Back to Journals » Cancer Management and Research » Volume 13
Network Pharmacology-Based Approach to Investigate the Molecular Targets of Sinomenine for Treating Breast Cancer
Authors Li XM, Li MT, Jiang N, Si YC, Zhu MM , Wu QY, Shi DC, Shi H, Luo Q, Yu B
Received 25 October 2020
Accepted for publication 18 January 2021
Published 9 February 2021 Volume 2021:13 Pages 1189—1204
DOI https://doi.org/10.2147/CMAR.S282684
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Sanjeev K. Srivastava
Xiao-Mei Li,1,2,* Mao-Ting Li,2,3,* Ni Jiang,1 Ya-Chen Si,3 Meng-Mei Zhu,2 Qiao-Yuan Wu,1 Dong-Chen Shi,4 Hui Shi,4 Qing Luo,1 Bing Yu2
1Cancer Research Laboratory, Affiliated Hospital of Zunyi Medical University, Zunyi, Guizhou Province, 563003, People’s Republic of China; 2Department of Cell Biology, Center for Stem Cell and Medicine, Navy Medical University (Second Military Medical University), Shanghai, 200433, People’s Republic of China; 3Student Brigade, Second Military Medical University, Shanghai, People’s Republic of China; 4Department of Respiratory and Critical Care Medicine, Shanghai Changhai Hospital, Shanghai, 200433, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Bing Yu; Qing Luo Email [email protected]; [email protected]
Purpose: Sinomenine has been known to inhibit the proliferation of breast cancer cells. However, its targets have not been found yet. This study aimed to search for molecular targets of sinomenine for treating breast cancer via network pharmacology.
Methods: Potential targets of sinomenine or breast cancer were separately screened from indicated databases. The common targets of both sinomenine and breast cancer were considered as the targets of sinomenine for treating breast cancer. A sinomenine-target-pathway network was constructed based on the obtained results from Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The putative targets of sinomenine were further determined by using protein–protein interaction (PPI) analysis and molecular docking. Finally, the putative targets were verified in vitro and in vivo.
Results: Twenty predicted targets were identified through network pharmacological analysis. Gene Ontology (GO) and KEGG pathway enrichment indicated that these predicted targets enriched in the process of MAP kinase activity, VEGF signaling pathway, Relaxin signaling pathway, Growth hormone synthesis, secretion and action. MAPK1, NOS3, NR3C1, NOS1 and NOS2 were further identified as the putative targets by using PPI and molecular docking analysis. Expression of MAPK1, NR3C1, NOS1, NOS2 and NOS3 genes were significantly regulated by sinomenine in both MCF-7 cells and MDA-MB-231 cells. Furthermore, the expression of NR3C1 in human breast cancer specimens was lower than that in para-tumor normal tissues. Meanwhile, the expression of NR3C1 in xenograft tumors was up-regulated after sinomenine treatment.
Conclusion: MAPK1, NR3C1, NOS1, NOS2 and NOS3 were identified as the putative targets of sinomenine for treating breast cancer. NR3C1 was preliminarily confirmed as a target of sinomenine in two breast cancer cell lines, xenograft tumor models and human breast cancer specimens. These data indicated that the network pharmacology-based prediction of sinomenine targets for treating breast cancer could be reliable.
Keywords: sinomenine, breast cancer, network pharmacology, targets screen, NR3C1
Introduction
Breast cancer is the most frequent malignancy in women all over the world and becomes a leading cause of death. Among females, breast cancer is diagnosed in 24.2% of all 8.6 million new cases and leads as 15.0% of all 4.2 million cancer deaths.1 In recent decades, the incidences of breast cancer, for both developing and developed countries, are clearly showing with both a growing trend and a trend to even younger ages. Surgery, radiation therapy, chemotherapy, hormone therapy and targeted therapy are still the main treatment for breast cancer presently.2 Among them, chemotherapy is often used to control the tumor cell population after surgery or under the situation that is not available for surgery. However, chemotherapy can often cause many adverse effects, such as fatigue, alopecia, nausea, premature ovarian failure, weight gain and cardiac dysfunction, etc., thus reducing the quality of life (QOL) for patients.3 In addition, drug resistance can be usually induced after long time exposed to chemotherapeutic agents, for both reducing the therapeutic effect and leading to death of patients.4 Therefore, it is very necessary to develop the new anti-breast cancer drugs for improving the clinical treatment of breast cancer.
Recently, Traditional Chinese Medicine (TCM) is paid more and more attention on treating cancers, due to the significant clinical efficacy, the low adverse effects and the improvement on QOL.5 Up to now, many plant-derived drugs, such as ginsenoside Rg3,6 bacopaside,7 epigallocatechin gallate,8 quercetin,9 curcumin,10 sulforaphane11 and sinomenine12–17 have been shown to possess the capacity to inhibit the proliferation of breast cancer cells. Among them, sinomenine is the 7,8-didehydro-4-hydroxy-3,7-dimethoxy-17-methylmorphinan-6-one, an alkaloid monomer extracted from the medicinal plant Sinomenium acutum, having chemical structure to contain four rings, A, B, C and D (Figure 1A).18 Sinomenine is known for its obvious pharmacological effects, such as anti-inflammation, anti-immunity and anti-arrhythmia, which is widely used in clinical treatments of rheumatoid diseases and arrhythmia. Its inhibitory effects on the proliferation, migration, invasion and metastasis of breast cancer cells have been recently paid considerable attention. Sinomenine can significantly inhibit the proliferation of ER (-)/PR (-) MDA-MB-231 and ER (+)/PR (+) MCF-7 cell lines,12–14 suggesting that sinomenine may have broad-spectrum of anti-breast cancer properties and may be used to treat both triple negative breast cancer (TNBC) and non-NTBC. Therefore, sinomenine has a potential application prospect for treatment of breast cancer. However, its cellular targets have not been found yet until now.
