Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 14
No Association Between ADIPOQ or MTHFR Polymorphisms and Gestational Diabetes Mellitus in South African Women
Authors Dias S , Adam S , Rheeder P , Pheiffer C
Received 27 November 2020
Accepted for publication 16 January 2021
Published 24 February 2021 Volume 2021:14 Pages 791—800
DOI https://doi.org/10.2147/DMSO.S294328
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Konstantinos Tziomalos
Stephanie Dias,1,2 Sumaiya Adam,2 Paul Rheeder,3 Carmen Pheiffer1,4
1Biomedical Research and Innovation Platform (BRIP), South African Medical Research Council, Cape Town, 7505, South Africa; 2Department of Obstetrics and Gynecology, University of Pretoria, Pretoria, 0001, South Africa; 3Department of Internal Medicine, Faculty of Health Sciences, University of Pretoria, Pretoria, 0001, South Africa; 4Division of Medical Physiology, Faculty of Health Sciences, Stellenbosch University, Cape Town, 7505, South Africa
Correspondence: Carmen Pheiffer
Biomedical Research and Innovation Platform (BRIP), South African Medical Research Council, Tygerberg, 7505, South Africa
Tel +27 21 938 0292
Email [email protected]
Purpose: Gestational diabetes mellitus (GDM) is a growing public health concern. GDM affects approximately 14% of pregnancies globally, and without effective treatment, is associated with short- and long-term complications in mother and child. Lower serum adiponectin (ADIPOQ) concentrations and aberrant DNA methylation have been reported during GDM. The aim of this study was to investigate the association between the ADIPOQ − 11377C>G and − 11391G>A, and methylenetetrahydrofolate reductase (MTHFR) 677C>T polymorphisms and GDM in a population of black South African women.
Materials and Methods: DNA was isolated from the peripheral blood of 447 pregnant women with (n=116) or without (n=331) GDM, where after ADIPOQ (rs266729 and rs17300539) and MTHFR (rs1801133) polymorphisms were genotyped using TaqMan Quantitative Real-Time PCR analysis.
Results: Women with GDM had a higher body mass index (p=0.012), were more insulin resistant (p< 0.001) and had lower adiponectin levels (p=0.013) compared to pregnant women with normoglycemia. Genotypic, dominant and recessive genetic models showed no association between ADIPOQ rs266729 and rs17300539 and MTHFR rs1801133 polymorphisms and GDM. Intriguingly, the risk G allele of ADIPOQ rs266729 was associated with higher fasting glucose and insulin concentrations, while the T allele in MTHFR rs1801133 was associated with higher fasting insulin concentrations only.
Conclusion: ADIPOQ rs266729 and rs17300539 and MTHFR rs1801133 polymorphisms are not associated with GDM in a population of black South African women. These findings suggest that these single nucleotide polymorphisms (SNPs) do not individually increase GDM risk in the African population. However, the role of these SNPs in possible gene-gene or gene-environment interactions remain to be established.
Keywords: SNP genotyping, molecular biomarkers, adiponectin, ADIPOQ, methylenetetrahydrofolate reductase, MTHFR, gestational diabetes mellitus, GDM
Introduction
Gestational diabetes mellitus (GDM), is defined as glucose intolerance that develops during pregnancy and usually returns to normoglycemia after birth.1 Globally, it is estimated that approximately 14% of pregnancies are complicated by GDM,2 although the prevalence varies between <1% and 28% according to the population studied and the diagnostic criteria employed.3 GDM is associated with adverse perinatal outcomes such as pre-eclampsia, caesarean section, fetal macrosomia, shoulder dystocia, hyperinsulinemia, hypoglycemia, hyperbilirubinemia and respiratory distress syndrome,4–8 and an increased risk of developing future metabolic disease such as obesity, type 2 diabetes (T2D), and cardiovascular disease in both mother and child in later life.9–13 Although the etiology of GDM is not yet fully elucidated, it is widely accepted that factors such as age, high body mass index (BMI), excessive gestational weight gain, family history of diabetes mellitus, previous pregnancies complicated by GDM and genetic polymorphisms increase susceptibility to GDM.14,15
