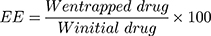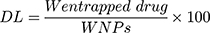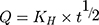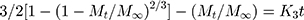Back to Journals » International Journal of Nanomedicine » Volume 16
New Acaciin-Loaded Self-Assembled Nanofibers as MPro Inhibitors Against BCV as a Surrogate Model for SARS-CoV-2
Authors Mohamad SA , Zahran EM , Abdel Fadeel MR, Albohy A , Safwat MA
Received 29 December 2020
Accepted for publication 16 February 2021
Published 2 March 2021 Volume 2021:16 Pages 1789—1804
DOI https://doi.org/10.2147/IJN.S298900
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Soad A Mohamad,1,* Eman Maher Zahran,2,* Maha Raafat Abdel Fadeel,3 Amgad Albohy,4 Mohamed A Safwat5
1Department of Pharmaceutics, Faculty of Pharmacy, Deraya University, Universities Zone, New Minia City, 61111, Egypt; 2Department of Pharmacognosy, Faculty of Pharmacy, Deraya University, Universities Zone, New Minia City, 61111, Egypt; 3Veterinary Serum and Vaccine Research Institute (VSVRI), Cairo, Egypt; 4Department of Pharmaceutical Chemistry, Faculty of Pharmacy, The British University in Egypt (BUE), El-Sherouk City, 1837, Egypt; 5Department of Pharmaceutics, Faculty of Pharmacy, South Valley University, Qena, 83523, Egypt
*These authors contributed equally to this work
Correspondence: Eman Maher Zahran
Deraya University, Universities Zone, New Minia City, 61111, Egypt
Email [email protected]
Background: SARS-COVID-2 has recently been one of the most life-threatening problems which urgently needs new therapeutic antiviral agents, especially those of herbal origin.
Purpose: The study aimed to load acaciin (ACA) into the new self-assembled nanofibers (NFs) followed by investigating their possible antiviral effect against bovine coronavirus (BCV) as a surrogate model for SARS-COV-2.
Methods: ACA was identified using 1H-NMR and DEPT-Q 13C-NMR spectroscopy, the molecular docking study was performed using Autodock 4 and a modification of the traditional solvent injection method was applied for the synthesis of the biodegradable NFs. Different characterization techniques were used to inspect the formation of the NFs, which is followed by antiviral investigation against BCV as well as MTT assay using MDBK cells.
Results: Core/shell NFs, ranging between 80– 330 nm with tiny thorn-like branches, were formed which attained an enhanced encapsulation efficiency (97.5 ± 0.53%, P< 0.05) and a dual controlled release (a burst release of 65% at 1 h and a sustained release up to > 24 h). The antiviral investigation of the formed NFs revealed a significant inhibition of 98.88 ± 0.16% (P< 0.05) with IC50 of 12.6 μM against BCV cells.
Conclusion: The results introduced a new, time/cost-saving strategy for the synthesis of biodegradable NFs without the need for electric current or hazardous cross-linking agents. Moreover, it provided an innovative avenue for the discovery of drugs of herbal origin for the fight against SARS-CoV-2 infection.
Keywords: acaciin, molecular docking, nanofibers, core-shell, BCV/SARS-COV-2
Introduction
The known compound acaciin, isolated from the Lamiaceous plant Ocimum menthiifolium,1,2 is a bioactive flavonoid with reported anti-oxidant and hepatoprotective activities.3 Since several plant-derived flavonoids as apigenin, quercetin, and kaempferol proved diverse antiviral properties,4 acaciin was proposed to follow the same behavior5 and was subsequently investigated against SARS-CoV-2. Recently, the serious viral infection with SARS-CoV-2 has caused a global outbreak of coronavirus illness (SARS-COV-2)6 which needs a development of diagnostic and detection approaches.7,8 Noteworthy, recent strategies for fast determination and monitoring of pathogenic SARS-Cov-2 viruses and diagnosis of virus-induced diseases as DNA-sensors based on DNA-modifying enzymes were reported.9 Coronaviruses are categorized into 4 subfamilies: alpha, beta, gamma and deltacoronaviruses, where SARS-CoV-2 belongs to the beta category.10 Betacoronaviruses maintain their integrity via four proteins: The E (envelop), M (membrane), N (nucleocapsid) and S (spike) proteins.11 The latter with its binding sites helps viral attachment to the host cells via the cellular receptor (ACE2) (a key receptor for coronavirus entry) as well as the epithelial Sialic acid receptors12 on the epithelium of the respiratory tract, thus promoting its clustering and pathogenesis. The second step includes adaptation of the virus to its host by genome encoding which facilitates the expression of the genes that encode necessary accessory proteins. Fortunately, a high similarity percentage was observed between SARS-COV-2 and bovine coronavirus (BCV), both of which belong to betacoronaviruses. Supporting phylogenetic analysis of their genomes13 revealed that the difference was in their hosts, where BCOV causes gastrointestinal and respiratory tract disease in cattle14 while COVID-19 causes the same illness in human, both of which are directed towards the respiratory system.12 They attain the same backbone and binding site sequence of the main protease (Mpro) target,13 which is considered a key protein in their pathogenesis playing a critical role in their replication and maturation. Accordingly, the structural similarity and the common Mpro supported studying BCV as a surrogate model for SARS-CoV-2,15,16 especially with the presence of governmental and ethical issues.17 Our study started with a computer-aided drug design for the supposed antiviral potential of ACA which targeted the Mpro, which was followed by a nanoformulation to improve the bioavailability of the lipophilic ACA. The NFs were fabricated via a modification of the solvent injection method, which is reported here for the first time, depending on the electrostatic interaction between the negatively charged lecithin phospholipid and the positively charged chitosan polysaccharide. The main principle of their choice was due to their biocompatibility, biodegradability and absence of toxicity.18 The process utilizes the self-assembly approach which allows both lecithin and chitosan to assemble spontaneously by electrostatic interactions without using electric current, hazardous organic solvents or cross-linking agents.19 Finally, the ACA-loaded NFs were tested against BCV to reveal their effect on the virus development which depends on the mechanistic hindrance of viral entry to the cells, as well as inhibition of RNA replication.
