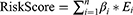Back to Journals » International Journal of General Medicine » Volume 15
Non-Canonical NF-κB Signaling Stratifies LGG into Subtypes with Distinct Molecular and Cellular Characteristic and Survival Expectancy
Authors Lin M, Huang T, Wang X, Li X , Ma J , Su L, Wu J
Received 2 December 2021
Accepted for publication 17 March 2022
Published 5 April 2022 Volume 2022:15 Pages 3677—3686
DOI https://doi.org/10.2147/IJGM.S347654
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Minhua Lin,1 Tianxiang Huang,2 Xuan Wang,1 Xuenan Li,3 Jingjiao Ma,3 Lan Su,3 Jun Wu2
1Department of Neurosurgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, People’s Republic of China; 2Department of Neurosurgery, Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China; 3Beijing Genetron Health, Co. Ltd, Beijing, 102206, People’s Republic of China
Correspondence: Jun Wu, Department of Neurosurgery, Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China, Tel +86 13508480515, Fax +86 731-89753039, Email [email protected]
Introduction: NF-κB signaling is involved in a wide range of biological processes including cell proliferation, cell survival and immunity. Meanwhile, as one of the major oncogenic pathways, its upregulation has been observed in many cancer types. Compared with canonical NF-κB signaling, its non-canonical branch was much less studied in cancerous context.
Methods: In this study, we leveraged multi-omics data across multiple platforms to investigate the activity of non-canonical NF-κB signaling in low-grade glioma (LGG) and explore its connection with molecular characteristics of LGG.
Results: We found that non-canonical NF-κB signaling could classify LGG patients into subgroups with significant survival difference. Non-canonical NF-κB-low group enriched with oligodendroglioma featured by CIC mutations and 1p19q co-deletion. On the another hand, LGG in non-canonical NF-κB-high group showed high frequency of EGFR mutations but relatively low frequency of IDH mutations. In addition, LGG in this group reflected immunosuppressive environment characterized by high level of cytotoxic T cell exhaustion and macrophage M2 infiltration. More comprehensive evaluation implied that LGG in non-canonical NF-κB-high group reflected significantly higher immunogenicity. Through a series of feature selection technique, we developed a model that can predict the prognosis of LGG patients in a cost-effective way.
Conclusion: Our analysis demonstrated the prognostic value of non-canonical NF-κB signaling in LGG. The survival difference between non-canonical NF-κB stratified groups may be explained by their distinct molecular characteristics as well as cellular context. Our prognostic model may help in offering better therapeutic strategy and clinical management.
Keywords: lower grade glioma, non-canonical NF-κB signaling, immune microenvironment, T cell exhaustion, M2 macrophages, prognosis
Introduction
Low-grade glioma (LGG) accounts for approximately 25% of primary brain tumors. Despite advances in treatment development, more than half of LGG tumors progress into therapy-resistant high-grade gliomas with poor prognosis.1 Several studies have revealed that LGG is genetically heterogeneous.2 Histopathological classification combined with the analysis of recurrently mutated genes such as isocitrate dehydrogenase 1 (IDH1), IDH2, TP53, EGFR, and ATRX is used to determine the prognosis of LGG patients.3,4 In addition, co-deletion of chromosome arms 1p and 19q (1p/19q codeletion) and O-6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status have been identified as prognostic factors for LGG.5 However, LGG patients with the same risk factors have different prognosis based on these markers, indicating that more prognostic factors remain unrevealed.