Accumulating studies report that polymorphisms in genes involved in metabolic adaptation during pregnancy may increase the risk for developing GDM. At least 34 single nucleotide polymorphisms (SNPs) were reported to be associated with GDM in at least two populations, with variants in the adiponectin (ADIPOQ) gene widely investigated during GDM.16 The expression of ADIPOQ, an adipocyte-derived hormone, is decreased during pregnancy and is associated with the development of insulin resistance and glucose intolerance.17–19 Accordingly, ADIPOQ is intensely researched as a biomarker to predict the risk of developing GDM.20 SNPs within the ADIPOQ gene may regulate adiponectin expression during pregnancy and increase the risk for GDM.21 To date, three ADIPOQ SNPs have been investigated during GDM. The rs266729 (−11377C>G) variant in the promoter region of ADIPOQ has been associated with an increased risk of GDM in Polish, Bulgarian and Asian populations, and a decreased risk of developing GDM in American populations.22–25 In a recent meta-analysis conducted in 12 studies, the rs2241766 (45T>G) variant in exon 2 of ADIPOQ were reported to be associated with an increased risk of GDM in Iranian, Malaysian, Brazilian and Asian populations,26 while variant rs1501299 (276G>T) in intron 2 of ADIPOQ was not associated with GDM in Polish, Bulgarian and Asian populations.23,24,26
Aberrant DNA methylation is associated with the development of GDM.27–31 DNA methylation is the most widely studied and best characterized epigenetic mechanism, that occurs due to the addition of a methyl group to the fifth carbon position of a cytosine nucleotide, generally leading to transcriptional repression.32,33 Methylenetetrahydrofolate reductase (MTHFR) is an enzyme in the transmethylation pathway and plays a critical role in regulating DNA methylation in response to environmental cues.34,35 Two MTHFR polymorphisms, 677C>T (rs1801133) and 1298A>C (rs1801131), has been shown to impair enzyme function and consequently dysregulate DNA methylation.36 These polymorphisms are widely studied during metabolic disease,37–39 and has been associated with insulin resistance,40 and an increased risk of developing cardiovascular disease,41,42 T2D43 and major depressive disorder44 in South African populations. Recently, the MTHFR rs1801133 polymorphism has been linked to higher folate concentrations in early pregnancy and an increased risk of developing GDM in Chinese women,45 and has been associated with GDM-related pregnancy complications such as gestational hypertension, pre-eclampsia and intrauterine fetal growth restriction.46,47
This study aimed to investigate the relationship between the ADIPOQ −11377C>G and −11391G>A, and MTHFR 677C>T polymorphisms and GDM in a population of South African women. We hypothesized that polymorphisms in these genes may underlie the differences in adiponectin expression48 and DNA methylation31 previously reported in this population. To our knowledge this is the first study to investigate the association between ADIPOQ and MTHFR polymorphisms and GDM in an African population.
Materials and Methods
Study Participants
Ethical approval for this study was granted by the University of Pretoria Health Sciences Ethics Committee (180/2012). The study was conducted according to the Declaration of Helsinki and all women gave written informed voluntary consent after the procedures had been fully explained in the language of their choice. This case-control study is nested within a prospective cohort study where 1000 pregnant women were recruited at a primary care clinic in Johannesburg, South Africa.49 Black African women with singleton pregnancies, who did not have pre-existing diabetes (type 1 (T1D) and T2D) were enrolled in the study. Random glucose and glycated hemoglobin (HbA1c) concentrations were measured in all participants. Women with random glucose and HbA1c concentrations >11.1 mmol/L and >6.5%, respectively, were excluded. Women who were included were asked to return in a fasted state for GDM testing and blood collection within 2 weeks.
Anthropometric, Biochemical and Clinical Data on Study Participants
At recruitment, age, gestational age (weeks), height (cm) and weight (kg) were obtained using standard procedures, and BMI was calculated as weight (kg)/height squared (m2). GDM was diagnosed using the 75-g 2-hr oral glucose tolerance test (OGTT) at 24–28 weeks of pregnancy according to the International Association of Diabetes in Pregnancy Study Group (IADPSG) criteria, and diagnosed if at least one glucose value was met (fasting plasma glucose ≥ 5.1mmol/L, 1 hr OGTT ≥ 10 mmol/L or 2 hr OGTT ≥ 8.5 mmol/L).50 Serum and whole blood samples were collected from participants in serum separator tubes (SST) and ethylenediaminetetraacetic acid (EDTA) tubes and stored at −80 °C until further analysis. To assess the relationship between GDM and inflammation in these women, C-reactive protein (CRP) levels were measured. CRP and insulin concentrations were measured using the turbidimetric and microparticle enzyme immunoassays (AxSYM, Abbott), respectively, in an accredited laboratory (Vermaak and Partners/Pathcare laboratories, South Africa), while adiponectin concentrations were measured using the human adiponectin enzyme-linked immunosorbent assay (ELISA) (Merck, Darmstadt, Germany). The homeostatic model assessment (HOMA), a measure of insulin resistance, was calculated using the equation: (fasting plasma glucose x fasting serum insulin)/22.5.