Thus, the aim of our study is to investigate the anti-SARS-CoV-2 potential of the isolated flavonoid, ACA, loaded on NFs, via a modification of some parameters to enhance morphological and biological properties (Scheme 1). The resultant NFs attained a large surface area to volume ratio, a sustained drug release, a high production rate, a high safety with low cost.
Materials and Methods
Instruments Used
Brüker Avance 400 MHz NMR spectrometer (Germany) was used for NMR analysis. For NF characterization, a microscope with camera Leica, model: DM1000 (US listed microscope) with (LEICA EC3 digital camera), a UV spectrophotometer (Jasco, Japan), a Nano Zeta Sizer (ZS) (Malvern Instruments, Malvern, UK), a particle sizer (Zeta sizer 3000 HAS; Malvern Instruments Ltd, Worcestershire, UK), a rotation viscometer (Brookfield-DVBT) and a DSC 131 evo (SETARAM Inc., France) were utilized. For biological study, a confocal laser scanning microscope (CLSM) (model SP2; Leica, Mannheim, Germany) equipped with an environmental chamber was used.
Plant Material
Aerial parts of O. menthiifolium were collected from the National garden of Jazan in Jazan, KSA. The air-dried aerial parts (500 g) were extracted by maceration with 95% ethanol, followed by concentration under pressure to a syrupy consistency (70 g). It was then suspended in water and successively extracted with petroleum ether, dichloromethane, ethyl acetate and butanol. The butanol fraction (6 g) was chromatographed on silica gel using ethyl acetate:methanol (70:30), followed by sephadex using methanol to give 4 fractions, from which the compound was precipitated, purified and analyzed.
Identification of the Isolated Compound
The compound was subjected to H1 and DEPT-Q-C13 NMR analysis (Figure S1 a, b), and the obtained data were compared to previously reported data,20 as well as previous metabolomic investigation results.2
ADME Analysis
On the basis of canonical SMILES of the selected ligands obtained from PubChem, ADME properties of the studied compound were calculated using online SwissADME program.21 This software computes physicochemical as well as pharmacokinetic properties and the drug-like nature of compounds, to detect their bioavailability via Lipinski’s rule of five.22 The values of the observed properties are presented in Figure S2.
In silico Docking Study
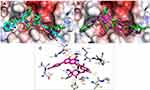
Docking of ACA in the active site of SARS-CoV-2 Mpro was performed using 6LU7 PDB code.23 The grid box used of 25*25*25Å3 was centered on the co-crystalized ligand with exhaustiveness of 16.0, and the 3D images were generated using PyMOL.24 The structure of ACA was downloaded from PubChem and subjected to energy minimization using 1000 steps following the steepest descent method, which was followed by 1000 steps of conjugate gradient algorithms on Avogadro software.24 Water molecules and non-protein residues in each enzyme were removed with the addition of hydrogens using PyMOL, followed by preparation of the proteins using Make Macromolecule command on PyRx.25,26 The RMSD value was reported using DockRMSD server,27 3D images were generated using PyMOL, while the 2D interaction maps were generated using LigPlot Plus.28
Synthesis of Self-Assembled ACA-L/Cs NFs
PVA, CMC, PPA and chitosan were tested with lecithin and the formed nanoformulations were investigated using a light microscope (Figure S3). NFs were formed only with the lecithin/chitosan combination, where nine different formulations, differing in the L/ACA concentration, were prepared according to the method described by29 with some parameter modifications. Lecithin 25, 50 and 70% (w/v) were dissolved in a 1:20 DMSO/water solution, in which ACA (5, 10 and 15%) was dissolved to obtain different weight ratios. The chitosan aqueous solution was prepared by diluting a standard solution of 20% chitosan (w/v) in acetic acid (0.1% v/v), from which 2 mL were slowly injected into 8 mL of ACA/lecithin solution using a metal needle tube. The internal diameter of the syringe was nearly 0.1 mm, about 1–2 mm distant from the collector, where the injection rate was adjusted at 1.8 mL h−1 under mechanical stirring of 1500 rpm for 15 minutes. The resulting suspension was filtered through a filter membrane (0.8 μm) where α-tocopherol was added to the resultant filtrate (Table 1).
 |
Table 1 Composition of Different Formulations of Acaciin-Loaded NFs Nanofibers |
Light Electron Microscopy (LEM)
LEM is used for preliminary examinations of NF materials during manufacturing,30 where the polymer samples can be monitored even in the swollen state, the same as they appear in both in vitro and in vivo experiments.
Characterization of Synthesized ACA-NFs
UV Spectroscopy
The UV absorption spectrum of both ACA/NFs and free ACA was obtained by a spectrophotometer in the range of 200–700 nm at 1.0 nm intervals. Samples were measured in triplicates at 25 °C in a rectangular quartz cuvette with a 1 cm path length.
Zeta Potential and Particle Size Measurement
Zeta potential measures the electrical charges of the particles in nano colloids using a Nano Zeta Sizer through electrophoretic light scattering of the nanoparticles in an aqueous solution. Smoluchowski equation was employed to calculate the Ʒ-potential at 25°C, collecting scattered light at 15° every 10 times for each sample.31 Ʒ-potential values were expressed as mean and standard deviation of the nine measures obtained for the nine different formulations. Dynamic light scattering (DLS) was used to determine the particle size of the NFs using a particle sizer at a fixed angle of 90° with a He-Ne laser of 633 nm at 25°C.
Viscosity
The viscosity of the polymer solutions was determined by a rotation viscometer at room temperature.
