Back to Journals » Drug Design, Development and Therapy » Volume 16
Network Pharmacology and Experimental Verification to Explore the Potential Mechanism of Yin-Huo-Tang for Lung Adenocarcinoma Recurrence
Authors Liu D, Lin S, Li Y, Zhou T, Hu K, Li Q
Received 14 October 2021
Accepted for publication 5 February 2022
Published 17 February 2022 Volume 2022:16 Pages 375—395
DOI https://doi.org/10.2147/DDDT.S343149
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Tin Wui Wong
Dianna Liu,1 Shicheng Lin,2 Yuan Li,2 Tian Zhou,1 Kaiwen Hu,1 Quanwang Li1
1Oncology Department, Dongfang Hospital, Beijing University of Chinese Medicine, Beijing, 100071, People’s Republic of China; 2Graduate School, Beijing University of Chinese Medicine, Beijing, 100029, People’s Republic of China
Correspondence: Kaiwen Hu; Quanwang Li, Email [email protected]; [email protected]
Purpose: Yin-Huo-Tang (YHT) is a classic traditional Chinese prescription, used to prevent lung adenocarcinoma (LUAD) relapse by “nourishing yin and clearing heat”. In this study, the mechanism of YHT in LUAD recurrence was investigated.
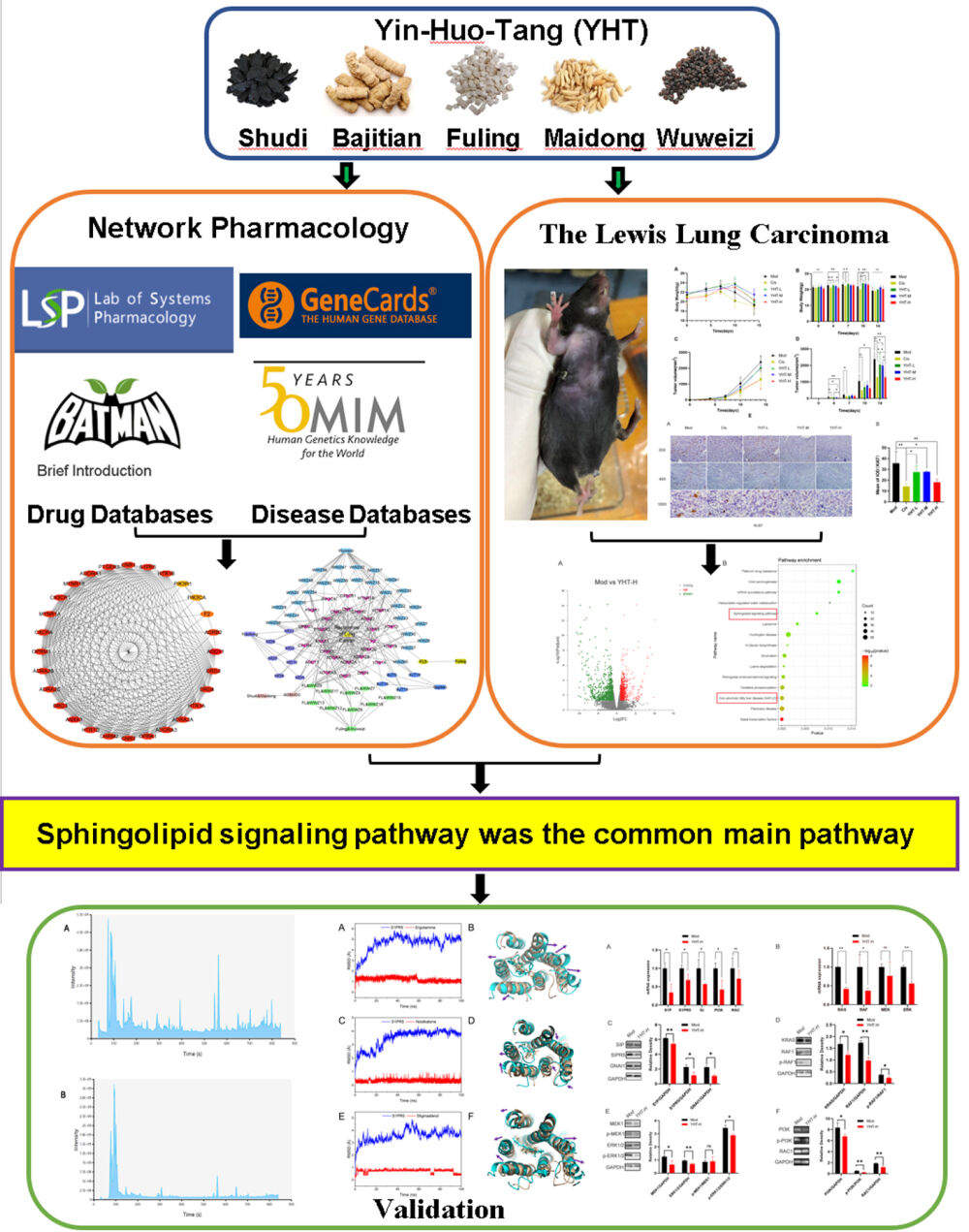
Methods: Firstly, the bioactive compounds and targets of YHT, as well as related targets of LUAD recurrence, were collected from public databases. The protein–protein interaction network, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed to find the pivotal compounds, hub genes, functional annotation and main pathways. Subsequently, RNA sequencing of recurrent tumor tissues from Lewis lung carcinoma mice treated with YHT was used to explore the main pathways. At the same time, pathways screened by network pharmacology and RNA sequencing analysis were considered the most important pathways. Finally, liquid chromatography mass spectrometry was used to validate the pivotal active ingredients. Molecular docking technology was performed to validate the binding association between the hub genes and the pivotal active ingredients. PCR and WB analysis were used to validate the main pathways.
Results: There were 128 active compounds and 419 targets interacting with YHT and LUAD recurrence. Network analysis identified 4 pivotal compounds, 28 hub genes and 30 main pathways. Sphingolipid signaling pathway was the common main pathway in network pharmacology and RNA sequencing results. The hub gene related to the sphingolipid signaling pathway was S1PR5. Qualitative phytochemical analysis confirmed the presence of 3 pivotal compounds, namely stigmasterol, nootkatone and ergotamine. The molecular docking verified that the pivotal compounds could good affinity with S1PR5. The PCR and WB analysis verified YHT suppressed Lewis lung cancer cells proliferation and migration by inhibiting the sphingolipid signaling pathway.