DNA Extraction and Genotyping
Genomic DNA was extracted from 2 mL of whole blood, using the QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) as previously described.51 DNA concentration was measured using the Qubit Fluorometer and the Quanti-iT dsDNA Broad Range assay kit (ThermoFisher, Massachusetts, USA). Genotyping was conducted using quantitative Real-Time PCR (qRT-PCR) with TaqMan genotyping assays52 (Table 1) on the QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems, California, USA). The Quantstudio 7 Real-Time PCR software v1.3 analysis tool was used for base-calling and visualization of the genotyping data (Supplementary Figure S1). Briefly, qRT-PCR was performed using 9.5 ng of DNA, 5 µL of TaqPath ProAmp Master Mix and 0.25 µL of 40X TaqMan SNP Genotyping Assay in a total volume of 10 µL, according to the manufacturer’s instructions (Applied Biosystems). The following PCR conditions were used: 10 min at 95 °C (initial denaturation/enzyme activation), 15 sec at 95 °C (denaturation) and 60 sec at 60 °C (annealing/extension) for 40 cycles. For quality control, 20% of samples were randomly selected and genotyped in duplicate. Positive and negative controls were included on all plates. Nine samples were randomly selected, and genotyping validated by DNA sequencing (Central Analytical Facilities, Cape Town, South Africa). Details of sequencing primers are shown in Supplementary Table S1. Primers for sequencing were designed on NCBI using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast). The minor allele frequency (MAF) for all SNPs were obtained from Ensemble 1000 genomes project (http://www.ensembl.org/Homo_sapiens/Info/Index). The African (AFR) MAF was determined using published data from seven sub-populations, including Yoruba in Ibadan, Nigeria, Luhya in Webuye, Kenya, Gambian in Western Divisions in the Gambia, Mende in Sierra Leone, Esan in Nigeria, Americans of African Ancestry in SW USA and African Caribbean in Barbados.
 |
Table 1 Details of ADIPOQ rs266729 and rs17300539 and MTHFR rs1801133 Single Nucleotide Polymorphisms Assays |
Statistical Analysis
Participant characteristics are expressed as the median and interquartile range (25th and 75th percentiles) since data were skewed. Testing for normality was conducted using the Shapiro–Wilk test in STATA 14 (StataCorp, College Station, USA). Univariable and multivariable logistic regression were performed to assess the association between genotype and participant characteristics. Data are expressed as the odds ratio (OR) and 95% confidence interval (CI), and were adjusted for confounding factors age, BMI and gestational age. The ADIPOQ rs266729 and rs17300539, and MTHFR rs1801133 genotype and allele frequencies, and the dominant and recessive models of inheritance were compared in GDM and non-GDM groups, using the Chi-squared (X2) test or Fisher’s exact test (frequency < 5). A p ≤ 0.05 was considered statistically significant. The Pearson’s X2 test was performed to determine whether the genotype frequencies at ADIPOQ rs266729 and rs17300539 and MTHFR rs1801133 were in Hardy-Weinberg equilibrium (HWE) (p > 0.05).
Results
Clinical and Biochemical Data
The clinical and biochemical data of the study participants are shown in Table 2. BMI (p=0.012), random (p<0.001) and fasting (p<0.001) glucose concentrations, 1 hr (p<0.001) and 2 hr (p<0.001) OGTT values, fasting insulin (p=0.03) and HbA1c (p=0.005) concentrations, and HOMA (p<0.001) were higher in women with GDM compared to women with normoglycemia, while gestational age (p=0.007) and serum adiponectin concentrations (p=0.013) were lower in women with GDM. CRP levels did not differ between women with GDM compared to women with normoglycemia.
 |
Table 2 Participant Characteristics According to GDM Status |
ADIPOQ Genotype Distribution and Association with GDM
The genotype and allele frequency distribution for ADIPOQ rs266729 and rs17300539 did not differ in women with GDM nor in women with normoglycemia (Table 3). The genotype frequency distribution of the ADIPOQ rs17300539 polymorphism was in accordance with HWE (Chi squared = 0.009; p=0.924), while the ADIPOQ rs266729 polymorphisms deviated from HWE (Chi squared = 24.518; p<0.001). Both dominant and recessive genetic models showed that the rs266729 and rs17300539 polymorphisms were not associated with GDM (Table 4). The number of women with the GG genotype (rs266729) and the AA genotype (rs17300539) were low or not observed at all, therefore the heterozygous and homozygous genotypes, CG+GG and GA+AA for rs266729 and rs17300539, respectively, were combined for further analysis. Regression analysis showed that the rs266729 CG+GG genotypes were associated with higher fasting glucose concentrations in women with normoglycemia (p=0.04) and higher fasting insulin concentrations in all women (p=0.004) and in women with GDM (p=0.009), while no association was observed for rs17300539 (Table 5). The association between rs266729 and fasting glucose and insulin concentrations remained significant after adjusting for age, BMI and gestational age. The rs17300539 GA+AA genotypes were not observed in women with GDM, therefore logistic regression could not be conducted.