Entrapment Efficiency (EE) and Drug Loading (DL)
The NF solution was centrifuged at 4500 rpm for 60 min, then the amount of the entrapped drug in the supernatant liquid was detected using a UV spectrophotometer (Jasco, Japan) at a wavelength of 240 nm. The EE and DL of ACA/NF were calculated according to equations 1 and 2 respectively:
Where W initial drug represents the initially added amount of the drug and W entrapped drug represents the amount of drug entrapped in the NFs. WNP represents the total weight of all components in the NFs.32
DSC Studies of the Free ACA and ACA-NFs
Differential scanning calorimetry (DSC) analysis of ACA, compared to ACA-NFs (1% w/w) was carried out, where the instrument was calibrated using the standards: Mercury, Indium, Tin, Lead, Zinc and Aluminum and employing Nitrogen and Helium as the purging gases. The test (for samples 1–7) was programmed at a heating zone from 25°C to 500°C and a heating rate of 10 °C/min (the heating zone was −50 to 200 °C for samples 8–10). The thermogram results were processed using CALISTO Data processing software v.149.32
In vitro Drug Release
The in vitro release of ACA from the corresponding NFs was performed using the dialysis membrane method.32 One milliliter of NF suspension was transferred into dialysis bags with a molecular mass cutoff of 12,000 Da. The bags were then suspended in 40 mL of ethanol: phosphate buffer saline at the ratio of 3:7 (v/v) at 37°C ± 0.5°C, which was performed in a shaking water bath at pH 6.8 ± 0.2 (JSSB-Series Water Bath) operating at 50 rpm. At time intervals of 0.5, 1, 2, 4, 6, 8, 10, 12 and 24 hr following application, 1 mL of receptor solution was taken out and 1 mL of blank medium at the same temperature was added into the receptor compartment. The results were compared to those obtained from the corresponding suspensions of ACA in DMSO/water (1:10).33 The mechanism of ACA release was determined following the kinetic models:
Hixson–Crowell cube root law: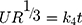 28,38
28,38
Whereas R, Q and Mt/M∞ refer to the fraction of the drug released at time t, K or KH is the rate constant related to each model, UR expresses the unreleased fraction of the drug while n is the diffusional exponent characterizing the release mechanism during the dissolution process.
Scanning Electron Microscopy (SEM)
High-resolution SEM was performed to investigate the NF morphology, which allows the high depth of sharpness about structures at various distances from the scanning level.39 The samples were diluted (1:10) in ultrapure water, then 20 µL of slurries were spread on amorphous polycarbonate grids and left to dry at room temperature. They were extra dried using CO2, sputter coated with gold in a metallizer, then examined under an SEM with accelerating voltage at 200 kV.32
Antiviral Assay
Viruses and Cells
Madin Darby bovine kidney cells (MDBK), obtained from VSVRI institute, were grown in Dulbecco’s modified Eagle’s medium (Gibco, Bethesda, Md.) supplemented with 10% fetal calf serum (Gibco) at 37°C and 5% CO2. The stocks of BCV (strain Mebus) were kindly received from VSVRI Institute. BCV, propagated in MDBK cells,40 supplemented with 10% fetal bovine serum (FBS) by following the standard tissue culture procedure41. The cells were incubated at 37 °C in a humidified incubator with 5% CO2 then sub-cultured at 80–90% confluency.
Infection of Cells
MDBK cells were grown for 2 days in petri dishes with a glass slide bottom (Mattek, Ashland, Mass.) for confocal time-lapse microscopy on coverslips. Dishes, coverslips, and sapphire disks were covered with 8–10 nm thick carbon obtained by evaporation under high-vacuum conditions to enhance cell growth. Then, cells were infected with BCV at a multiplicity of infection (MOI) of 1, 5, and 10 and kept at 4°C for 1 h to admit adsorption prior to incubation at 37°C for up to 48 h.
Confocal Microscopy
Time-lapse microscopy was performed with a confocal laser scanning microscope (CLSM) equipped with an environmental chamber at 37°C and 5% CO2. For nuclear analyses, cells were fixed with 4% paraformaldehyde and stained with DAPI in phosphate-buffered saline (1 μg/mL). Samples were embedded in a fluorescence mounting medium (DakoCytomation, Glostrup, Denmark) and analyzed by CLSM. Images were deconvolved with a blind deconvolution algorithm using the Huygens Essential program suite (SVI, Hilversum, The Netherlands).42
Virus Titration (Infectivity Test)
Conventional micro-titration cell culture assay for titration of the viral cells was performed on MDBK cells. First, cells were seeded in 96-well and then incubated at 37 °C in a humidified atmosphere containing 5% CO2 for 24–48 h. Then, 10 folds serial dilutions of viral stock were prepared, and cells were infected with 100 μL of the virus. After 2 hours, excess virus was removed, then the cytopathic effect was recorded daily for 5–6 days during incubation, and the IC50, CC50 and SI were calculated.
Cytotoxicity Assay of ACA-NFs by MTT (Tetrazolium) Assay
The MTT assay was measured for the ACA-NFs using MDBK cell lines, which is essential for determination of the cytotoxicity of both ACA and ACA-NFs prior to testing their anti-viral potential.43 The test starts with performing a chemical reduction for tetrazole, using mitochondrial enzymes to form the formazan crystals, which were then dissolved by adding acidified alcohol. Then, MDBK cell lines were prepared by trypsinization and suspending in minimal medium using trypan blue as exclusion dye. After dying and cell preparation, they were seeded in a tissue culture plate (96 well) with adjusting cell concentration to 5 × 105 cells/well. The wells were incubated overnight at 37 °C in a humidified incubator with 5% CO2; then ACA and ACA/NFs were added to the wells in different concentrations and incubated at the same conditions for 8–10 h. After ending the incubation period, a 10 μL (5 mg mL−1) MTT reagent was added in each well to complete incubation up to 4 h, then acidified isopropanol (100 μL of 0.1 N) was added to each well and kept for 30 min in dark at room temperature, followed by additional 1 min on a shaker.