The tumor microenvironment (TME) mediates cancer progression, responses to therapy and prognosis. Tumor-infiltrating immune cells (TIICs) are critical components of the TME and are associated with clinical outcomes.6 TIICs mainly include tumor-infiltrating lymphocytes (TILs), tumor-associated macrophages (TAMs), natural killer (NK) cells, dendritic cells, and myeloid-derived suppressor cells (MDSCs). TAMs consist of a small portion of M1 macrophages and a large portion of M2 macrophages. Interferon-γ (IFN–γ) and/or lipopolysaccharides (LPS) induce M1 macrophage polarization, while IL-4, IL-13 or TGFb1 induces M2 macrophage polarization.7 M1 macrophages promote the generation of T helper type 1 lymphocytes, thus fighting tumors, while M2 macrophages promote tumor progression. M2 macrophages are associated with poor prognosis in high-grade glioma.8 TILs are usually associated with favorable outcomes; in contrast, immunosuppressive cells such as regulatory T (Treg) cells, MDSCs and M2 macrophages are associated with poor outcomes.9 These findings suggest that TIICs might be the determining factors for LGG heterogeneity. Therefore, elucidating the underlying TIIC mechanisms is essential to provide actionable targets for immunotherapy to achieve precise treatment for LGG.
The NF-κB signaling is a major pathway that mediates TIICs.10 The NF-κB family has five core members: RelA (p65), RelB, Rel, NF-κB1 (p50 and its precursor p105), and NF-κB2 (p52 and its precursor p100). These core members form dimers to regulate two distinct NF-κB signaling activation mechanisms: the canonical NF-κB signaling (p50/RelA) and the non-canonical NF-κB signaling (p52/RelB). An atypical NF-κB signaling lacks both RelA and RelB. Inhibiting NF-κB activation in TAMs converts them from M2 to M1 macrophages.11 IL-1β activates MDSCs via NF-κB signaling.12 NF-κB activation is strongly associated with T cell infiltration in human lung cancer.13 Although less studied comparing with canonical NF-κB signaling, the non-canonical NF-κB signaling has been known to regulate immune response as well as the development of immune related organs. For instance, non-canonical NF-κB involves thymus development by regulating the development of thymic epithelial cells.14 Non-canonical NF-κB also plays a role in the development of secondary lymphoid organs, including spleen and lymph node.15 In terms of immune response, non-canonical NF-κB signaling participates in the generation as well as maintenance of effector and memory T cells.16,17 Dendritic cells (DCs) is the primary antigen presenting cells linking innate immunity and adaptive immunity. Its maturation has been also found to be regulated by non-canonical NF-κB signaling.18 While the role of non-canonical NF-κB signaling in regulating immunity is being uncovered, the interaction between dysregulated non-canonical NF-κB signaling and immune environment in cancerous context remains unknown.
Here, we investigated the activity of non-canonical NF-κB signaling in LGG by leveraging multi-omics data from The Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA).19 We found that non-canonical NF-κB signaling can divided LGG into molecularly distinct groups. Particularly, non-canonical NF-κB-high group enriched with LGG featured by oncogenic mutations and immunosuppressive environment, consistently aligned with the poor survival of LGG patients in this groups. Our analysis provided potential patient stratification strategy for better therapeutic strategy as well as prognosis.
Materials and Methods
Data Retrieval and Preprocessing
The RNA-Seq data, somatic mutation data and clinical information of LGG tissue samples were downloaded from The Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA) data portal. Samples that met the following criteria were included in our study: 1) primary tumor samples; 2) both somatic mutation and expression data were available; 3) patients’ clinical information included both prognosis and previous biomarker test results is available. The expression data were normalized as fragments per kilobase million (FPKM). Somatic mutation data was downloaded in MAF format. The clinical data were integrated and downloaded through cBioPortal (http://www.cbioportal.org).
Estimation of NF-κB Signaling Activity
The gene sets of canonical NF-κB signaling, non-canonical NF-κB signaling and atypical NF-κB signaling were retrieved from the Molecular Signatures Database (MSigDB).20 The pathway enrichment score was calculated using single sample gene set enrichment analysis (ssGSEA, v4).20 Survival analysis was conducted using Kaplan-Meier estimator. Patients stratification was done using X-Tile.21
Characterization of Tumor-Infiltrating Immune Cells (TIICs) and Immunogenicity Estimation
ESTIMATE (Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data: https://bioinformatics.mdanderson.org/estimate/ https://www.gsea-msigdb.org/gsea/index.jsp) was used to estimate the relative fraction of tumor cells, stromal cells and immune cells in LGG sample. ImmunecellAI (http://bioinfo.life.hust.edu.cn/ImmuCellAI) was used for immune cell decomposition.
