) at King Saud bin Abdulaziz University for Health Sciences (KSAU-HS) for their continued support and Dr. Mohamed Boudjelal for the generous help during the experimental work.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas, have drafted or written, or substantially revised or critically reviewed the article, have agreed on the journal to which the article will be submitted, reviewed and agreed on all versions of the article before submission, during revision, the final version accepted for publication, and any significant changes introduced at the proofing stage, agreed to take responsibility and be accountable for the contents of the article.
Funding
The authors acknowledge financial support from King Abdullah International Medical Research Center (KAIMRC), Ministry of National Guard Health Affairs, Riyadh, Kingdom of Saudi Arabia; Grant # (SP19.271.R).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Pearce A, Haas M, Viney R, et al. Incidence and severity of self-reported chemotherapy side effects in routine care: a prospective cohort study. PLoS One. 2017;12(10):e0184360. doi:10.1371/journal.pone.0184360
3. Azaizeh H, Saad B, Cooper E, Said O. Traditional Arabic and Islamic medicine, a re-emerging health aid. Evid Based Complement Alternat Med. 2010;7(4):419–424. doi:10.1093/ecam/nen039
4. Saad B, Azaizeh H, Said O. Tradition and perspectives of Arab herbal medicine: a review. Evid Based Complement Alternat Med. 2005;2(4):475–479. doi:10.1093/ecam/neh133
5. Omeish AF, Abbadi W, Ghanma IM, et al. Hospital-based study on the use of herbal medicine in patients with coronary artery disease in Jordan. J Pak Med Assoc. 2011;61(7):683–687.
6. Tang J, Song X, Zhu M, Zhang J. Study on the pharmacokinetics drug-drug interaction potential of Glycyrrhiza uralensis, a traditional Chinese medicine, with lidocaine in rats. Phytother Res. 2009;23(5):603–607. doi:10.1002/ptr.2450
7. Nagulapalli Venkata KC, Swaroop A, Bagchi D, Bishayee A. A small plant with big benefits: fenugreek (Trigonella foenum-graecum Linn.) for disease prevention and health promotion. Mol Nutr Food Res. 2017;61(6):1600950. doi:10.1002/mnfr.201600950
8. Pietta PG. Flavonoids as antioxidants. J Nat Prod. 2000;63(7):1035–1042. doi:10.1021/np9904509
9. Kaviarasan S, Vijayalakshmi K, Anuradha CV. Polyphenol-rich extract of fenugreek seeds protect erythrocytes from oxidative damage. Plant Foods Hum Nutr. 2004;59(4):143–147. doi:10.1007/s11130-004-0025-2
10. Nair S, Nagar R, Gupta R. Antioxidant phenolics and flavonoids in common Indian foods. J Assoc Physicians India. 1998;46(8):708–710.
11. Feki A, Jaballi I, Cherif B, et al. Therapeutic potential of polysaccharide extracted from fenugreek seeds against thiamethoxam-induced hepatotoxicity and genotoxicity in Wistar adult rats. Toxicol Mech Methods. 2019;29(5):355–367. doi:10.1080/15376516.2018.1564949
12. Hannan JM, Ali L, Rokeya B, et al. Soluble dietary fibre fraction of Trigonella foenum-graecum (fenugreek) seed improves glucose homeostasis in animal models of type 1 and type 2 diabetes by delaying carbohydrate digestion and absorption, and enhancing insulin action. Br J Nutr. 2007;97(3):514–521. doi:10.1017/S0007114507657869
13. Sauvaire Y, Petit P, Broca C, et al. 4-Hydroxyisoleucine: a novel amino acid potentiator of insulin secretion. Diabetes. 1998;47(2):206–210. doi:10.2337/diab.47.2.206
14. Fuller S, Stephens JM. Diosgenin, 4-hydroxyisoleucine, and fiber from fenugreek: mechanisms of actions and potential effects on metabolic syndrome. Adv Nutr. 2015;6(2):189–197. doi:10.3945/an.114.007807
15. Basch E, Ulbricht C, Kuo G, Szapary P, Smith M. Therapeutic applications of fenugreek. Altern Med Rev. 2003;8(1):20–27.