Determination of in vitro Antiviral Activity
Determination of the antiviral potential was performed44 by adding a 25 μL of viral suspension (106 TCD50 mL−1) of MDBK cells to 25 μL of different concentrations of ACA-NFs in a PCR tube which was incubated for 1 min at 37 °C. The viral suspension (25 μL 106 TCD50 mL−1) was added to different tissue culture well plates containing MDBK cells with a concentration of 5 × 105 cells mL−1, then the wells were shaken and placed in a CO2 incubator for 1 h. Different concentrations of ACA-NFs were added to the wells and further incubated up to 72 h at 37 °C. Two controls were kept parallel for incubation with treated wells: Triple control containing cells and viral suspension free from NFs and triple control containing only cells free from NFS or viral suspension. Then, detection of antiviral activity was determined by daily inspection of tissue cultures for viral cytopathogenicity and after 72 h applying the MTT assay. The CC50 (cytotoxic concentration for 50% of cells) and IC50 (inhibitory concentration for 50% of infected cells) values were estimated from concentration–effect curves after linear regression analysis. The mean values of three independent experiments as well as the selectivity index (SI=CC50/IC50) were then calculated.45
Statistical Analysis
All results were expressed as mean ± standard deviation and analyzed with one-way analysis of variance followed by a multi-parametric Tukey’s post hoc test in Graph Prism® (GraphPad Software Inc., San Diego, CA), with statistical significance established at p < 0.05.
Results and Discussion
Identification of the Isolated Compound
The compound, isolated for the first time from O. menthiifolium, was identified as acaciin (linarin, acacetin-7-O-rutinoside)20 (Figure 1), and is considered one of the common taxonomic markers of the genus Ocimum, family Lamiaceae.46 Acaciin: off white, solid; 1HNMR (DMSO-d6) ppm: δH 12.9 (1H, br. s, OH-5), 8.04 (2H, d, J = 9.2 Hz, H-2ʹ, H-6ʹ), 7.17 (2H, d, J = 9.2 Hz, H-3ʹ, H-5ʹ), 6.95 (1H, s, H-3), 6.8 (1H, s, H-8), 6.46 (1H, s, H-6), 5.07 (1H, d, J = 9.2 Hz, H-1ʹ’), 4.53 (1H, s, H-1ʹ’’), 3.89 (3H, s, OMe-4ʹ), 3.65 (1H, d, J = 8.5 Hz, H-6ʹ’), 1.08 (3H, d, J = 6.3 Hz, H-6ʹ’’); 13C-NMR (DMSO-d6) ppm: δC 182.5 (C-4), 164.4 (C-7), 161.6 (C-9), 157.5 (C-5), 130.7 (C-6), 128.9 (C-2ʹ,6ʹ), 123.1 (C-1ʹ), 115.2 (C-3ʹ,5ʹ), 105.9 (C-10), 104.3 (C-3), 100.4 (C-1ʹ’), 100.1 (C-1ʹ’’), 95.3 (C-8), 76.1 (C-3ʹ’, C-5ʹ’), 73.5 (C-2ʹ’), 72.5 (C-4ʹ’’), 70.8 (C-2ʹ’’), 70.1 (C-4ʹ’), 68.8 (C-5ʹ’’), 66.5 (C-6ʹ’), 56.1 (OMe-4ʹ), 18.2 (C-6ʹ’’).
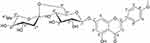 |
Figure 1 Chemical structure of acaciin. |
ADME Analysis
Lipinski’s rule of five states that for any compound to be selected as a potential drug, it cannot have more than 1 violation of the following: (a) Molecular mass < 500 Dalton (b) high lipophilicity (expressed as Log P<5) (c) less than 5 hydrogen bond donors (d) Less than 10 hydrogen bond acceptors. If a compound of interest possesses three or more of the aforementioned criteria then the compound is likely to be a potential candidate for drug development.22 Accordingly, ACA attained a relatively low solubility, inability to cross the blood–brain barrier and absence of any effect on Cytochromes P450. Noteworthy, the aglycon itself lacked violations from Lipinski’s rule of five, but the inclusion of the disaccharide moiety caused some related violations (Figure S2).
In silico Docking Study
ACA was docked against the four main targets of SARS-CoV-2 and attained a docking score of −9.0, −6.7, −7.1 Kcal/mol with nsp16/10, ACE2-PD and RBD-S-protein, respectively, and −9.1 Kcal/mol with Mpro. The internal ligand attained −8.2 and −7.8 Kcal/mol when docked against nsp16/10 and Mpro, respectively. The docking procedure gave a good accuracy with an RMSD value of 1.839 between docked and co-crystalized ligands as calculated by DockRMSD server47 (Figure 2). ACA overlapped with the co-crystalized ligand, followed the same behavior and occupied the same binding pocket with the same interactions (Figure 2), but with a better docking score. It formed H-bonding via the C=O with Ser 144 which is reported as one of the key residues of Mpro,48 and via 5-OH, C=O and 4ʹ-OMe with His163, Leu141 and Thr26, respectively, while the sugar part formed H-bonding with His164, Thr190, Tyr54 and Asp187 (Figure 2). The co-crystalized ligand was reported to interact with most of these residues.49
Nanofibers Formation
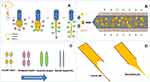
The solvent injection method was applied with modifying some parameters which are expected to affect the shape of the formed nanoformulation (Figure 3). It was observed that when PVA, CMC, PPA and lecithin were tested and microscopically examined, only lecithin formed nanofibers while the other polymers formed nanoparticles (Figure S3). The principle of our new technique for NF formation utilizes the electrostatic interaction between the positively charged chitosan and negatively charged lecithin.50 This resulted in decreasing the surface tension which led to elongation and chain entanglement of the nanoparticle to form a short nanofiber.