Network pharmacology was established by Hopkins AL in 2007, which has been proven as a powerful tool to explore the complexity and comprehensiveness of activation mechanism for TCM.19 It illustrates the intricate interactions among genes, proteins and metabolites related to diseases and/or drugs by integrating multidisciplinary concepts including biochemical, bioinformatics and systematic biology, which are consistent with multi-component and multi-target properties of TCM.20 Its studies are mainly based on big data set analyses to collect information about herbal active components, targets and diseases in order to build component-target interactions and target-disease networks. Tissue–target interaction is used to clarify the molecular mechanism of herbal medicine, and target-disease network is used to discover the relationship between target proteins and diseases.21 With the application of network pharmacology in TCM, the traditional paradigm of “one target, one drug” is changing to the strategy of “multi-target, multi-component drug”.22 Presently, combination of network pharmacological prediction and experimental verification has become an important method to realize the main molecular targets and action mechanism of herbal medicine.20
In this study, we applied network pharmacology analysis to predict the potential therapeutic targets and signaling pathways of sinomenine for the treatment of breast cancer. Next, we validated the predicted potential targets in two breast cancer cell lines treated with sinomenine in vitro and in vivo. The results of our study were expected to obtain the new intervention on both targets and signaling pathways for the treatment of breast cancer.
Materials and Methods
Screening of Targets of Sinomenine
Three databases, including STITCH (http://stitch.embl.de/), SwissTargetPrediction (http://www.swisstargetprediction.ch/) and TCM potential target database (TCM-PTD, http://tcm.zju.edu.cn/), were used to identify the sinomenine targets. The SMILE string of the sinomenine (Compound CID: 5459308) obtained from the PubChem database with “sinomenine” was used as the key word in the above three databases to get the corresponding target sets of sinomenine with the indicated filter parameters. The filtering parameters of each database were set as: STITCH database (low confidence 0.150, the rest of the values are default), the SwissTargetPrediction database (probability >0.2, the rest of the values are default) and the TCM-PTD database (Score>0.5, the rest of the values are default). It should be noted that the TCM-PTD database can only be accessed with the permission of the Department of Traditional Chinese Medicine and Engineering of Zhejiang University. Finally, the union of the three target subsets obtained from these databases was recognized as the potential targets of sinomenine.
Screening of Targets of Breast Cancer
Four databases including GeneCards (http://www.genecards.org/), PharmGKB database (https://www.pharmgkb.org/), Therapeutic Target Database (TTD, https://db.idrblab.org/ttd/) and the Comparative Toxicogenomics Database (CTD; http://ctdbase.org/) were used to identify the targets of breast cancer. “Breast Cancer” was used as keyword in the GeneCards database to predict the targets of breast cancer and the top 1000 scored genes were identified as the targets of breast cancer. “Breast Neoplasms” was used as keyword in the PharmGKB database to obtain the targets of breast cancer. “Breast Cancer” was used as keyword in the TTD and CTD databases to get the targets of breast cancer. Finally, the union of the four target subsets obtained from these four databases was recognized as the targets of breast cancer.
GO and KEGG Pathway Enrichment Analysis
The ClusterProfiler package (version 3.16.1) in R software (version 4.0.2) was hired for GO and KEGG pathway enrichment analysis.23 The GO terms and the KEGG pathways were considered statistically significant when q-value ≤0.05. Then, each top 10 GO terms of molecular function (MF), cellular components (CC) and biological process (BP) and the top 20 KEGG pathways were selected for further analysis.
Construction of the Drug-Targets-Pathway Network
To further probe the relationships among sinomenine, targets and pathways in the treatment of breast cancer, the data of interaction between the targets and the pathways, retained in the results of KEGG enrichment analysis, were visualized to construct the drug-targets-pathway network by using the merge function of Cytoscape (version 3.6.0), an open-source bioinformatics software platform for visualizing molecular interaction networks.24
PPI Network Analysis
The PPI for the predicted targets were calculated on the web of the Retrieval of Interacting Genes (STRING) database (https://string-db.org/).25 The cut-off criterion of minimum required interaction score was set as ≥0.4 and the other parameters were default. Then, PPI networks were visualized by Cytoscape.
Molecular Docking Simulation
Molecular docking has been widely used to describe the strength of binding interactions between molecules. AutoDock Vina (version 1.1.2)26 was used to perform molecular docking experiments on sinomenine and putative therapeutic targets to predict their interaction activity. Firstly, the SMILES structure of sinomenine (compound CID: 5459308) was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/), and the three-dimensional structure of sinomenine was prepared in the Open Babel (version 3.0.0), including the following steps: add hydrogen atoms to the molecular structure, select PH=7.2, MMFF94 force field to add charge and minimize energy. Secondly, the crystal structures of the putative therapeutic targets of sinomenine and its original ligands were obtained from the protein data bank of NCBI (RCSB PDB). The accuracy of molecular docking was verified by the root mean square deviation (RMSD) value from the interaction between the putative targets of sinomenine and its original ligands,27 which was calculated by Kabsch algorithm using AutoDock Vina (version 1.1.2). When the RMSD value is less than 2.0 Å, the result is considered accurate.28 Then, the size and center of the docking box were defined as the center of the original ligand of the protein crystal structure, and the size encompasses the key residues of the active site where the original ligand is located. Finally, the interaction activities between sinomenine and its putative therapeutic targets were predicted by AutoDock Vina. The specific approaches are as follows: 1) separate the target protein from its ligand. 2) add hydrogen atoms. 3) calculate the charge. The docking scoring (Affinity) and the sinomenine-protein complexes were extracted from AutoDock Vina. Ligand-protein interactions were analyzed with LigPlus (version 2.24) software and two-dimensional figures were obtained.27 Then, the PyMoL (version 4.5.0) software is used to make three-dimensional figures.