 |
Table 3 Genotype and Allele Frequency of ADIPOQ rs266729 and rs17300539 Polymorphisms in GDM and Non-GDM Groups |
 |
Table 4 Association Between ADIPOQ rs266729 and rs17300539 Polymorphisms and GDM in Dominant and Recessive Genetic Models |
 |
Table 5 Participant Characteristics According to ADIPOQ rs266729 and rs17300539 Genotype Carriers |
MTHFR Genotype Distribution and Association with GDM
The genotype and allele frequency for MTHFR rs1801133 did not differ in women with GDM nor in women with normoglycemia (Table 6). The genotype frequency distribution of MTHFR rs1801133 polymorphism deviated from HWE (Chi-squared = 71.934; p<0.001). Both dominant and recessive genetic models showed that the rs1801133 polymorphism was not associated with GDM (Table 7). The number of women with the rs1801133 TT genotype was low, thus, heterozygous and homozygous genotypes, CT+TT were combined for further analysis. Regression analysis showed that the rs1801133 CT+TT genotypes were associated with higher fasting insulin concentrations in all women (p=0.04) and in women with normoglycemia (p=0.04) (Table 8). However, after adjusting for age, BMI and gestational age, only the association between rs1801133 CT+TT genotype and fasting insulin concentrations in women with normoglycemia remained significant. MTHFR genotypes were not associated with global DNA methylation levels (Supplementary Table S2).
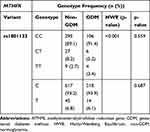 |
Table 6 Genotype and Allele Frequency of MTHFR rs1801133 Polymorphisms in GDM and Non-GDM Groups |
 |
Table 7 Association Between MTHFR rs1801133 Polymorphisms and GDM in Dominant and Recessive Genetic Models |
 |
Table 8 Participant Characteristics According to MTHFR rs1801133 Genotype Carriers |
Discussion
This study investigated the association between ADIPOQ and MTHFR polymorphisms, and GDM in a population of black South African women. Results of this study showed that women with GDM had a higher body mass index, were more insulin resistant and had lower adiponectin levels compared to pregnant women with normoglycemia. Genotypic, dominant and recessive genetic models showed no association between ADIPOQ rs266729 and rs17300539 and MTHFR rs1801133 polymorphisms and GDM in this population. Intriguingly, the risk G allele in ADIPOQ rs266729 was associated with higher fasting glucose in women with normoglycemia, and higher fasting insulin in all women and in women with GDM, while the T allele in MTHFR rs1801133 was associated with higher fasting insulin, in women with normoglycemia.
Studies investigating the association between ADIPOQ rs266729 polymorphisms and GDM have reported conflicting results. Consistent with our findings, Gueuvoghlanian-Silva et al, found no association between ADIPOQ rs266729 and GDM in a sample of 248 pregnant Brazilian women.53 However, these authors found that the CC genotype was associated with higher serum adiponectin concentrations, whereas this association was not demonstrated in our study. Contradictory to our study, four studies reported that the ADIPOQ rs266729 polymorphism is associated with GDM. The G allele was associated with GDM in studies conducted in 130 Chinese women,22 411 Polish women,24 and 562 Iranian women.54 In contrast to these findings, Beltcheva et al reported that the C allele was associated with GDM in a Bulgarian population.23 Discrepancies between these studies may be due to BMI, stage of pregnancy, sample size and ethnicity, which are critical limitations of GDM associated genetic studies.25 Well-designed, large scale, studies that are conducted in diverse populations are required to further explore the role of this polymorphism in the development of GDM. Moreover, different risk allele frequencies of SNPs could further explain the differences observed between studies. As such, the rs266729 G allele occurs at a lower frequency in Africans (9.0%) compared to Europeans (28.1%), East Asians (27.6%), South Asians (28.4%) and Americans (24%). However, in our sample of 447 black South African women, the G allele occurred at a frequency of 22.1% (99 women), a frequency almost double than previously reported in the African population. These result suggest that the G allele in rs266729 may not be a significant risk factor for GDM in the South African population. GDM is a multifactorial disorder that occurs due to the interplay between genetic and environmental factors, thus, the interaction between diet, physical activity, smoking, alcohol consumption and genetics may influence susceptibility to GDM.55–57 Although, many studies have investigated the association between ADIPOQ rs17300539 polymorphisms and metabolic disease,58–62 ours is the first to investigate this polymorphism during GDM. The A allele was associated with T2D in French58 and German59 Caucasian populations, however, studies in Pakistani60 and African American populations,61 failed to show an association between the rs17300539 polymorphisms and T2D. Interestingly, Olckers et al showed that the G allele was associated with T2D in a black South African population.62 Although T2D and GDM are both associated with insulin resistance and glucose intolerance, the pathophysiologic mechanisms that underlie these conditions during pregnancy may differ, possibly explaining the differences observed between our study and Olckers et al.62
The MTHFR rs1801133 polymorphism was not associated with GDM in our study and it is consistent with findings by Khan et al and Franzago et al who similarly failed to see an association between rs1801133 and GDM in a South Indian63 and Italian population,64 respectively. Previously, the T allele was shown to be associated with HOMA and serum insulin concentrations in non-pregnant Iranian women at high risk of developing insulin resistance,40 and with Chinese participants at risk of metabolic syndrome.39 In our study, the T allele was associated with higher fasting insulin concentrations. Thus, these results suggest that MTHFR polymorphism may be associated with pathophysiological mechanisms underlying the development of GDM. However, further work is required to explore the potential mechanism associated with MTHFR polymorphisms and serum insulin levels in the South African population.