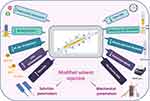 |
Figure 3 Cartoon representation of the modified solvent injection method. |
Parameters Affecting the NFs Formation
The key parameter for manufacturing the NFs (Figure 3) utilizes the electrostatic interaction between chitosan and lecithin as reported,51 that is why the other tested polymers (PVA, CMC and PPA) gave NPs instead of NFs (Figure S3). Accordingly, such interaction generates electrostatic forces which overcome the surface tension of the polymer solution (Figure 4), besides using a metallic nozzle which generated more electrostatic friction, all of which made Self-assembly and phase separation,52 that led to the formation of core/shell fibers (Figure 4). Another parameter which controls the morphology and diameter of fibers is the molecular weight of polymers as it affects rheological properties of the solution as well as the core/shell fiber formation caused by the higher mobility organizing the polymers into a core-shell rather than a continuous structure. Imagining a drop on the tip of the nozzle, the inner polymer is covered with the continuous phase of the other polymer, and when the electrical forces are sufficiently robust, a continuous phase entraps and sucks the drop into a core-shell jet. Noteworthy, core-shell NFs are regarded as one of the most significant breakthroughs in the area of NF formation which significantly affects the dual drug release profiles and reduces to a minimum the direct contact of bioactive agents in an aqueous core solution with the potentially dangerous solvents of a shell, leaving only the core-shell interface.53 Moreover, viscosity is also a parameter; it supports the core/shell formation, where the more viscous phase (acaciin-dissolved lecithin) forms the core, and the less viscous (chitosan) wraps that core forming the shell, that was confirmed with the high positive Ʒ-potential of the surface.54 Furthermore, increasing the L/Cs ratio enhances the high viscosity and subsequently generates extra charges which overcome the surface tension and boost the chain entanglement54 (Figure 4). Additionally, adjusting a minimum flow rate sends less solution through the gap between the needle tip and the collector, which permits longer stretching of the ejected droplets, more solvent evaporation as well as jet formation. Thus, a stable cone jet is formed alternately with an unstable receded jet which leads to variations of the NF diameter (Figure 4).55 Besides, shortening the distance between the needle tip and the collector hinders fibers from too much stretching and slightly shortens the formed fibers and widens their diameter.56 Remarkably, the orifice diameter directly determines the flow of the solution and the initial diameter of the fiber, resulting in a fiber formation when it is adequately narrow.57 Finally, reducing the rotational speed is reported to eliminate beads and achieve a better ductility with a high entrapment efficiency.55 In conclusion, imagining a spherical droplet held in vacuum at the needle tip, accompanied with adjusting the previous parameters, can virtually produce self-assembled, stable and bead-free core/shell short NFs.
Characterization of the Formed NFs
LEM investigation of the nine formulations (Table 1) revealed the formation of short NFs, but formulation 3 attained the best characterization results and was further selected for SEM analysis (Table S1). The significantly high Ʒ-potential of >+50 mV (Figure 5), which is directly proportional to the increased L/Cs ratio and a high drug content, indicates a high stability of the formed NFs. It is reported that the size and polydispersity index (PDI) influence loading, release and stability of the drug inside NFs. The smaller the particle size, lower drug concentration and the lower PDI (≤0.4), the greater is the exposed surface area, causing a faster release of the encapsulated drugs and a lower aggregation.18 Such low PDI value (0.2±0.02, P<0.05) induces a mechanical effect through electrical stabilization,58 as well as a bio-inhibitory effect through interaction between the NFs positive charges and negatively charged surfaces of microbial cell.51 Considering the particle size, it ranged from 80–330 nm which is inversely proportional to both increasing L/Cs ratio and decreasing the drug content being valuable in antimicrobial evaluation, where smaller particle size causes increased uptake of vehicles into microbial cell.51 DSC thermogram (Figure 5) detects any variation of crystalline properties of the loaded aca into the NFs, and it is known that when the drug–polymer interaction is suitable, the presence of the drug does not greatly affect the Tg temperature of the polymer.59 Accordingly, comparing DSC thermogram of pure ACA and ACA-loaded NFs showed relative similarity, with the exothermic band fluctuating around a glass transition temperature (Tg) of 120° and confirming the absence of chemo-physical variations as a result of preparation. When the drug release profile was investigated (Figure 5), it showed a dual-controlled release which is typical for the core/shell structure,60 with initial burst release of 65% after 1 hr, followed by a gradual release extending up to 24 hr showing an excellent release profile. This is expected to achieve a high therapeutic index and a long-lasting biological effect. Besides, the observed high encapsulation efficiency (EE) (>97.3 ± 0.5%, p <0.05) (Figure 5) indicated that a large portion of ACA was preferably inserted into the lipid nuclei of the NFs, and only a small portion was lost in the aqueous phase during preparation process.18
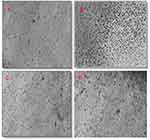
Finally, the SEM examination showed glabrous, bead-free, spindle-shaped, core/shell NFs with a diameter range of 250 nm–3 µm and a length range of 5–20 µm. The morphology of both the plain NFs (Figure 6) and ACA-NFs (Figure 6) was approximately similar with slight widening of the latter as a result of ACA entrapment into the core. Moreover, the previously discussed difference between the stable and receded jets is reported to make some diameter variations,61 illustrated in Figure 6. Furthermore, the core/shell structures showed a shell thickness range of 300–500 nm and a core diameter range of 500 nm–3 µm (Figure 7).
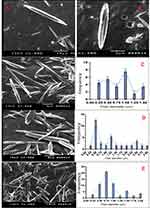 |
Figure 6 Nanofibers of (A) free ACA and (B) ACA-NFs, c-e) express NF diameter variations |
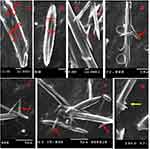 |
Figure 7 (A, B) Different core/shell NFs, (C–F) thorn-like branches of the NFs (red arrows), (G) immature thorn-like branches (yellow arrow). |
Surprisingly, thorn-like branches were obtained on the outer surfaces of the NFs with a width range of 200–400 nm and a length range of 800 nm-2 µm (Figure 7), and with some immature branches of about 200–300 nm width and up to 500 nm length (Figure 7). Such branches are reported to be produced via electrospinning post-temperature treatment62 or successive ionic layer adsorption and reaction.63 They are generally fabricated for supportive purposes to provide a higher surface/volume ratio, a higher mechanical establishment64 as well as improved matrix adhesion due to numerous and sufficient contact areas with the tested microbes in biocidal investigations.63 Obtaining such branches can be tentatively explained by the relatively high temperature and humidity (The operation room temperature was 43° C with 60% humidity).