16. Alsemari A, Alkhodairy F, Aldakan A, et al. The selective cytotoxic anti-cancer properties and proteomic analysis of Trigonella foenum-graecum. BMC Complement Altern Med. 2014;14:114. doi:10.1186/1472-6882-14-114
17. Chatterjee S, Kumar M, Kumar A. Chemomodulatory effect of Trigonella foenum graecum (L.) seed extract on two stage mouse skin carcinogenesis. Toxicol Int. 2012;19(3):287–294. doi:10.4103/0971-6580.103670
18. Khalil MI, Ibrahim MM, El-Gaaly GA, Sultan AS. Trigonella foenum (Fenugreek) induced apoptosis in hepatocellular carcinoma cell line, HepG2, mediated by upregulation of p53 and proliferating cell nuclear antigen. Biomed Res Int. 2015;2015:914645. doi:10.1155/2015/914645
19. Al-Timimi LAN. Antibacterial and anticancer activities of fenugreek seed extract. Asian Pac J Cancer Prev. 2019;20(12):3771–3776. doi:10.31557/APJCP.2019.20.12.3771
20. Ahmed SI, Hayat MQ, Zahid S, et al. Isolation and identification of flavonoids from anticancer and neuroprotective extracts of Trigonelle foenum-graecum. Tropical J Pharm Res. 2017;16(6):1391–1398. doi:10.4314/tjpr.v16i6.25
21. Binarová P, Tuszynski J. Tubulin: structure, functions and roles in disease. Cells. 2019;8(10):1294. doi:10.3390/cells8101294
22. Field JJ, Díaz JF, Miller JH. The binding sites of microtubule-stabilizing agents. Chem Biol. 2013;20(3):301–315. doi:10.1016/j.chembiol.2013.01.014
23. Naaz F, Haider MR, Shafi S, et al. Anti-tubulin agents of natural origin: targeting taxol, vinca, and colchicine binding domains. Eur J Med Chem. 2019;171:310–331. doi:10.1016/j.ejmech.2019.03.025
24. Nagireddy PKR, Kommalapati VK, Siva Krishna V, Sriram D, Tangutur AD, Kantevari S. Imidazo[2,1-b]thiazole-coupled natural noscapine derivatives as anticancer agents. ACS Omega. 2019;4(21):19382–19398. doi:10.1021/acsomega.9b02789
25. Ali R, Samman N, Al Zahrani H, et al. Isolation and characterization of a new naturally immortalized human breast carcinoma cell line, KAIMRC1. BMC Cancer. 2017;17(1):803. doi:10.1186/s12885-017-3812-5
26. Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi:10.1038/srep42717
27. Filimonov D, Lagunin A, Gloriozova T, et al. Prediction of the biological activity spectra of organic compounds using the PASS online web resource. Chem Heterocycl Compd. 2014;50(3):444–457.
28. Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–341. doi:10.1016/j.ddtec.2004.11.007
29. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26. doi:10.1016/s0169-409x(00)00129-0
30. Leo A, Hansch C, Elkins D. Partition coefficients and their uses. Chem Rev. 1971;71(6):525–616.
31. Jorgensen WL, Duffy EM. Prediction of drug solubility from Monte Carlo simulations. Bioorg Med Chem Lett. 2000;10(11):1155–1158. doi:10.1016/s0960-894x(00)00172-4
32. Ajay BGW, Murcko MA. Designing libraries with CNS activity. J Med Chem. 1999;42(24):4942–4951. doi:10.1021/jm990017w
33. Melo PS, De Azevedo MB, Zullo MA, Fabrin-Neto JB, Haun M. Cytotoxicity of the phytosterol diosgenin and its derivatives in rat cultured hepatocytes and V79 fibroblasts. Hum Exp Toxicol. 2004;23(10):487–493. doi:10.1191/0960327104ht474oa
34. Skalsta H. Chemical constituents. In: Georgios AP, editor. Fenugreek, The Genus Trigonella. New York: Taylor & Francis Inc; 2002:132.
35. Li G, Luan G, He Y, et al. Polyphenol stilbenes from fenugreek (Trigonella foenum-graecumL.) seeds improve insulin sensitivity and mitochondrial function in 3T3-L1 adipocytes. Oxid Med Cell Longev. 2018;2018:7634362. doi:10.1155/2018/7634362
36. Torres F, Quintana J, Estévez F. 5,7,3ʹ-trihydroxy-3,4ʹ-dimethoxyflavone-induced cell death in human leukemia cells is dependent on caspases and activates the MAPK pathway. Mol Carcinog. 2010;49(5):464–475. doi:10.1002/mc.20619
37. Beutler JA, Cardellina JH
38. Liu E, Kuang Y, He W, Xing X, Gu J. Casticin induces human glioma cell death through apoptosis and mitotic arrest. Cell Physiol Biochem. 2013;31(6):805–814.
39. Lin Y, Sun H, Dang Y, Li Z. Isoliquiritigenin inhibits the proliferation and induces the differentiation of human glioma stem cells. Oncology Rep. 2018;39(2):687–694.
40. Butina D, Segall MD, Frankcombe K. Predicting ADME properties in silico: methods and models. Drug Discovery Today. 2002;7(11):S83–S88.
41. Mukhtar E, Adhami VM, Sechi M, Mukhtar H. Dietary flavonoid fisetin binds to β-tubulin and disrupts microtubule dynamics in prostate cancer cells. Cancer Letters. 2015;367(2):173–183.
42. Torres F, Quintana J, Estévez F. 5, 7, 3′‐trihydroxy‐3, 4′‐dimethoxyflavone inhibits the tubulin polymerization and activates the sphingomyelin pathway. Mol Carcinog. 2011;50(2):113–122.
43. Gupta K, Panda DJB. Perturbation of microtubule polymerization by quercetin through tubulin binding: a novel mechanism of its antiproliferative activity. Biochemistry. 2002;41(43):13029–13038.
44. Beutler JA, Hamel E, Vlietinck AJ, et al. Structure− activity requirements for flavone cytotoxicity and binding to tubulin. J Med Chem. 1998;41(13):2333–2338.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