To the best of our knowledge, this is the first study to investigate the association between the ADIPOQ rs266729 and rs17300539, and MTHFR rs1801133 polymorphisms and GDM in a South African population. A strength of our study is that GDM was diagnosed using the IADPSG criteria,50 which is widely advocated to improve diagnosis of GDM. Women were recruited at a primary health care facility, which supports generalizability of our study findings to the community. Furthermore, genotyping results were validated by DNA sequencing, minimizing the possibility of genotyping error. However, a few limitations should be considered when interpreting the study results. Due to limited serum samples and the cross-sectional nature of the study, lipid profiles and gestational weight gain during and after pregnancy,65,66 known to affect GDM risk, were not assessed in this study, and may have been associated with ADIPOQ polymorphisms. Our sample size was moderate, and although it is larger than many previous studies investigating SNPs during GDM, a lower risk allele frequency compared to previous studies may have led to our study being underpowered to detect significant associations between the investigated SNPs and GDM.67 Replication of this analysis in a larger sample size is required to definitively rule out the association between the investigated polymorphisms and GDM. Furthermore, genotype frequencies of ADIPOQ rs266729 and rs1801133 deviated from HWE, suggesting that these SNPs may be under possible selection pressure. We recommend that technologies such as the H3A array, which contains SNPs specific to the African population be conducted to improve the ability to detect genetic susceptibility loci for genetic association studies in the African population. Importantly, gene-gene and gene-environment interactions68 may also contribute to the risk of GDM and should be accounted for in genetic association studies.55–57
Conclusion
ADIPOQ rs266729 and rs17300539 and MTHFR rs1801133 polymorphisms are not associated with GDM in a population of black South African women. These findings suggest that these SNPs do not individually increase GDM risk in the African population. However, the role of these SNPs in possible gene-gene or gene-environment interactions remain to be established.
Abbreviations
GDM, gestational diabetes mellitus; ADIPOQ, adiponectin gene; MTHFR, methylenetetrahydrofolate reductase gene; SNP, single nucleotide polymorphism; SA, South Africa; T2D, type 2 diabetes; T1D, type 1 diabetes; HbA1c, glycated hemoglobin; OGTT, oral glucose tolerance test; IADPSG, International Association of Diabetes in Pregnancy Study Group; CRP, c-reactive protein; HOMA, homeostatic model assessment; ELISA, enzyme-linked immunosorbent assay; EDTA, ethylenediaminetetraacetic acid; HWE, Hardy-Weinberg Equilibrium; BMI, body mass index.
Data Sharing Statement
The data analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
The authors are grateful to the study subjects who voluntarily participated in the study, and would like to thank Prof Craig Kinnear, an associate professor at Stellenbosch University and specialist scientist at the South African Medical Research Council, for his assistance with the data analysis in the final revised version.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was funded by the National Research Foundation, South Africa (Unique Grant no. 99391) (SD) and the South Africa Medical Research Council (CP).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Metzger BE, Buchanan TA, Coustan DR, et al. Summary and recommendations of the fifth international workshop-conference on gestational diabetes mellitus. Diabetes Care. 2007;30(Supplement 2):S251–S260. doi:10.2337/dc07-s225
2. International Diabetes Federation. IDF diabetes atlas - Across the globe; 2017. Available from: http://diabetesatlas.org/across-the-globe.html.
3. Jiwani A, Marseille E, Lohse N, et al. Gestational diabetes mellitus: results from a survey of country prevalence and practices. J Matern Fetal Neonatal Med. 2012;25(6):600–610. doi:10.3109/14767058.2011.587921
4. Hadar E, Hod M. Maternal complications of GDM. Diapedia. 2013. doi:10.14496/dia.41040851413.13
5. Moore LE. Fetal and neonatal consequences of maternal diabetes. In: Moore LE, editor. Diabetes in Pregnancy: The Complete Guide to Management. Cham: Springer International Publishing; 2018:7–16.