Antiviral Assay
The possible toxic effects of the nine formulations of ACA-NFs on MBDK cells were assessed via the MTT cell viability assay (the gold standard for cytotoxicity65) as well as microscopic examination.66 The antiviral activity of the ACA-NFs was evaluated based on the ability of ACA to prevent the cytopathic effects (CPE) of the virus on MBDK cell culture and the results were recorded (Table S2). The antiviral activity dramatically increased in a dose-dependent manner up to 78.14%, 87.16%, and 98.88% with concentrations 1, 3 and 6 ppm of ACA-NFs with a minor cytotoxicity of 6.30 ± 4.31%, 5.00 ± 0.023% and 8.3 ± 0.16% (P<0.05), respectively (Figure 8). Increasing the concentration up to 10 ppm had no effect on the activity and slightly increased cytotoxicity up to 9.21 ± 0.01%, P<0.05, all of which was compared to free ACA (12.5 µg/mL) with 75.40% activity, and a marked low toxicity of 10.8 ± 0.1% (P<0.05).
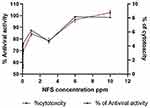 |
Figure 8 A dual-axis diagram of effect of different NF concentrations on the antiviral activity and cytotoxicity. |
CTEM (scanning transmission electron microscope) (Figure 9) showed the plaque variations among the normal cells (a), negative control (b) and the treated cells of both 6 μL/mL ACA-NFs (C) and free ACA (D). The ACA-NFs alleviated the cell damage and reduced plaque formation and cell fusion, which indicated a significant viral inhibition and cell protection up to 48 h. Since it is reported that a promising antiviral activity can be achieved if SI≥4,45 acaciin gave SI of 4.2 (IC50= 12.6 µM, CC50= 52.8 µM) which indicated a favorable antiviral potential.
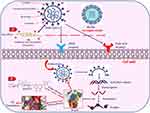
The possible antiviral effect is predicted to be started by electrostatic interaction between the positively charged-coated NFs and the negatively charged lipid bilayer on the microbial envelop (Figure 10). The biological effect might proceed via two pathways: The first is the binding affinity of ACA (predicted by in silico studies) with the Mpro target which inhibited the translation of polyproteins from the viral RNA and interfered with the early stages of viral replication. The second one is the mechanical action of ACA-loaded NFs which inhibited the viral receptor binding via electrostatic interaction and prevented the viral entry to the host cells.
Conclusion
The current study evoked, for the first time, the remarkable antiviral potential of acaciin as a flavonoid isolated from herbal origin. The study produced short core/shell nanofibers with excellent physicochemical properties and with the absence of electric current or toxic cross-linking agents. Our new technique is safe and more favorable than the common methods owing to the usage of well-accepted biodegradable candidates (lecithin and chitosan) in the synthesis of the NFs. Moreover, the application of the self-assembly approach protected the encapsulated drug from harsh stress due to electric current used in common methods as electrospinning. The new NFs attained branched surfaces for good mechanical properties as well as core/shell structures which achieved a dual controlled release, regarded as one of the most significant breakthroughs in the area of NF formation. Our procedure successfully produced uniform NFs with perfect surface characteristics and could save time and money spent in other techniques with additional hazardous steps. Moreover, and from a biological point of view, the obtained NFs could mechanically inhibit the viral entry to the host cells, combined with the Mpro inhibiting activity of ACA itself, all of which could hinder the progress of BCV which has commonly been used as a surrogate model for SARS-COV-2. Our ground-breaking findings provide a deep insight for exploring and developing natural-based antiviral drugs, especially with the incorporation of nanoformulation, with improved mechanical and functional characteristics to support the global battle against current SARS-CoVID-2 disease.
Acknowledgment
The authors are grateful to Dr. Fahad AbdelAziz, Professor of Botany and the head director of the Jazan Garden who authenticated, approved and permitted the plant for work. Deep thanks are also extended to VSVRI Institute for supplying the cell line cells and viral strains for antiviral assay, and to D. Usama Ramadan Abdelmohsen, associate professor of pharmacognosy, Deraya University, Minia, Egypt for kind help.
Disclosure
The authors report no conflict of interest in this work.
References
1. Zahran EM, Abdelmohsen UR, Hussein AS, et al. Antiulcer potential and molecular docking of flavonoids from Ocimum forskolei Benth., family Lamiaceae. Nat Prod Res. 2019;1–5.
2. Zahran EM, Abdelmohsen UR, Shalash MM, et al. Local anaesthetic potential, metabolic profiling, molecular docking and in silico ADME studies of Ocimum forskolei, family Lamiaceae. Nat Prod Res. 2020;1–7.
3. Li Y, Guang C, Zhao N, Feng X, Qiu F. LC–MS/MS method for simultaneous determination of linarin and its metabolites in rat plasma and liver tissue samples: application to pharmacokinetic and liver tissue distribution study after oral administration of linarin. Molecules. 2019;24(18):3342. doi:10.3390/molecules24183342
4. Jo S, Kim S, Shin DH, Kim M-S. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem. 2020;35(1):145–151.
5. Mazraedoost S, Behbudi G, Mousavi SM, Hashemi SA. Covid-19 treatment by plant compounds. Adv Applied NanoBio Technol. 2020;2(1):23–33.
6. Bradburne A, Bynoe M, Tyrrell D. Effects of a” new” human respiratory virus in volunteers. Br Med J. 1967;3(5568):767.
7. Mousavi SM, Hashemi SA, Parvin N, et al. Recent biotechnological approaches for treatment of novel COVID-19: from bench to clinical trial. Drug Metab Rev. 2020;1–30.