Tumor mutation burden (TMB) is defined as the average amount of somatic mutations per Mb within the protein coding region. Somatic mutations were counted only if they meet the following criteria: 1) mutant allele frequency > 5%; 2) mutant does not included in common_dbSNP of the dbSNP knowledgebase; 3) population frequency of mutant < 1% in 1000 Genomes Project. Immunophenogram22,23 was used to analyze the mRNA expression data of the TCGA-LGG dataset to estimate Immunophenotypescore (IPS), Briefly, immunogenicity considers four categories of immune-relevant gene sets representing effector cells (ECs), suppressive cells (SCs), major histocompatibility complex molecules (MHCs), and immunomodulators or checkpoints (CPs), The IPS was scaled to 0–10 based on weighted averaged Z-scores of the determinants from the above four categories and increased with higher immunogenicity, as previously described.22 Similarly, T cell–inflamed GEP was used to estimate the immunogenicity of LGG samples.24
Survival Analysis and Construction of Prognostic Model
Survival analysis was performed with the R package survival analysis. For univariate survival analysis, Log rank test was performed to determine statistical difference. In terms of multivariate survival analysis, Cox proportional hazards model was applied to determine the independence of prognostic predictor by adjusting potential confounders. P value less than 0.05 was considered as statistically significant.
To constructing the prognostic model, TCGA-LGG patients were randomly divided into a training set (n=273) and an internal testing set (n=137) at ratio of 2:1. Through univariate regression, 91 genes (Supplementary Table 2) were prioritized from 165 genes (Supplementary Table 1) that were initially collected from MSigDB. To better generalize our prognostic model, we performed least absolute shrinkage and selection operator (LASSO) regression to select 9 genes (Supplementary Table 3) with greater prognostic power. To account for potential confounders, such as age, sex, grade, MGMT promoter methylation status, 1p19q status, and IDH, we further used multivariate Cox regression to shrink the final gene list to nine genes whose expression levels were used to construct a risk score system based on the following formula:
where “n” is the total number of prognostic genes; βi and Ei represent the LASSO regression coefficient of gene i and log2 transformed relative expression value of gene i. Based on the median risk score, LGG patients were divided into a high-risk group and low-risk group. A multivariate Cox model was used to assess the impact of the model on the survival of LGG patients. To validate the prognostic capability of our model, an independent dataset was retrieved from the CGGA dataset and subjected to our model after preprocessing. The ROC was generated using R package “timeROC”.
Statistical Analysis
R language (v3.6.1) was used for statistical analysis. Between groups comparison was conducted using either Mann–Whitney U-test or Kruskal–Wallis test depending on the number of groups. Correlation between continuous variables was measured using Spearman correlation coefficient. Considering the large sample size subjected to statistical analysis, in addition to P-value (*p<0.05, **p<0.01 and ***p<0.001), fold change or Pearson coefficient was applied to reflect the degree of difference or correlation.
Results
Non-Canonical NF-κB Signaling Pathway Was Upregulated in LGG and Associated with Poor Patient Prognosis
In this study, we first examined the expression level of the core members (NF-κB1, NF-κB2, RELA, RELB and REL) of NF-κB signaling pathway. As shown in Figure 1A, these core members were universally upregulated in LGG relative to healthy tissues. Given that these core genes overlappingly involve in three NF-κB signaling branches: canonical NF-κB signaling, non-canonical NF-κB signaling and atypical NF-κB signaling, we further evaluated the activity in each branch using ssGSEA. Sample-wise correlation in Figure 1B showed that the expression of core genes highly correlated with activity of non-canonical NF-κB signaling (R=0.82) while only moderately correlated with canonical NF-κB signaling and atypical NF-κB signaling (R=0.57 and R=0.61, Supplementary Figure 1A and B), implying that the activation of NF-κB signaling is mainly attributed by its non-canonical branch. We also explored the association of NF-κB signaling with LGG patients’ survival. 410 LGG patients were stratified into low-, medium- and high non-canonical NF-κB signaling groups using X-Tile. Kaplan-Meier estimator showed that, among all three NF-κB signaling branches, only non-canonical NF-κB signaling stratified LGG patients into subgroups with differentiated survival outcome. (Figure 1C, p-value=0.016 and Supplementary Figure 1C and D). Specifically, LGG patients with high non-canonical NF-κB signaling activity own median survival time of 33.96 month, significantly shorter than LGG patients with medium or low NF-κB signaling activity. These results suggested that, compared with other branches, non-canonical NF-κB signaling imposes greater impact on the progression of LGG.