Conclusion: The potential mechanism and therapeutic effect of YHT against the recurrence of LUAD may be ascribed to inhibition of the sphingolipid signaling pathway.
Keywords: sphingolipid signaling pathway, traditional Chinese medicine, S1PR5, stigmasterol, nootkatone, ergotamine
Graphical Abstract:
Introduction
Lung adenocarcinoma (LUAD) is one of the most common malignancies with a rise in new cases worldwide each year. Early diagnosis and surgical treatment are of great importance.1 Undergoing surgery for early-stage patients and performing adjuvant treatments according to the conditions of the patient are recommended, but about 30–50% of early-stage lung cancer patients would die within 5 years of recurrent diseases.2 The pathogenesis of LUAD recurrence is closely related to lipid metabolism alterations,3 immune infiltration,4,5 gene mutations,6,7 cell proliferation,5 enhanced stemness, DNA repair deficiency, bacterial microbiome8 and angiogenesis.9,10 It is worth mentioning that the role of sphingolipid metabolism in tumor growth has attracted more and more attention in recent years.11–13 The inhibition of S1P-S1P receptor signaling provided opportunity to develop new anticancer therapeutic strategy.
In the theory of traditional Chinese medicine (TCM), LUAD belongs to the category of “lung obstruction”, which can be treated by “nourishing yin and clearing heat”. Various Chinese herbal medicines and decoctions, such as Rabdosia rosthornii (Diels) H. Hara,14 Tripterygium Hook.f.,15 Ze-Qi-Tang16 and Fei-Liu-Ping ointment17 have been reported to inhibit autophagy and apoptosis of tumor cell, angiogenesis and epithelial-to-mesenchymal transition (EMT) outside the tumor.18,19 Thus, TCM has an advantage in suppressing recurrence in lung cancer postoperative patients.20,21
Yin-Huo-Tang (YHT) is famous for clearing “heat” and nourish “yin” in TCM, which was first published in Chen shiduo’s “Bian Zheng Qi Wen” of the Chinese Qing Dynasty. It consists of the Rehmannia glutinosa (Gaertn.) DC. (Chinese name: Shudi), the Morinda officinalis F.C. How (Chinese name: Bajitian), the Poria cocos (Schw.) Wolf (Chinese name: Fuling), the Ophiopogon japonicus (Thunb.) Ker Gawl. (Chinese name: Maidong) and the Schisandra chinensis (Turcz.) Baill. (Chinese name: Wuweizi). Currently, YHT is used to treat lung cancer22–24 based on the function of “nourishing yin and clearing heat”. However, the mechanism of YHT for treatment of LUAD remains unclear, impeding progress in clinical usage of YHT. Further studies are therefore necessary to explore the mechanisms involved.