8. Hashemi SA, Behbahan NGG, Bahrani S, et al. Ultra-sensitive viral glycoprotein detection NanoSystem toward accurate tracing SARS-CoV-2 in biological/non-biological media. Biosens Bioelectron. 2020;171:112731.
9. Dronina J, Bubniene US, Ramanavicius A. The application of DNA polymerases and Cas9 as representative of DNA-modifying enzymes group in DNA sensor design. Biosens Bioelectron. 2020;112867.
10. Helmy YA, Fawzy M, Elaswad A, Sobieh A, Kenney SP, Shehata AA. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J Clin Med. 2020;9(4):1225. doi:10.3390/jcm9041225
11. Han Y, Král P. Computational Design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14(4):5143–5147. doi:10.1021/acsnano.0c02857
12. Morniroli D, Giannì ML, Consales A, Pietrasanta C, Mosca F. Mosca F. Human Sialome and Coronavirus Disease-2019 (COVID-19) pandemic: an understated correlation? Front Immunol. 2020;11:1480. doi:10.3389/fimmu.2020.01480
13. Chandel V, Raj S, Rathi B, Kumar D. In Silico Identification of Potent COVID-19 Main Protease Inhibitors from FDA Approved Antiviral Compounds and Active Phytochemicals through Molecular Docking: a Drug Repurposing Approach. 2020.
14. Szczepanski A, Owczarek K, Bzowska M, et al. Canine respiratory coronavirus, bovine coronavirus, and human coronavirus OC43: receptors and attachment factors. Viruses. 2019;11(4):328. doi:10.3390/v11040328
15. Joshi RS, Jagdale SS, Bansode SB, et al. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J Biomol Struct Dyn. 2020;1–16.
16. Yoshizawa N, Ishihara R, Omiya D, Ishitsuka M, Hirano S, Suzuki T. Application of a photocatalyst as an inactivator of bovine coronavirus. Viruses. 2020;12(12):1372. doi:10.3390/v12121372
17. Matsumoto M, Mukai T, Furukawa S, Ohori H. Inhibitory effects of epigallocatechin gallate on the propagation of bovine coronavirus in Madin‐Darby bovine kidney cells. Animal Sci J. 2005;76(5):507–512. doi:10.1111/j.1740-0929.2005.00297.x
18. Souza MP, Vaz AF, Correia MT, Cerqueira MA, Vicente AA, Carneiro-da-cunha MG. Quercetin-loaded lecithin/chitosan nanoparticles for functional food applications. Food Bioprocess Technol. 2014;7(4):1149–1159. doi:10.1007/s11947-013-1160-2
19. Ma Q, Gao Y, Sun W, et al. Self-Assembled chitosan/phospholipid nanoparticles: from fundamentals to preparation for advanced drug delivery. Drug Deliv. 2020;27(1):200–215. doi:10.1080/10717544.2020.1716878
20. Li Y-L, Li J, Wang N-L, Yao X-S. Flavonoids and a new polyacetylene from Bidens parviflora Willd. Molecules. 2008;13(8):1931–1941. doi:10.3390/molecules13081931
21. Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1):42717. doi:10.1038/srep42717
22. Rane JS, Chatterjee A, Kumar A, Ray S. Targeting SARS-CoV-2 Spike Protein of COVID-19 with naturally occurring phytochemicals: an in silico study for drug development. ChemRxiv. 2020:1–11.
23. Ebada SS, Al-Jawabri NA, Youssef FS, et al. In vivo antiulcer activity, phytochemical exploration, and molecular modelling of the polyphenolic-rich fraction of Crepis sancta extract. Inflammopharmacology. 2020;28(1):321–331. doi:10.1007/s10787-019-00637-x
24. Schrodinger L. The PyMOL molecular graphics system. Version. 2010;1(5):20.
25. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;1–4.
26. Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;1–6.
27. Bell EW, Zhang Y. DockRMSD: an open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. J Cheminform. 2019;11(1):40. doi:10.1186/s13321-019-0362-7
28. Laskowski RA, Swindells MB. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. ACS Publications; 2011.
29. Tan Q, Liu W, Guo C, Zhai G. Preparation and evaluation of quercetin-loaded lecithin-chitosan nanoparticles for topical delivery. Int J Nanomedicine. 2011;6:1621.
30. Amiraliyan N, Nouri M, Kish MH. Electrospinning of silk nanofibers. I. An investigation of nanofiber morphology and process optimization using response surface methodology. Fibers Polymers. 2009;10(2):167–176. doi:10.1007/s12221-009-0167-9
31. Iraolagoitia XLR, Martini MF. Ca2+ adsorption to lipid membranes and the effect of cholesterol in their composition. Colloids Surf B Biointerfaces. 2010;76(1):215–220. doi:10.1016/j.colsurfb.2009.10.037
32. Liu D, Chang PR, Chen M, Wu Q. Chitosan colloidal suspension composed of mechanically disassembled nanofibers. J Colloid Interface Sci. 2011;354(2):637–643. doi:10.1016/j.jcis.2010.11.041
33. Shah VP, Tsong Y, Sathe P, Liu J-P. In vitro dissolution profile comparison—statistics and analysis of the similarity factor, f2. Pharm Res. 1998;15(6):889–896. doi:10.1023/A:1011976615750
34. Sood A, Panchagnula R. Drug release evaluation of diltiazem CR preparations. Int J Pharm. 1998;175(1):95–107. doi:10.1016/S0378-5173(98)00268-3
35. Carbinatto FM, de Castro AD, Evangelista RC, Cury BSF. Insights into the swelling process and drug release mechanisms from cross-linked pectin/high amylose starch matrices. Asian j Pharm Sci. 2014;9(1):27–34. doi:10.1016/j.ajps.2013.12.002
36. Higuchi T. Theoretical analysis of rate of release of solid drugs dispersed in solid matrices. J Pharm Sci. 1963;52(12):1145–1149.