Cell Lines, Sinomenine and Human Breast Cancer Specimens
Human breast cancer MCF-7 and MDA-MB-231 cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). MCF-7 and MDA-MB-231 cells were grown in Roswell Park Memorial Institute 1640 medium (RPMI 1640) (Corning, Manassas, VA, USA) supplemented with 10% fetal bovine serum (Gibco BRL, Carlsbad, MD, USA) and 1% penicillin-streptomycin solution. Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2. Sinomenine was purchased from Selleckchem. The purity of sinomenine was 99.88%, offered by its manufacturer. It was dissolved in dimethyl sulfoxide (DMSO) to prepare 190 mM stock solution for cell experiment in vitro, or dissolved in 0.9% saline to prepare 20 mg/mL (60.72 mM) for intraperitoneal injection in vivo. Twelve matched human breast cancer specimens and adjacent normal tissues were obtained from the Affiliated Hospital of Zunyi Medical University (Guizhou, China), with written informed consent from the patients and approval from the Institutional Review Board of Zunyi Medical University ([2020] 1–177).
Real-Time PCR Analysis
MDA-MB-231 and MCF-7 cells were treated with sinomenine at the concentrations of their respective IC50 values. Total RNA was extracted from cells using Trizol Reagent (Invitrogen, Grand Island, NY) according to the manufacturer’s protocol. To access mRNA expression, 1 μg total RNA was reverse transcribed to first strand cDNA using the M-MLV Reverse Transcriptase (Promega, USA). Real-time PCR was performed on StepOnePlusTM Real-time PCR Systems (Thermo Fisher Scientific) with ChamQ Universal SYBR qPCR Master Mix (Vazyme, #P122) in 20 μL reaction mixture following the manufacturer’s protocol. All samples were examined in triplicate. The sequences of the primers were shown in Supplementary Table S1. The relative expression levels of targets were calculated using the 2−ΔΔCt method. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control.
CCK8 Assays
For CCK8 assays, 5 × 103 MDA-MB-231 cells or MCF-7 cells per well were plated on 96-well plates and incubated for 24 h. MDA-MB-231 cells or MCF-7 cells were treated with indicated concentration sinomenine for 48 hours. CCK8 reagent (10 μL) was added to each well and incubated for 2 h at 37°C, then the optical density value was measured at a wavelength of 450 nm by Molecular Devices Spectramax 190 Microplate Reader. To eliminate the effect of DMSO, the cell viability was determined by the value of OD 450 nm of sinomenine treated cells/the value of OD 450 nm of control cells treated with the equal volume of DMSO.
Immunochemistry Staining
Immunochemistry staining was performed as our previous procedure.29 NR3C1 antibody (24,050-1-AP) was purchased from Proteintech Group (USA). Hematoxylin solution was purchased from Yuxiu Biotech (Shanghai, China).
Western Blot
Western blots were performed as described previously.30 The proteins were extracted with RIPA and quantified with BCA assay kit. Twenty micrograms of total protein per lane were separated by SDS-PAGE electrophoresis and were transferred to PVDF membrane. The membranes were blocked with 5% skimmed milk powder for 1h, then incubated with NR3C1 antibody (diluted 1:3000) at 4°C overnight. Then, membranes were washed with TBST, and incubated with the Peroxidase-conjugated AffiniPure Goat Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch) (diluted 1:6000) at 37°C for 1h. The immune complexes were visualized by using Pierce™ ECL Western Blotting Substrate (Thermo Fisher).
Establishment of Xenograft Tumor Model
MDA-MB-231 cells (5 × 105/mice) were mixed with Matrigel and injected subcutaneously into the right groin of the 4-week-old female BALB/c nude mice (n=12). Two weeks after cell injection, mice were randomly divided into two groups: one group received intraperitoneal injection of 0.9% saline as control every day, and the other group received intraperitoneal injection of sinomenine (100 mg/kg body weight) every day as previously reported.31 The tumor volume was measured with vernier caliper every 4 days and the tumor growth curve was plotted. After 4 weeks of sinomenine administration, the mice were sacrificed and the tumors were removed to be further analyzed. BALB/c nude mice were obtained from the Animal Centre of the Chinese Academy of Medical Sciences, and were kept in standard pathogen-free conditions. All animal experiments were conducted according to the Guidance for the Care and Use of Laboratory Animals, approved by the Institutional Ethics Committee of the Affiliated Hospital of Zunyi Medical University (KLLY(A)-2019-001).
NR3C1 mRNA Expression and Survival Analyses Through Gene Expression Profiling Interactive Analysis (GEPIA)
The mRNA expression of NR3C1 in human breast cancer specimens was analyzed on the web of GEPIA (http://gepia.cancer-pku.cn/index.html).32 We utilized the data set of breast invasive carcinoma (BRCA) deposited in the Cancer Genome Atlas (TCGA) to analyze the mRNA expression of NR3C1 and its correlation with survival of breast cancer patients.