Non-Canonical NF-κB Signaling Stratifies LGG Patients into Molecularly Distinct Groups
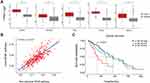
We thought to further characterize the molecular difference between LGG groups. Analysis on Whole Exome Sequencing (WES) reflected mutational spectrum in Figure 2A. We only displayed driver genes in LGG defined in Francisco Martínez-Jiménez et al’s study.25 Non-canonical NF-κB signaling stratified groups were labeled at the top alone with major clinical information and genomic events in LGG. As expected, IDH1, ATRX and TP53 were prevalently mutated across all groups. EGFR was mutated in increasing trend from non-canonical NF-κB-low to -high groups (Fisher’s exact test, p-value < 0.01) and appeared in a mutually exclusive manner with IDH1 mutations. IDH mutations has been known as a prevalent genomic event correlated with favorable outcome of LGG patients.26 On the other hand, mutations in EGFR may upregulate non-canonical NF-κB signaling and, along with other EGF induced oncogenic signaling pathways, promote tumor cell proliferation.27 Thus, it is possible that the high frequency of EGFR mutants while low frequency of IDH1 mutation attribute to the poor survival of LGG patients in non-canonical NF-κB-high group. Besides, we found that CIC mutations and 1p19q co-deletion were enriched in NF-κB-low group (Fisher’s exact test, p-value < 0.01). Consistently, these two markers of oligodendroglioma highlighted the dominancy of oligodendroglioma in NF-κB-low group.28
Non-Canonical NF-κB Signaling Associated with Immunosuppressive Environment and Inhibition of Cytotoxic T Cells
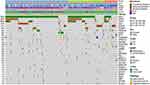
TME mediates tumor progression, relapse and response to therapy29,30 and NF-κB signaling has been known to modulate the TME to a great extent.31,32 Therefore, we explored the association between activity of non-canonical NF-κB signaling and the landscape of Tumor-infiltrating immune cells (TIICs) in LGG samples. We first estimated the relative fraction of immune cell infiltration of each LGG sample. As Figure 3A shown, the activity of non-canonical NF-κB signaling positively correlated with overall degree of TIICs (Kruskal–Wallis test, p-value <0.05). This observation is consistent with our current knowledge of non-canonical NF-κB signaling in enhancing the immune response. TIICs represent a mixture of numerous immune cell types corresponding to different, even opposite, biological function. To gain a higher resolution of cellular composition of LGG samples under the influence of non-canonical NF-κB signaling, we further dissected TIIC subpopulations through deconvolution of RNA expression (See Method). We found that cytotoxic T cells demonstrated elevated infiltration from non-canonical NF-κB-low to -high group (Figure 3B). However, subsequent analysis found that the fraction of exhausted T cell showed the same trend (Figure 3C), implying that most cytotoxic T cells in NF-κB-high group may have lost their cytotoxic capacity against tumor cells. T cells exhaustion has been believed to be induced by some tumor cells capable of shaping its surrounding into immunosuppressive environment. Another frequently observed immune cell type with such capacity is macrophage type 2 (M2).We found that M2 was dominant across all three groups and showed elevated trend from non-canonical NF-κB-low to -high group (Figure 3D). Moreover, we analyzed correlation between the non-canonical NF-κB signaling and M2 markers. As shown in Figure 3E, the expression of M2 marker genes consistently correlated with activity of non-canonical NF-κB signaling. The above analysis results regarding cytotoxic T cells, exhausted T cells and macrophage were later validated in CGGA LGG dataset (Supplementary Figure 2). Together, we suggested that although the activity of nc NF-κB signaling positively correlates with immune infiltration, the elevated immune infiltration in nc NF-κB group displayed a detrimental immune microenvironment which may be a factor contributing to the poor prognosis of this group.