6. Hod M, Merlob P, Friedman S, et al. Gestational diabetes mellitus: a survey of perinatal complications in the 1980s. Diabetes. 1991;40(Supplement_2):74–78. doi:10.2337/diab.40.2.S74
7. Soufizadeh N, Farhadifar F, Soufizadeh N, et al. Fetal macrosomia: risk factors, maternal, and perinatal outcome. Ann Med Health Sci Res. 2013;3(4):546–550. doi:10.4103/2141-9248.122098
8. Young BC, Ecker JL. Fetal macrosomia and shoulder dystocia in women with gestational diabetes: risks amenable to treatment? Curr Diab Rep. 2013;13(1):12–18. doi:10.1007/s11892-012-0338-8
9. Kim C, Newton KM, Knopp RH. Gestational diabetes and the incidence of type 2 diabetes: a systematic review. Diabetes Care. 2002;25(10):1862–1868. doi:10.2337/diacare.25.10.1862
10. Clausen TD, Mathiesen ER, Hansen T, et al. High prevalence of type 2 diabetes and pre-diabetes in adult offspring of women with gestational diabetes mellitus or type 1 diabetes: the role of intrauterine hyperglycemia. Diabetes Care. 2008;31(2):340–346. doi:10.2337/dc07-1596
11. Zhao P, Liu E, Qiao Y, et al. Maternal gestational diabetes and childhood obesity at age 9–11: results of a multinational study. Diabetologia. 2016;59(11):2339–2348. doi:10.1007/s00125-016-4062-9
12. Bellamy L, Casas J-P, Hingorani AD, et al. Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet. 2009;373(9677):1773–1779. doi:10.1016/S0140-6736(09)60731-5
13. Poon LC, McIntyre HD, Hyett JA, et al. The first-trimester of pregnancy - A window of opportunity for prediction and prevention of pregnancy complications and future life. Diabetes Res Clin Pract. 2018;145:20–30. doi:10.1016/j.diabres.2018.05.002
14. Pons RS, Rockett FC, de Almeida Rubin B, et al. Risk factors for gestational diabetes mellitus in a sample of pregnant women diagnosed with the disease. Diabetol Metab Syndr. 2015;7(S1):A80. doi:10.1186/1758-5996-7-S1-A80
15. Wu L, Cui L, Tam WH, et al. Genetic variants associated with gestational diabetes mellitus: a meta-analysis and subgroup analysis. Sci Rep. 2016;6(1):30539. doi:10.1038/srep30539
16. Dias S, Pheiffer C, Abrahams Y, et al. Molecular Biomarkers for Gestational Diabetes Mellitus. Int J Mol Sci. 2018:19. doi:10.3390/ijms19102926.
17. Mohammadi T, Paknahad Z. Adiponectin concentration in gestational diabetic women: a case-control study. Clin Nutr Res. 2017;6(4):267–276. doi:10.7762/cnr.2017.6.4.267
18. Worda C, Leipold H, Gruber C, et al. Decreased plasma adiponectin concentrations in women with gestational diabetes mellitus. Am J Obstet Gynecol. 2004;191(6):2120–2124. doi:10.1016/j.ajog.2004.04.038
19. Bozkurt L, Göbl CS, Baumgartner-Parzer S, et al. Adiponectin and Leptin at Early Pregnancy: association to Actual Glucose Disposal and Risk for GDM-A Prospective Cohort Study. Int J Endocrinol. 2018;2018:5463762. doi:10.1155/2018/5463762
20. Lobo TF, Torloni MR, Mattar R, et al. Adipokine levels in overweight women with early-onset gestational diabetes mellitus. J Endocrinol Invest. 2019;42(2):149–156. doi:10.1007/s40618-018-0894-0
21. Smetnev S, Klimushina M, Kutsenko V, et al. Associations of SNPs of the ADIPOQ Gene with Serum Adiponectin Levels, Unstable Angina, and Coronary Artery Disease. Biomolecules. 2019;9(10):9. doi:10.3390/biom9100537
22. Liang Z, Dong M, Cheng Q, et al. Gestational diabetes mellitus screening based on the gene chip technique. Diabetes Res Clin Pract. 2010;89(2):167–173. doi:10.1016/j.diabres.2010.04.001
23. Beltcheva O, Boyadzhieva M, Angelova O, et al. The rs266729 single-nucleotide polymorphism in the adiponectin gene shows association with gestational diabetes. Arch Gynecol Obstet. 2014;289(4):743–748. doi:10.1007/s00404-013-3029-z
24. Pawlik A, Teler J, Maciejewska A, et al. Adiponectin and leptin gene polymorphisms in women with gestational diabetes mellitus. J Assist Reprod Genet. 2017;34(4):511–516. doi:10.1007/s10815-016-0866-2
25. Bai Y, Tang L, Li L, et al. The roles of ADIPOQ rs266729 and MTNR1B rs10830963 polymorphisms in patients with gestational diabetes mellitus: a meta-analysis. Gene. 2020;730:144302. doi:10.1016/j.gene.2019.144302
26. Huang Q, Wang Y, Gu B, et al. Whether the risk of gestational diabetes mellitus is affected by TNF-α, IL-6, IL-10 or ADIPOQ polymorphisms: a meta-analysis. Diabetol Metab Syndr. 2020;12(1):1–8. doi:10.1186/s13098-020-00582-8
27. Enquobahrie DA, Moore A, Muhie S, et al. Early pregnancy maternal blood DNA methylation in repeat pregnancies and change in gestational diabetes mellitus status-A pilot study. Reproduct Sci. 2015;22(7):904–910. doi:10.1177/1933719115570903
28. Kang J, Lee C-N, Li H-Y, et al. Genome-wide DNA methylation variation in maternal and cord blood of gestational diabetes population. Diabetes Res Clin Practice. 2017;132:127–136. doi:10.1016/j.diabres.2017.07.034
29. Kang J, Lee C-N, Li H-Y, et al. Association of Interleukin-10 Methylation Levels With Gestational Diabetes in a Taiwanese Population. Front Genet. 2018;9:222. doi:10.3389/fgene.2018.00222
30. Wu P, Farrell WE, Haworth KE, et al. Maternal genome-wide DNA methylation profiling in gestational diabetes shows distinctive disease-associated changes relative to matched healthy pregnancies. Epigenetics. 2018;13(2):122–128. doi:10.1080/15592294.2016.1166321
31. Dias S, Adam S, Rheeder P, et al. Altered genome-wide DNA methylation in peripheral blood of South African women with gestational diabetes mellitus (In press). Int J Mol Sci. 2020.