Statistical Analysis
All experimental data were statistically analyzed using SPSS 19.0 software and the results are showed as the mean ± standard error of mean (SEM). Statistical methods were indicated in figure legends. p-values less than 0.05 were considered statistically significant.
Results
Screening the Candidate Targets of Sinomenine for Treating Breast Cancer Through Network Pharmacological Analysis
The STITCH database, the SwissTargetPrediction database and the TCM-PTD database were used together to screen the targets of sinomenine for treating breast cancer. Totally, 38, 15 and 10 potential targets of sinomenine were retrieved from each of the above three databases, respectively (Supplementary Table S2). After removing the repetitive targets, the total amount of obtained potential targets reduced to 58. On the other hand, the potential targets associated with breast cancer were searched from the GeneCards (Top 1000 relevance scores from large to small), PharmGKB, TTD and CTD databases. From each of the four databases, 1000, 92, 118, and 574 potential targets were obtained, respectively (Supplementary Table S3). After removing the repetitive targets, the total number of potential targets associated with breast cancer were adjusted to 1401 finally. Remarkably, there were 22 common targets (ESR1, AR, PGR, CREB1, NOS2, MAPK1, CD274, ABCB1, MAPK3, PTK2, MAPK14, BMP4, PXN, NR3C1, MAPK7, MAPK12, NOS3, NOS1, DEK, DRD4, DRD2, POR) between 58 potential targets of sinomenine and 1401 potential targets associated with breast cancer (Figure 1B). Because ESR1 and PGR were only expressed in non-TNBC but not in TNBC, they were all excluded from the group of candidate targets. The remained 20 candidate targets were finally listed as the predicted targets of sinomenine for treating breast cancer.
Characteristics and Functions of Predicted Targets for Treating Breast Cancer are Elucidated Through GO and KEGG Pathway Enrichments
To realize both characteristics and functions of these 20 predicted targets, analyses of GO enrichment and KEGG pathway enrichment were performed. Results were summarized in supplementary Table S4. The top 10 GO terms of CC, BP and MF are shown in Figure 1C. Most of the showed GO terms, such as MAP kinase activity and positive regulation of cyclase activity had correlations with breast cancer.33,34 Meanwhile, the first 20 KEGG pathways are shown in Figure 1D. The top 4 pathways significantly correlated with breast cancer were VEGF signaling pathway, Relaxin signaling pathway, Growth hormone synthesis, secretion and action, as well as Human cytomegalovirus infection, which were all reported to involve in the occurrence, development, metastasis and treatment of breast cancer.35–38
To further characterize the relationships among sinomenine, targets and signaling pathways associated with breast cancer, a Drug-Target-Pathway network was constructed by using Cytoscape. In this network, there are 41 nodes that represent sinomenine, targets and pathways, and 134 edges that encode the interactions among sinomenine, target and pathway (Figure 1E). The targets of MAPK1, MAPK3, MAPK14, MAPK12 have more edges. Here, MAPK signaling pathways were specially known to affect various diseases including tumorigenesis.39 Taken together, the results of GO and KEGG pathway enrichments supported that all of the 20 predicted targets had the characteristics and functions as the therapeutic targets of sinomenine for treating breast cancer.
Characterization of the Targets of Sinomenine by Using PPI Network and Molecular Docking Analyses
Next, 20 predicted targets were analyzed by using PPI network with STRING database (medium confidence, 0.400) in order to systemically realize the interactions among them along with relative functions. As shown in Figure 2A, a total of 56 relationships (edges) between 18 targets (nodes) were identified. The top 10 targets from high to low ranked edges were MAPK3, MAPK1, CREB1, MAPK14, NOS3, NR3C1, AR, MAPK12, NOS1 and NOS2 (Table 1).
 |
Table 1 The Node Degrees of Putative Targets from PPI Network |
Due to the lack of crystal structures for BMP4, the binding affinities between sinomenine and the other 19 predicted targets were evaluated by autodock Vina software. The accuracy of molecular docking was evaluated by RMSD and the autodock score of putative targets is shown in Table 2. The lower the autodock score, the higher the affinity between sinomenine and each specific target. The top 10 targets having higher affinity with sinomenine were NOS3, NR3C1, NOS1, NOS2, DRD4, ABCB1, MAPK1, POR, MAPK7 and DRD2. After the group of top 10 ranked targets that were screened from PPI network were matched with another group of top 10 ranked targets that were identified by molecular docking analysis, total of 5 common targets (MAPK1, NOS3, NR3C1, NOS1 and NOS2) for both groups were identified. The two-dimensional and three-dimensional molecular docking diagrams of the 5 common targets with sinomenine and its original ligand are shown in Figure 2B–F, respectively. Meanwhile, the amino acid residues of targets which interacted with sinomenine and original ligands via hydrogen bonds and hydrophobic contact are summarized in Table 3.