We hypothesized that the activation of the non-canonical NF-κB signaling pathway might be involved in inhibiting or exhausting cytotoxic T cells. The heatmap of the relative expression levels of T cell checkpoint, including PD-L1, PD-L2, TIM3, CTLA4 and SERPINB9 were elevated in the non-canonical NF-κB-high group (Figure 3F). The TIM3 and PD-L1 pathways have been identified as markers of T cell exhaustion.33,34 These observations, alone with higher mutation burden in non-canonical NF-κB-high group (Figure 3G), implied that LGG patients of this group may be more likely to benefited from immunotherapy. We further assessed the immunogenicity in a more comprehensive manner. Immunophenoscore (IPS) reflects tumor’s potential responsiveness to immune checkpoint inhibitors (ICIs) by simultaneously measuring the expression of key genes involved in immune-effective cells, immunosuppressive cells, MHC complex and immunomodulators.22 As Figure 3H shown, LGG samples in non-canonical NF-κB-high group demonstrated significantly higher immunogenicity. Similarly, T cell–inflamed GEP is a rigorously validated model which measures the potential of antitumor response.35 As expected, samples in non-canonical NF-κB-high group showed much higher GEP score among three groups (Figure 3I). In the past, clinical trials of immunotherapy in brain cancer have not been as successful as in non-CNS cancer partially due to anatomical structure and specialized immune environment of brain.36 However, accurate molecular stratification may help identify LGG patients who are more likely to benefit from ICIs.
Non-Canonical NF-κB Signaling Pathway is an Independent Prognostic Factor for LGG Patients
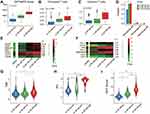
Our previous analysis indicated that non-canonical NF-κB signaling significantly associated with worse OS of LGG patients (Figure 1C, Log rank test, p-value=0.016), we thought to explore the feasibility of constructing a simplified prognostic model that predict the likelihood of patient being alive after specific period of time (See Method). We randomly divided 410 patients in the TCGA-LGG dataset into the training set (n =273) and the testing set (n =137). Using training set, we prioritized nine feature genes for model construction through a series of feature selection technique. Figure 4A showed the ranked sample risk alone with the expression of prioritized feature genes. LGG patients were divided into low- and high-risk group based on the median risk score. The Kaplan-Meier curve showed significant survival difference with p-value less than 0.01 (Figure 4B and C). This survival difference was validated in testing dataset (Figure 4D and E). AUROC of the model was 0.85, 0.83 and 0.71 for 1, 3, and 5 years, respectively (Figure 4F). To test the independence of the model, potential covariates (tumor grade, age, gender, MGMT promoter methylation status, 1p19q status and IDH) were incorporated into Cox proportional-hazards model. Results indicated that, in addition to known survival predictors IDH and tumor grade, the risk model can independently predict survival of LGG patients (Figure 4G). Lastly, the prognostic independence of non-canonical NF-κB signaling as well as the robustness of the model was validated in an external validation set was retrieved from CGGA (Supplementary Figure 3).
Discussion
In this study, we identified that the non-canonical NF-κB signaling is upregulated in LGG and its activity is positively correlated with increased M2 macrophage infiltration and T cell exhaustion. More specifically, LGG patients with high non-canonical NF-κB activity display immune suppressive microenvironment, more complexed mutational landscape and poor survival outcome. We showed that activity of non-canonical NF-κB signaling is a validated prognostic signature in predicting the survival of LGG patients.