32. Christensen BC, Marsit CJ. Epigenomics in Environmental Health. Front Genet. 2011;2:2. doi:10.3389/fgene.2011.00084
33. Ling C, Rönn T. Epigenetics in human obesity and type 2 diabetes. Cell Metab. 2019;29(5):1028–1044. doi:10.1016/j.cmet.2019.03.009
34. Goyette P, Sumner JS, Milos R, et al. Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat Genet. 1994;7(2):195–200. doi:10.1038/ng0694-195
35. Froese DS, Huemer M, Suormala T, et al. Mutation Update and Review of Severe Methylenetetrahydrofolate Reductase Deficiency. Hum Mutat. 2016;37(5):427–438. doi:10.1002/humu.22970
36. Lievers KJ, Boers GH, Verhoef P, et al. A second common variant in the methylenetetrahydrofolate reductase (MTHFR) gene and its relationship to MTHFR enzyme activity, homocysteine, and cardiovascular disease risk. J Mol Med. 2001;79(9):522–528. doi:10.1007/s001090100253
37. Maeda M, Fujio Y, Azuma J. MTHFR gene polymorphism and diabetic retinopathy. Curr Diabetes Rev. 2006;2(4):467–476. doi:10.2174/1573399810602040467
38. Chen A-R, Zhang H-G, Wang Z-P, et al. C-reactive protein, vitamin B12 and C677T polymorphism of N-5,10-methylenetetrahydrofolate reductase gene are related to insulin resistance and risk factors for metabolic syndrome in Chinese population. Clin Invest Med. 2010;33(5):E290. doi:10.25011/cim.v33i5.14354
39. Yang B, Fan S, Zhi X, et al. Associations of MTHFR C677T and MTRR A66G Gene Polymorphisms with Metabolic Syndrome: a Case-Control Study in Northern China. Int J Mol Sci. 2014;15(12):21687–21702. doi:10.3390/ijms151221687
40. Kheradmand M, Maghbooli Z, Salemi S, et al. Associations of MTHFR C677T polymorphism with insulin resistance, results of NURSE Study (Nursing Unacquainted Related Stress Etiologies). J Diabetes Metab Disord. 2017;16:16. doi:10.1186/s40200-017-0303-9
41. Ramkaran P, Phulukdaree A, Khan S, et al. Methylenetetrahydrofolate reductase C677T polymorphism is associated with increased risk of coronary artery disease in young South African Indians. Gene. 2015;571(1):28–32. doi:10.1016/j.gene.2015.06.044
42. Scholtz CL, Odendaal HJ, Thiart R, et al. Analysis of two mutations in the MTHFR gene associated with mild hyperhomocysteinaemia-Hetergenous distribution in the South African population. South African Med J. 2002;92(6):464–467.
43. Matsha TE, Pheiffer C, Mutize T, et al. Glucose Tolerance, MTHFR C677T and NOS3 G894T Polymorphisms, and Global DNA Methylation in Mixed Ancestry African Individuals. J Diabetes Res. 2016;2016:1–8. doi:10.1155/2016/8738072
44. Delport D, Schoeman R, van der Merwe N, et al. Significance of dietary folate intake, homocysteine levels and MTHFR 677 C>T genotyping in South African patients diagnosed with depression: test development for clinical application. Metab Brain Dis. 2014;29(2):377–384. doi:10.1007/s11011-014-9506-7
45. Liu PJ, Liu Y, Ma L, et al. Associations Between Gestational Diabetes Mellitus Risk and Folate Status in Early Pregnancy and MTHFR C677T Polymorphisms in Chinese Women. Diabetes Metab Syndr Obes. 2020;13:1499–1507. doi:10.2147/DMSO.S250279
46. Del Gobbo GF, Price EM, Hanna CW, et al. No evidence for association of MTHFR 677C>T and 1298A>C variants with placental DNA methylation. Clin Epigenetics. 2018;10(1). doi:10.1186/s13148-018-0468-1
47. Jankovic‐Karasoulos T, Furness DL, Leemaqz SY, et al. Maternal folate, one-carbon metabolism and pregnancy outcomes. Matern Child Nutr. 2021;17(1):e13064. doi:10.1111/mcn.13064
48. Adam S, Pheiffer C, Dias S, et al. Association between gestational diabetes and biomarkers: a role in diagnosis. Biomarkers. 2018;23(4):386–391. doi:10.1080/1354750X.2018.1432690
49. Adam S, Rheeder P. Screening for gestational diabetes mellitus in a South African population: prevalence, comparison of diagnostic criteria and the role of risk factors. South African Med J. 2017;107(6):523–527. doi:10.7196/SAMJ.2017.v107i6.12043
50. Metzger BE, Persson B, Buchanan TA, et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33(7):676–682. doi:10.2337/dc10-0719