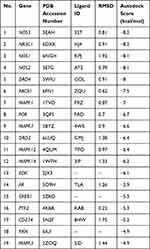 |
Table 2 The Autodock Score of Putative Targets with Sinomenine from Molecular Docking Analysis |
 |
Table 3 The Amino Acid Residue of Targets Which Interacted with Sinomenine and Original Ligands via Hydrogen Bonds and Hydrophobic Contact |
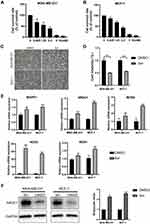
Sinomenine Regulated the Expression of 5 Predicted Targets in vitro
To verify sinomenine affecting on 5 predicted targets in MDA-MB-231 cells and MCF-7 cells, as two breast cancer cell lines, were treated with sinomenine. Results of proliferation assay showed that the inhibitory effect of sinomenine on cell growth had the manner of dose-dependent, observed with a feature of the higher concentration of sinomenine and the more obvious inhibitory effect (Figure 3A and B). The IC50 of sinomenine on MDA-MB-231 cells and MCF-7 cells were 1.6 and 1.9 mM, respectively. It was also confirmed that the growths of both MDA-MB-231 cells and MCF-7 cells were inhibited for about 50% when the sinomenine treatment was at a IC50 concentration for 48 hours (Figure 3C and D). Then, expression levels of these 5 predicted target genes (MAPK1, NOS3, NR3C1, NOS1 and NOS2) were analyzed by using Real-time PCR assay. Results showed that the expression levels of genes of the 5 predicted targets in both two breast cancer cell lines had significant changes after sinomenine treatment (Figure 3E). MAPK1 and NOSs were previously reported to involve in occurrence, development, metastasis and treatment of breast cancer.12,40–43 So, the expression level of NR3C1 protein was selected to be further examined by Western blot assay. Results showed that the expression level of NR3C1 protein was significantly up-regulated after sinomenine treatment in the two breast cancer cells (Figure 3F), consistent with the transcriptional level results from Real-time PCR assay. These results suggested that NR3C1 may be one of the targets of sinomenine.
Sinomenine Suppresses the Cell Growth of Breast Cancer Through the Up-Regulation of the Expression Level of NR3C1 in vivo
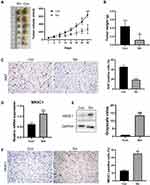
To explore whether sinomenine had in vivo anti-tumor effects through a regulation on expression of NR3C1, the breast cancer xenografts were formed by injection of MDA-MB-231 subcutaneously into nude mice (n=12) for 2 weeks. The nude mice with breast cancer xenografts were treated with sinomenine (intraperitoneal administration of 100 mg/kg bodyweight, dissolved in 0.9% saline) every day. After sinomenine treatment for 28 days, both tumor volume and weight were significantly reduced (Figure 4A and B). Here, Ki67 was used as evaluation marker, because it is universally expressed in all proliferating cells and is an important predictive marker of breast cancer survival and recurrence.44 Results of immunochemistry staining of Ki67 indicated that the capacity of cell proliferation for breast cancer xenografts in vivo was significantly inhibited by sinomenine (Figure 4C). Furthermore, expression of NR3C1 was examined in the xenograft tumors after treatment of sinomenine and 0.9% saline of control, separately. Results of Real-time PCR showed that the mRNA expression level of NR3C1 was significantly upregulated after treated with sinomenine (Figure 4D). Meanwhile, results of both Western blot and immunochemistry staining also showed that the protein levels of NR3C1 were also increased in the sinomenine treated xenografts (Figure 4E and F).
NR3C1 is Differently Downregulated in Human Breast Cancer Specimens
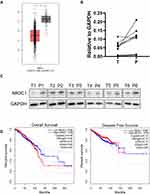
The above results showed that expression levels of NR3C1 were upregulated in breast cancer cells after treated with sinomenine both in vivo and in vitro, suggesting that NR3C1 could be potentially used as a therapeutic target of breast cancer. Here, the expression pattern of NR3C1 was specially analyzed in TCGA-BRCA dataset through GEPIA. Results revealed that the mRNA expression level of NR3C1 in tissues of breast cancer was significantly lower than that in normal tissues (Figure 5A). Then, the expression levels of NR3C1 were examined in the human breast cancer specimens. The results of Real-time PCR showed that the expression level of NR3C1 mRNA in breast cancer tissues was significantly lower than that in the matched para-tumoral tissues (Figure 5B). These findings on NR3C1 gene transcriptional level were further validated on NR3C1 gene translational level by using Western blot (Figure 5C). Furthermore, the correlation between NR3C1 mRNA expression and survival of breast cancer patients was analyzed. Results indicated that low expression of NR3C1 correlated with overall survival and disease-free survival of patients with breast cancer (Figure 5D). Together, these results supported that NR3C1 could be potentially used as a therapeutic target for treatment of breast cancer.
Discussion
Network pharmacology has been successfully used as a unique innovative method for studying the novel targets of TCM, especially multi-component and multi-target interactions, based on the interactions of “disease-gene-target-drug”. Sinomenine is a kind of alkaloid extracted from the medicinal plant of Sinomenium acutum, which has multiple targets and can affect a variety of signal pathways. In this study, we retrieved two groups of targets: 1) 58 targets of sinomenine from three public databases including STITCH, SwissTargetPrediction and TCM-PTD databases; and 2) 1401 targets of breast cancer from GeneCards, PharmGKB, TTD and CTD databases. Totally, 22 predicted targets were obtained as potential targets of sinomenine for treating of breast cancer, based on that they are both targets of sinomenine and targets of breast cancer. Afterward, 20 predicted targets were further realized as candidate targets after excluding ER and PR for no expression in non-TNBC. Finally, MAPK1, NOS3, NR3C1, NOS1 and NOS2 were identified as the targets of sinomenine for treating breast cancer from network pharmacological analysis and molecular docking analysis.
Because of absence in TNBC, ER and PR were regarded as the therapeutic targets in non-TNBC.45–47 Here, sinomenine was found in our study to significantly inhibit the proliferation of both MDA-MB-231 cells (TNBC cell line) and MCF-7 cells (non-TNBC cell line) in vitro, suggesting that sinomenine can be used as a broad-spectrum anti breast cancer drug for treating both TNBC and non-TNBC, and that ER and PR should not be the targets of sinomenine in MDA-MB-231 cells (TNBC cell line). Thus, ER and PR were excluded from the list of targets of sinomenine for treating breast cancer. Whether ER and PR are the targets of sinomenine in MCF7 cells (non-TNBC cell line) will be determined in further study.