Our study showed that the non-canonical NF-κB signaling was upregulated in LGG and associated with poor OS rate of LGG patients. This result aligned with previous studies that non-canonical NF-κB promotes glioma cell invasion independently of the canonical NF-κB signaling.37,38 Meanwhile, LGG patients in non-canonical NF-κB-high group displayed significantly higher frequency of EGFR mutation. Previous studies have demonstrated that constitutive activation of EGFR can trigger NF-κB signaling through activation of IκB kinase (IKK).27 The EGF- dependent NF-κB activation, alone with other EGF induced oncogenic signaling, facilitates cancer cell proliferation and consequently resulted in poor survival of LGG patients. While patients in NF-κB-low and NF-κB-medium did not show survival difference, they differed in histology. Compared with NF-κB-medium group, NF-κB-low group was dominated by oligodendroglioma featured by high frequency of 1p19q co-deletion as well as CIC truncating mutations.
LGG is thought to be an immune desert relative to other cancer types. However, our analysis demonstrated that immune cells were infiltrated in LGG. In particular, the dominant infiltration of M2 macrophage was consistent with previous studies.39,40 The fraction of M2 macrophages was the highest in the non-canonical NF-κB-high group. Additionally, the expression of M2 macrophage markers were remarkably upregulated in the non-canonical NF-κB-high group, suggesting that the activation of the non-canonical NF-κB signaling correlated with the immunosuppressive microenvironment in LGG.
T cells are critical in antitumor immunity. Non-canonical NF-κB signaling is essential for the generation and maintenance of effector T cells and memory T cells.16 We found that the activation of the non-canonical NF-κB signaling positively correlated with the overexpression of T cell checkpoint PD-L1, PD-L2, CTLA4, TIM3 and SERPIN9B. PD-L1, PD-L2 and CTLA4 are well-studied immune checkpoints that inhibit cytotoxic function of T cells. TIM3 functions as an immune checkpoint by prompting T cell exhaustion in human and mouse tumors.41 Previous studies demonstrated that cotargeting TIM3 and PD1 can induce tumor regression by reversing T cell exhaustion and restoring antitumor immunity in mouse colon carcinoma,42 Furthermore, researches have shown that TAMs are able to upregulate PD-L1 expression42 and targeting TAMs is one of the strategy under clinical investigation.43,44 In our study, the molecular characterization suggested possible efficacy of those therapeutic approaches in treating LGG patient in non-canonical NF-κB-high group. Of course, further in-vitro experimental investigation need to be carried out to validate the therapeutic rationale.
Conclusion
Our results demonstrated that a prognostic signature based on the non-canonical NF-κB signaling is an independent prognostic factor for LGG patients. Two characteristics revealed by our study may explain the poor prognosis of LGG patients with elevated non-canonical NF-κB signaling: the distinct mutational landscape of this group may confer to the overcapacity of tumor cell proliferation; The immunosuppressive microenvironment featured with elevated M2 macrophage infiltration and cytotoxic T cell exhaustion may protect the tumor from immune response and further facilitate tumor growth. These molecular and cellular characteristics suggest potential response to immunotherapy of patients in this group. Meanwhile, our analysis demonstrated that, through feature selection technique, the prognostic model based on non-canonical NF-κB signaling can predict survival of LGG patients in a cost-effective way.
Ethics Statement
The research project was reviewed and approved by the Ethic Committee of the Xiangya Hospital of Central South University. The research design and methods are in accordance with the requirements of regulations and procedures regarding to human subject protection laws such as GCP and ICH-GCP.
Funding
The authors received no specific funding for this work.
Disclosure
Xuenan Li, Jingjiao Ma, and Lan Su are employees of Beijing Genetron Health, Co. Ltd. The authors report no other potential conflicts of interest for this work and declare that they have no competing financial interests.