37. Baker R. Controlled release: mechanisms and rates. Controlled Release Biol Active Agents. 1974;15.
38. Chawla V, Tiwary AK, Gupta S. Characterization of polyvinylalcohol microspheres of diclofenac sodium: application of statistical design. Drug Dev Ind Pharm. 2000;26(6):675–680.
39. Širc J, Hobzova R, Kostina N, et al. Morphological characterization of nanofibers: methods and application in practice. J Nanomater. 2012;2012.
40. Lüscher-Mattii M. Polyanions—a lost chance in the fight against HIV and other virus diseases? Antivir Chem Chemother. 2000;11(4):249–259.
41. Ammerman NC, Beier‐Sexton M, Azad AF. Growth and maintenance of Vero cell lines. Curr Protoc Microbiol. 2008;11(1):
42. Brightman A, Rajwa B, Sturgis J, McCallister M, Robinson J, Voytik‐Harbin S. Time‐lapse confocal reflection microscopy of collagen fibrillogenesis and extracellular matrix assembly in vitro. Biopolymers. 2000;54(3):222–234.
43. Mosmann T, Fong T. Specific assays for cytokine production by T cells. J Immunol Methods. 1989;116(2):151–158.
44. Sametband M, Shukla S, Meningher T, et al. Effective multi-strain inhibition of influenza virus by anionic gold nanoparticles. MedChemComm. 2011;2(5):421–423.
45. Savi LA, Caon T, de Oliveira AP, et al. Evaluation of antirotavirus activity of flavonoids. Fitoterapia. 2010;81(8):1142–1146.
46. Grayer RJ, Veitch NC, Kite GC, Paton AJ, Garnock-Jones PJ. Scutellarein 4′-methyl ether glycosides as taxonomic markers in Teucridium and Tripora (Lamiaceae, Ajugoideae). Phytochemistry. 2002;60(7):727–731.
47. Bell EW, Zhang Y. DockRMSD: an open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. J Cheminform. 2019;11(1):1–9.
48. Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412.
49. Jin Z, Du X, Xu Y, et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:1–5.
50. Sonvico F, Cagnani A, Rossi A, et al. Formation of self-organized nanoparticles by lecithin/chitosan ionic interaction. Int J Pharm. 2006;324(1):67–73.
51. Ilk S, Saglam N, Özgen M. Kaempferol loaded lecithin/chitosan nanoparticles: preparation, characterization, and their potential applications as a sustainable antifungal agent. Artif Cells, Nanomed Biotechnol. 2017;45(5):907–916.
52. Fang S-P, Jao P, Senior DE, Kim K-T, Yoon Y-K. Study on high throughput nanomanufacturing of photopatternable nanofibers using tube nozzle electrospinning with multi-tubes and multi-nozzles. Micro Nano Sys Lett. 2017;5(1):10.
53. Qian W, Yu D-G, Li Y, Liao Y-Z, Wang X, Wang L. Dual drug release electrospun core-shell nanofibers with tunable dose in the second phase. Int J Mol Sci. 2014;15(1):774–786.
54. Fahami A, Fathi M. Fabrication and characterization of novel nanofibers from cress seed mucilage for food applications. J Appl Polym Sci. 2018;135(6):45811.
55. Obregon N, Agubra V, Pokhrel M, et al. Effect of polymer concentration, rotational speed, and solvent mixture on fiber formation using forcespinning®. Fibers. 2016;4(2):20.
56. Nune SK, Rama KS, Dirisala VR, Chavali MY. Electrospinning of collagen nanofiber scaffolds for tissue repair and regeneration. Nanostruc Novel Therapy Elsevier. 2017;281–311.
57. Zhang Z, Sun J. Research on the development of the centrifugal spinning. EDP Sci. 2017;07003.
58. Saeed RM, Dmour I, Taha MO. Stable chitosan-based nanoparticles using polyphosphoric acid or hexametaphosphate for tandem ionotropic/covalent crosslinking and subsequent investigation as novel vehicles for drug delivery. Original research. Front Bioeng Biotechnol. 2020;8(4). doi:10.3389/fbioe.2020.00004
59. Sarmento B, Ferreira D, Veiga F, Ribeiro A. Characterization of insulin-loaded alginate nanoparticles produced by ionotropic pre-gelation through DSC and FTIR studies. Carbohydr Polym. 2006;66(1):1–7.
60. Bimbo LM, Denisova OV, Makila E, et al. Inhibition of influenza A virus infection in vitro by saliphenylhalamide-loaded porous silicon nanoparticles. ACS Nano. 2013;7(8):6884–6893.
61. Jiang Y, Fang D, Song G, Nie J, Chen B, Ma G. Fabrication of core–shell nanofibers by single capillary electrospinning combined with vapor induced phase separation. New Jo Chem. 2013;37(9):2917–2924.
62. Meng F, Zhao R, Zhan Y, Liu X. Design of thorn-like micro/nanofibers: fabrication and controlled morphology for engineered composite materials applications. J Mater Chem. 2011;21(41):16385–16390.
63. Wu X, Cao L, Song J, Si Y, Yu J, Ding B. Thorn-like flexible Ag2C2O4/TiO2 nanofibers as hierarchical heterojunction photocatalysts for efficient visible-light-driven bacteria-killing. J Colloid Interface Sci. 2020;560:681–689. doi:10.1016/j.jcis.2019.10.119
64. Koombhongse S, Liu W, Reneker DH. Flat polymer ribbons and other shapes by electrospinning. J Polym Sci B Polym Phys. 2001;39(21):2598–2606.
65. Arkoun M, Daigle F, Heuzey M-C, Ajji A. Mechanism of action of electrospun chitosan-based nanofibers against meat spoilage and pathogenic bacteria. Molecules. 2017;22(4):585. doi:10.3390/molecules22040585
66. El-Gaffary M, Bashandy M, Ahmed A, El-Borady OM. Self-assembled gold nanoparticles for in-vitro inhibition of bovine viral diarrhea virus as surrogate model for HCV. Mater Res Express. 2019;6(7):075075.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.