51. Dias S, Adam S, Van Wyk N, et al. Global DNA methylation profiling in peripheral blood cells of South African women with gestational diabetes mellitus. Biomarkers. 2019;24(3):225–231. doi:10.1080/1354750X.2018.1539770
52. Alharbi KK, Al-Sulaiman AM, Bin Shedaid MK, et al. MTNR1B genetic polymorphisms as risk factors for gestational diabetes mellitus: a case-control study in a single tertiary care center. Ann Saudi Med. 2019;39(5):309–318. doi:10.5144/0256-4947.2019.309
53. Gueuvoghlanian-Silva BY, Torloni MR, Mattar R, et al. Profile of inflammatory mediators in gestational diabetes mellitus: phenotype and genotype. Am J Reproductive Immunol. 2012;67(3):241–250. doi:10.1111/j.1600-0897.2011.01090.x
54. Nezamzadeh F, Esmailkhani A, Edalati E, et al. Link between single nucleotide polymorphism of rs266729 and rs2241766 in the ADIPOQ gene and gestational diabetes in an Iranian population. Gene Rep. 2019;14:72–75. doi:10.1016/j.genrep.2018.11.009
55. Bao W, Michels KB, Tobias DK, et al. Parental smoking during pregnancy and the risk of gestational diabetes in the daughter. Int J Epidemiol. 2016;45(1):160–169. doi:10.1093/ije/dyv334
56. Khan T, Macaulay S, Norris SA, et al. Physical activity and the risk for gestational diabetes mellitus amongst pregnant women living in Soweto: a study protocol. BMC Womens Health. 2016;16(1). doi:10.1186/s12905-016-0345-z
57. Mijatovic-Vukas J, Capling L, Cheng S, et al. Associations of diet and physical activity with risk for gestational diabetes Mellitus: a systematic review and meta-analysis. Nutrients. 2018;10(6):698. doi:10.3390/nu10060698
58. Vasseur F, Helbecque N, Dina C, et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11(21):2607–2614. doi:10.1093/hmg/11.21.2607
59. Schwarz PEH, Govindarajalu S, Towers W, et al. Haplotypes in the Promoter Region of the ADIPOQ gene are associated with increased diabetes risk in a german caucasian population. Horm Metab Res. 2006;38(7):447–451. doi:10.1055/s-2006-947842
60. Nadeem A, Mumtaz S, Naveed AK, et al. Association of ADIPOQ C-11377G (rs266729), TNF-α αG-308A(rs1800629) and TNF-α αG-238A(rs361525) single nucleotide polymorphisms with type 2 diabetes mellitus in Pakistani population. J Coll Physicians Surg Pak. 2017;27(10):625–630.
61. Bostrom MA, Freedman BI, Langefeld CD, et al. Association of adiponectin gene polymorphisms with type 2 diabetes in an African American population enriched for nephropathy. Diabetes. 2009;58(2):499–504. doi:10.2337/db08-0598
62. Olckers A, Towers GW, Van der Merwe A, et al. Protective effect against type 2 diabetes mellitus identified within the ACDC gene in a black South African diabetic cohort. Metabolism. 2007;56(5):587–592. doi:10.1016/j.metabol.2006.10.004
63. Khan IA, Shaik NA, Kamineni V, et al. Evaluation of gestational diabetes mellitus risk in South Indian women based on MTHFR (C677T) and FVL (G1691A) mutations. Front Pediatr. 2015;3. doi:10.3389/fped.2015.00034
64. Franzago M, Fraticelli F, Marchetti D, et al. Nutrigenetic variants and cardio-metabolic risk in women with or without gestational diabetes. Diabetes Res Clin Pract. 2018;137:64–71. doi:10.1016/j.diabres.2018.01.001
65. Alharbi KK, Khan IA, Eldesouky MH, et al. The genetic polymorphism in the STK11 does not affect gestational diabetes. Acta Biochimica Polonica. 2015;62(3):569–572. doi:10.18388/abp.2015_1025
66. Li G, Kong L, Zhang L, et al. Early pregnancy maternal lipid profiles and the risk of gestational diabetes mellitus stratified for body mass index. Reprod Sci. 2015;22(6):712–717. doi:10.1177/1933719114557896
67. Salanti G, Amountza G, Ntzani EE, et al. Hardy-Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur J Hum Genet. 2005;13(7):840–848. doi:10.1038/sj.ejhg.5201410
68. Shaat N, Groop L. Genetics of gestational diabetes mellitus. Curr Med Chem. 2007;14:569–583. doi:10.2174/092986707780059643
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