References
1. Louis DN, Perry A, Reifenberger G, et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1
2. Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade ii and iii gliomas. Nat Genet. 2015;47(5):458–468. doi:10.1038/ng.3273
3. Chan AK, Yao Y, Zhang Z, et al. Combination genetic signature stratifies lower-grade gliomas better than histological grade. Oncotarget. 2015;6(25):20885–20901. doi:10.18632/oncotarget.4928
4. Liu XY, Gerges N, Korshunov A, et al. Frequent atrx mutations and loss of expression in adult diffuse astrocytic tumors carrying idh1/idh2 and tp53 mutations. Acta Neuropathol. 2012;124(5):615–625. doi:10.1007/s00401-012-1031-3
5. Weller M, Stupp R, Hegi ME, et al. Personalized care in neuro-oncology coming of age: why we need mgmt and 1p/19q testing for malignant glioma patients in clinical practice. Neuro Oncol. 2012;14(Suppl 4):iv100–iv108. doi:10.1093/neuonc/nos206
6. Fridman WH, Galon J, Pagès F, Tartour E, Sautès-Fridman C, Kroemer G. Prognostic and predictive impact of intra- and peritumoral immune infiltrates. Cancer Res. 2011;71(17):5601–5605. doi:10.1158/0008-5472.Can-11-1316
7. Murray PJ. Macrophage polarization. Annu Rev Physiol. 2017;79:541–566. doi:10.1146/annurev-physiol-022516-034339
8. Sørensen MD, Dahlrot RH, Boldt HB, Hansen S, Kristensen BW. Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol Appl Neurobiol. 2018;44(2):185–206. doi:10.1111/nan.12428
9. Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–945. doi:10.1038/nm.3909
10. Taniguchi K, Karin M. Nf-κb, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi:10.1038/nri.2017.142
11. Porta C, Rimoldi M, Raes G, et al. Tolerance and m2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappab. Proc Natl Acad Sci USA. 2009;106(35):14978–14983. doi:10.1073/pnas.0809784106
12. Tu S, Bhagat G, Cui G, et al. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell. 2008;14(5):408–419. doi:10.1016/j.ccr.2008.10.011
13. Hopewell EL, Zhao W, Fulp WJ, et al. Lung tumor nf-κb signaling promotes t cell-mediated immune surveillance. J Clin Invest. 2013;123(6):2509–2522. doi:10.1172/jci67250
14. Dejardin E. The alternative nf-kappab pathway from biochemistry to biology: pitfalls and promises for future drug development. Biochem Pharmacol. 2006;72(9):1161–1179. doi:10.1016/j.bcp.2006.08.007
15. Sun SC. The noncanonical nf-κb pathway. Immunol Rev. 2012;246(1):125–140. doi:10.1111/j.1600-065X.2011.01088.x
16. Yu J, Zhou X, Nakaya M, Jin W, Cheng X, Sun SC. T cell-intrinsic function of the noncanonical nf-κb pathway in the regulation of GM-CSF expression and experimental autoimmune encephalomyelitis pathogenesis. J Immunol. 2014;193(1):422–430. doi:10.4049/jimmunol.1303237
17. Li Y, Wang H, Zhou X, et al. Cell intrinsic role of nf-κb-inducing kinase in regulating t cell-mediated immune and autoimmune responses. Sci Rep. 2016;6:22115. doi:10.1038/srep22115
18. Gerondakis S, Grumont R, Gugasyan R, et al. Unravelling the complexities of the nf-|[kappa]|b signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25(51):6781–6799. doi:10.1038/sj.onc.1209944
19. Zhao Z, Zhang KN, Wang Q, et al. Chinese glioma genome atlas (cgga): a comprehensive resource with functional genomic data from Chinese glioma patients - ScienceDirect. Genom Proteom Bioinform. 2021;19(1):12.
20. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
21. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi:10.1158/1078-0432.CCR-04-0713
22. Charoentong P, Finotello F, Angelova M, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi:10.1016/j.celrep.2016.12.019
23. Hakimi AA, Reznik E, Lee C-H, et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29(1):104–116. doi:10.1016/j.ccell.2015.12.004
24. Ott PA, Bang YJ, Piha-Paul SA, et al. T-cell–inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: keynote-028. J Clin Oncol. 2019;37(4):318–327. doi:10.1200/JCO.2018.78.2276
25. Martínez-Jiménez F, Muiños F, Sentís I, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20(10):555–572. doi:10.1038/s41568-020-0290-x
26. Yan H, Parsons DW, Jin G, et al. Idh1 and idh2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi:10.1056/NEJMoa0808710
27. Shostak K, Chariot A. Egfr and nf-κb: partners in cancer. Trends Mol Med. 2015;21(6):385–393. doi:10.1016/j.molmed.2015.04.001
28. Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in cic and fubp1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. doi:10.1126/science.1210557
29. Klemm F, Joyce JA. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 2015;25(4):198–213. doi:10.1016/j.tcb.2014.11.006
30. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi:10.1038/nm.3394
31. Bao B, Thakur A, Li Y, et al. The immunological contribution of nf-κb within the tumor microenvironment: a potential protective role of zinc as an anti-tumor agent. Biochimica et Biophysica Acta. 2012;1825(2):160–172. doi:10.1016/j.bbcan.2011.11.002
32. Hoesel B, Schmid JA. The complexity of nf-κb signaling in inflammation and cancer. Mol Cancer. 2013;12(1):1–15.
33. Jones RB, Ndhlovu LC, Barbour JD, et al. Tim-3 expression defines a novel population of dysfunctional t cells with highly elevated frequencies in progressive HIV-1 infection. J Exp Med. 2008;205(12):2763–2779. doi:10.1084/jem.20081398
34. Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting tim-3 and pd-1 pathways to reverse t cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207(10):2187–2194. doi:10.1084/jem.20100643
35. Ayers M, Lunceford J, Nebozhyn M, et al. Ifn-γ–related mRNA profile predicts clinical response to pd-1 blockade. J Clin Investig. 2017;127(8):2930–2940. doi:10.1172/JCI91190
36. Sampson JH, Gunn MD, Fecci PE, Ashley DM. Brain immunology and immunotherapy in brain tumours. Nat Rev Cancer. 2020;20(1):12–25. doi:10.1038/s41568-019-0224-7
37. Duran CL, Lee DW, Jung JU, et al. NIK regulates MT1-MMP activity and promotes glioma cell invasion independently of the canonical nf-κb pathway. Oncogenesis. 2016;5(6):e231. doi:10.1038/oncsis.2016.39
38. Tchoghandjian A, Jennewein C, Eckhardt I, Rajalingam K, Fulda S. Identification of non-canonical nf-κb signaling as a critical mediator of smac mimetic-stimulated migration and invasion of glioblastoma cells. Cell Death Dis. 2013;4(3):e564. doi:10.1038/cddis.2013.70
39. Vidyarthi A, Agnihotri T, Khan N, et al. Predominance of m2 macrophages in gliomas leads to the suppression of local and systemic immunity. Cancer Immunol Immunother. 2019;68(12):1995–2004. doi:10.1007/s00262-019-02423-8
40. Hussain SF, Yang D, Suki D, Aldape K, Grimm E, Heimberger AB. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro Oncol. 2006;8(3):261–279. doi:10.1215/15228517-2006-008
41. Wolf Y, Anderson AC, Kuchroo VK. Tim3 comes of age as an inhibitory receptor. Nat Rev Immunol. 2020;20(3):173–185. doi:10.1038/s41577-019-0224-6
42. Fourcade J, Sun Z, Benallaoua M, et al. Upregulation of tim-3 and pd-1 expression is associated with tumor antigen-specific cd8+ t cell dysfunction in melanoma patients. J Exp Med. 2010;207(10):2175–2186. doi:10.1084/jem.20100637
43. Anfray C, Ummarino A, Andón FT, Allavena P. Current strategies to target tumor-associated-macrophages to improve anti-tumor immune responses. Cells. 2020;9(1):46.
44. Belgiovine C, Digifico E, Anfray C, Ummarino A, Torres andón F. Targeting tumor-associated macrophages in anti-cancer therapies: convincing the traitors to do the right thing. J Clin Med. 2020;9(10):3226.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.