The expression of MAPK1, NOSs and NR3C1 were significantly affected by sinomenine in both two cell lines. MAPK signaling pathway was reported to regulate various biological processes involved in cancer progression, such as proliferation, apoptosis and immune escape. Therefore, MAPK signaling pathway is the therapeutic target for the treatment of many kinds of tumors.48 Li et al Showed that sinomenine hydrochloride inhibited the proliferation of human breast cancer cells by inducing G1/S cycle arrest, apoptosis, and DNA damage mediated by ATM/Chk2 and ATR/CHK1 in MDA-MB-231 and MCF-7 cells, which showed a dose-dependent upregulation on expression levels of phospho-MAPK3/1, phospho-JNK and phospho-MAPK14.12 Furthermore, MAPK signaling pathway was reported to be involved in treatment of collagen-induced arthritis by using sinomenine administration combined with acupuncture.49 Results from these reported studies are consistent with our results of Real-time PCR, indicating that MAPK1 may be the targets of sinomenine.
Nitric oxide synthase (NOS), as an enzyme to catalyzes the formation of nitric oxide (NO), has three main subtypes: nNOS (NOS1) mainly exists in neuronal tissue, iNOS (NOS2) can be induced in a variety of cells and tissues, and eNOS (NOS3) was first found in vascular endothelial cells.40 The NO synthesized by nNOS and eNOS plays its biological function through the cGMP-mediated pathway. On the other hand, the NO produced by iNOS exerts its biological and pathological functions via cGMP-independent pathways.50 Although the role of NO in oncology is still controversial, it has been suggested that NO of high concentrations had an anti-neoplastic function, whereas NO of low concentrations could be pro-angiogenic and pro-tumor formation.51 It is well known that nNOS and eNOS produce low level of fast-acting NO, while, iNOS produces large amounts of NO.52,53 In the present study, we found that the expression of three NOS isoforms was significantly increased in both MDA-MB-231 and MCF-7 cells after sinomenine treatment. Based on the previous findings of different effects of three NOS isoforms, it is possible that, in our study, sinomenine inhibited the proliferation of breast cancer cells through inducing the up-regulation of iNOS. However, the roles of NOS during sinomenine inhibiting proliferation of breast cancer cells have not been determined in this study and will be continuously studied further.
NR3C1 (also known as glucocorticoid receptor) involves in many biological effects, such as cell proliferation, differentiation, apoptosis, migration and immune regulation, and plays an important role in the occurrence, development, treatment and prognosis of various types of tumors.54 In this study, we found that the expression level of NR3C1 in breast cancer tissue samples was significantly lower than that in para-tumoral tissue samples, which was consistent with the expression patterns of NR3C1, which were found in both TCGA-BRCA dataset and reported in a previous study.55 Meanwhile, the patients with lower expression of NR3C1 had a lower disease-free survival rate. Tonsing-Carter E et al reported that glucocorticoid receptor modulation inhibited ER-positive breast cancer cell proliferation by suppressing ER chromatin occupancy at the shared ER-regulated enhancers, such as CCND1 (Cyclin D1).56 These findings suggested that NR3C1 is one of the promising targets for treating breast cancer. Here, in xenogeneic breast cancer model in vivo, we also found that Sinomenine upregulated the expression of NR3C1, which was consistent with our results from the studies on two breast cancer cell lines in vitro. However, how sinomenine inhibits the proliferation of breast cancer through NR3C1 has not been fully realized yet, which will be further studied in future.
Conclusion
We found that MAPK1, NR3C1, NOS1, NOS2 and NOS3 are the putative targets of sinomenine for treating breast cancer. Among these targets, NR3C1 was identified as a target of sinomenine for treating breast cancer through our findings on sinomenine inhibiting proliferations of breast cancer cells in vitro, xenograft tumor model in vivo and various types of human breast cancer specimens. These data indicated that the network pharmacology-based investigation could be well effectively used for finding the molecular targets of sinomenine for treating breast cancer. Our study not only provides a new method for additional research on the mechanism of anti-breast cancer of sinomenine, but also provides new molecular targets for treating breast cancer in future.
Acknowledgments
This study was supported by National Key R&D Program of China (2018YFA0107500), National Natural Science Foundation of China (31771511 and 81860469), Foundation strengthening program in technical field of China (2019-JCJQ-JJ-068), Second military medical university research project on translational application of precision medicine (2017JZ43) and Science Foundation Project of Guizhou health and Family Planning Commission (gzwjkj2017-1-024).
Bing Yu and Qing Luo are co-corresponding authors.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Tryfonidis K, Senkus E, Cardoso MJ, Cardoso F. Management of locally advanced breast cancer-perspectives and future directions. Nat Rev Clin Oncol. 2015;12(3):147–162. doi:10.1038/nrclinonc.2015.13
3. Azim HA
4. Niu L, Liu L, Yang S, Ren J, Lai P, Chen GG. New insights into sorafenib resistance in hepatocellular carcinoma: responsible mechanisms and promising strategies. Biochim Biophys Acta Rev Cancer. 2017;1868(2):564–570. doi:10.1016/j.bbcan.2017.10.002
5. Kim W, Lee WB, Lee JW, et al. Traditional herbal medicine as adjunctive therapy for breast cancer: a systematic review. Complement Ther Med. 2015;23(4):626–632. doi:10.1016/j.ctim.2015.03.011
6. Nakhjavani M, Palethorpe HM, Tomita Y, et al. Stereoselective anti-cancer activities of ginsenoside Rg3 on triple negative breast cancer cell models. Pharmaceuticals (Basel). 2019;12(3):117. doi:10.3390/ph12030117
7. Palethorpe HM, Smith E, Tomita Y, et al. Bacopasides I and II act in synergy to inhibit the growth, migration and invasion of breast cancer cell lines. Molecules. 2019;24(19). doi:10.3390/molecules24193539.
8. Fujiki H, Suganuma M, Kurusu M, et al. New TNF-alpha releasing inhibitors as cancer preventive agents from traditional herbal medicine and combination cancer prevention study with EGCG and sulindac or tamoxifen. Mutat Res. 2003;523–524:119–125. doi:10.1016/S0027-5107(02)00327-5
9. Schröder L, Marahrens P, Koch JG, et al. Effects of green tea, matcha tea and their components epigallocatechin gallate and quercetin on MCF‑7 and MDA-MB-231 breast carcinoma cells. Oncol Rep. 2019;41(1):387–396. doi:10.3892/or.2018.6789
10. Tajbakhsh A, Hasanzadeh M, Rezaee M, et al. Therapeutic potential of novel formulated forms of curcumin in the treatment of breast cancer by the targeting of cellular and physiological dysregulated pathways. J Cell Physiol. 2018;233(3):2183–2192. doi:10.1002/jcp.25961
11. Lewinska A, Adamczyk-Grochala J, Deregowska A, Wnuk M. Sulforaphane-induced cell cycle arrest and senescence are accompanied by DNA hypomethylation and changes in microRNA profile in breast cancer cells. Theranostics. 2017;7(14):3461–3477. doi:10.7150/thno.20657
12. Li X, Wang K, Ren Y, et al. MAPK signaling mediates sinomenine hydrochloride-induced human breast cancer cell death via both reactive oxygen species-dependent and -independent pathways: an in vitro and in vivo study. Cell Death Dis. 2014;5:e1356. doi:10.1038/cddis.2014.321
13. Zhang H, Ren Y, Tang X, et al. Vascular normalization induced by sinomenine hydrochloride results in suppressed mammary tumor growth and metastasis. Sci Rep. 2015;5(1):8888. doi:10.1038/srep08888
14. Li X, Li P, Liu C, et al. Sinomenine hydrochloride inhibits breast cancer metastasis by attenuating inflammation-related epithelial-mesenchymal transition and cancer stemness. Oncotarget. 2017;8(8):13560–13574. doi:10.18632/oncotarget.14593
15. Yang S, Peng LY, Peng W, et al. Anticancer potentials of sinomenine from sinomenium acutum: a mini-review. Trop J Pharm Res. 2018;17(12):2519–2526. doi:10.4314/tjpr.v17i12.30
16. Song L, Liu D, Zhao Y, et al. Sinomenine reduces growth and metastasis of breast cancer cells and improves the survival of tumor-bearing mice through suppressing the SHh pathway. Biomed Pharmacother. 2018;98:687–693. doi:10.1016/j.biopha.2017.12.065
17. Gao G, Liang X, Ma W. Sinomenine restrains breast cancer cells proliferation, migration and invasion via modulation of miR-29/PDCD-4 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):3839–3846. doi:10.1080/21691401.2019.1666861
18. Zhang YS, Han JY, Iqbal O, Liang AH. Research advances and prospects on mechanism of sinomenin on histamine release and the binding to histamine receptors. Int J Mol Sci. 2018;20(1):70. doi:10.3390/ijms20010070
19. Hopkins AL. Network pharmacology. Nat Biotechnol. 2007;25(10):1110–1111. doi:10.1038/nbt1007-1110
20. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi:10.1038/nchembio.118
21. Niu X, Zhang J, Ni J, et al. Network pharmacology-based identification of major component of angelica sinensis and its action mechanism for the treatment of acute myocardial infarction. Biosci Rep. 2018;38(6):6. doi:10.1042/BSR20180519
22. Zhang R, Zhu X, Bai H, Ning K. Network pharmacology databases for traditional Chinese medicine: review and assessment. Front Pharmacol. 2019;10:123. doi:10.3389/fphar.2019.00123
23. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
24. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
25. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–607D613. doi:10.1093/nar/gky1131
26. Trott O, Olson AJ. AutoDock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi:10.1002/jcc.21334
27. Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–2786. doi:10.1021/ci200227u
28. Huang SY, Zou X. Efficient molecular docking of NMR structures: application to HIV-1 protease. Protein Sci. 2007;16(1):43–51. doi:10.1110/ps.062501507
29. Yu B, Li H, Chen J, et al. Extensively expanded murine-induced hepatic stem cells maintain high-efficient hepatic differentiation potential for repopulation of injured livers. Liver Int. 2020;40(9):2293–2304. doi:10.1111/liv.14509
30. Yang LH, Wang Y, Qiao S, et al. Liver-enriched activator protein 1 as an isoform of CCAAT/enhancer-binding protein beta suppresses stem cell features of hepatocellular carcinoma. Cancer Manag Res. 2018;10:873–885. doi:10.2147/CMAR.S160172
31. He X, Maimaiti M, Jiao Y, Meng X, Li H. Sinomenine induces G1-phase cell cycle arrest and apoptosis in malignant glioma cells via downregulation of sirtuin 1 and induction of p53 acetylation. Technol Cancer Res Treat. 2018;17:1533034618770305. doi:10.1177/1533034618770305
32. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–98W102. doi:10.1093/nar/gkx247
33. Lu H, Tran L, Park Y, et al. Reciprocal regulation of DUSP9 and DUSP16 expression by HIF1 controls ERK and p38 MAP kinase activity and mediates chemotherapy-induced breast cancer stem cell enrichment. Cancer Res. 2018;78(15):4191–4202. doi:10.1158/0008-5472.CAN-18-0270
34. Moody TW, Nuche-Berenguer B, Jensen RT. Vasoactive intestinal peptide/pituitary adenylate cyclase activating polypeptide, and their receptors and cancer. Curr Opin Endocrinol Diabetes Obes. 2016;23(1):38–47. doi:10.1097/MED.0000000000000218
35. Cao WH, Liu XP, Meng SL, et al. USP4 promotes invasion of breast cancer cells via relaxin/TGF-β1/Smad2/MMP-9 signal. Eur Rev Med Pharmacol Sci. 2016;20(6):1115–1122.
36. Clarke R, Tyson JJ, Dixon JM, Endocrine resistance in breast cancer – an overview and update. Mol Cell Endocrinol. 2015;418(Pt 3):220–234. doi:10.1016/j.mce.2015.09.035
37. Subramani R, Nandy SB, Pedroza DA, Lakshmanaswamy R. Role of growth hormone in breast cancer. Endocrinology. 2017;158(6):1543–1555. doi:10.1210/en.2016-1928
38. Geisler J, Touma J, Rahbar A, Söderberg-Nauclér C, Vetvik KA. Review of the potential role of human cytomegalovirus (HCMV) infections in breast cancer carcinogenesis and abnormal immunity. Cancers (Basel). 2019;11(12):12. doi:10.3390/cancers11121842
39. Yuan J, Dong X, Yap J, Hu J. The MAPK and AMPK signalings: interplay and implication in targeted cancer therapy. J Hematol Oncol. 2020;13(1):113.
40. Huerta S, Chilka S, Bonavida B. Nitric oxide donors: novel cancer therapeutics (review). Int J Oncol. 2008;33(5):909–927.
41. Dávila-González D, Choi DS, Rosato RR, et al. Pharmacological inhibition of NOS activates ASK1/JNK pathway augmenting docetaxel-mediated apoptosis in triple-negative breast cancer. Clin Cancer Res. 2018;24(5):1152–1162. doi:10.1158/1078-0432.CCR-17-1437
42. Granados-Principal S, Liu Y, Guevara ML, et al. Inhibition of iNOS as a novel effective targeted therapy against triple-negative breast cancer. Breast Cancer Res. 2015;17(1):25. doi:10.1186/s13058-015-0527-x
43. Basudhar D, Somasundaram V, de Oliveira GA, et al. Nitric oxide synthase-2-derived nitric oxide drives multiple pathways of breast cancer progression. Antioxid Redox Signal. 2017;26(18):1044–1058. doi:10.1089/ars.2016.6813
44. Juríková M, Danihel Ľ, Polák Š. Ki67, PCNA, and MCM proteins: markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118(5):544–552. doi:10.1016/j.acthis.2016.05.002
45. Niu XL, Wang Y, Yao Z, et al. Autocrine interferon-γ may affect malignant behavior and sensitivity to tamoxifen of MCF-7 via estrogen receptor β subtype. Oncol Rep. 2015;34(6):3120–3130. doi:10.3892/or.2015.4294
46. Yip CH, Rhodes A. Estrogen and progesterone receptors in breast cancer. Future Oncol. 2014;10(14):2293–2301. doi:10.2217/fon.14.110
47. Zheng Y, Shao X, Huang Y, et al. Role of estrogen receptor in breast cancer cell gene expression. Mol Med Rep. 2016;13(5):4046–4050. doi:10.3892/mmr.2016.5018
48. Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120(22):3446–3456. doi:10.1002/cncr.28864
49. Xu M, Liu S, Wan R, Chen Y. Combined treatment with sinomenine and acupuncture on collagen-induced arthritis through the NF-κB and MAPK signaling pathway. Oncol Lett. 2018;15(6):8770–8776. doi:10.3892/ol.2018.8394
50. MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15(1):323–350. doi:10.1146/annurev.immunol.15.1.323
51. Chinje EC, Stratford IJ. Role of nitric oxide in growth of solid tumours: a balancing act. Essays Biochem. 1997;32:61–72.
52. Lechner M, Lirk P, Rieder J. Inducible nitric oxide synthase (iNOS) in tumor biology: the two sides of the same coin. Semin Cancer Biol. 2005;15(4):277–289. doi:10.1016/j.semcancer.2005.04.004
53. Dhar A, Brindley JM, Stark C, Citro ML, Keefer LK, Colburn NH. Nitric oxide does not mediate but inhibits transformation and tumor phenotype. Mol Cancer Ther. 2003;2(12):1285–1293.
54. Liu SH, Otal-Brun M, Webb TE. Glucocorticoid receptors in human tumors. Cancer Lett. 1980;10(3):269–275. doi:10.1016/0304-3835(80)90080-4
55. Nesset KA, Perri AM, Mueller CR. Frequent promoter hypermethylation and expression reduction of the glucocorticoid receptor gene in breast tumors. Epigenetics. 2014;9(6):851–859. doi:10.4161/epi.28484
56. Tonsing-Carter E, Hernandez KM, Kim CR, et al. Glucocorticoid receptor modulation decreases ER-positive breast cancer cell proliferation and suppresses wild-type and mutant ER chromatin association. Breast Cancer Res. 2019;21(1):82. doi:10.1186/s13058-019-1164-6
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.