The study aimed to explore the mechanism of YHT against the recurrence of LUAD. In recent years, network pharmacology has been widely used to reveal the mechanism of Chinese herbs.25,26 However, the number of bioactive compounds, targets and pathways predicted by network pharmacology is large. The workload of experimental verification is huge.27 In this study, network pharmacology was used to predict the initial bioactive compounds, targets and pathways. RNA sequencing was used to narrow the scope of predicted targets and pathways, liquid chromatography mass spectrometry was used to narrow the scope of bioactive compounds. Molecular docking technology was performed to validate the binding association between the hub genes and the pivotal active ingredients. PCR and WB analysis were used to validate the main pathways.
Materials and Methods
Candidate Compounds and Targets Screening
All of the chemical ingredients and targets of YHT were collected from the Traditional Chinese Medicine Systems Pharmacology database and Analysis Platform (TCMSP, http://tcmspw.com),28 Bioinformatics Analysis Tool for Molecular Mechanism of Traditional Chinese Medicine (BATMAN-TCM, http://bionet.ncpsb.org.cn/batman-tcm/)29 and wide-scale searches of the literature. The compounds and targets that met the criteria of oral bioavailability ≥30% and drug similarity ≥0.14 in TCMSP, and score cutoff >30, adjusted P value <0.05 in BATMAN-TCM were chosen for further analysis.30
Identification of Disease Targets
We retrieved LUAD recurrent protein targets from GeneCards (https://www.genecards.org/)31 and OMIM databases (https://omim.org/).32 After eliminating duplicates, the potential targets for LUAD recurrence were identified. Overlapping targets from drug and disease were obtained for further study. All the targets were standardized through UniProt database (https://sparql.uniprot.org/).33
Functional Annotation and Pathway Enrichment Analyses
R software version 4.1.1 (https://www.r-project.org/) and the “clusterProfiler”, “colorspace”, “stringi”, “stringr”, “ggplot2” packages were used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses.34 P values <0.05 were considered to be statistically significant.
Protein–Protein Interaction (PPI) Data
The Search Tool for the Retrieval of Interacting Genes database (STRING, https://www.string-db.org/)34 was used to construct a PPI network of the detected targets. Interaction score >0.9 was considered statistically significant. Furthermore, the result of the PPI network was imported into the Cytoscape (version 3.7.1) plugin to create network visualizations and screened the hub genes with cytoHubba plugin.35,36
Network Construction
Cytoscape (version 3.7.1) software was used to construct drug-active ingredients-hub genes-disease network diagrams.34
Preparation and Qualitative Phytochemical Analysis of YHT
YHT granule was purchased from Beijing Tcmages Pharmaceutical Co., LTD (Beijing, China). It consists of five common Chinese herbal extracts: Rehmannia glutinosa (Gaertn.) DC., the Morinda officinalis F.C. How, the Poria cocos (Schw.) Wolf, the Ophiopogon japonicus (Thunb.) Ker Gawl. and the Schisandra chinensis (Turcz.) Baill., as listed in Table 1. The five granules were mixed in water with a ratio of 30:10:10:5:2. Chromatographic separation was accomplished in a Thermo Ultimate 3000 system. We used the methods described in the Chinese Pharmacopoeia.37
 |
Table 1 Compositions of YHT |
Chemical Reagents
Cisplatin was obtained from QILU PHARMACEUTICAL (Shandong, China). Pentobarbital sodium salt was obtained from Sigma-Aldrich (St. Louis, MO, USA). Sodium chloride for injection was obtained from Tiancheng Pharmaceutical (Hebei, PR China). Absolute ethanol and isopropanol were obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
Cell Culture
Lewis lung cancer cell (LLC) lines LL/2 (LLC1) were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were maintained in a high-glucose DMEM (Gibco, Gaithersburg, MD) supplemented with 10% fetal bovine serum (Gibco, Gaithersburg, MD), penicillin (100 U/mL, Gibco) and streptomycin (100 μg/mL, Gibco) at 37°C in a CO2 incubator.
Animals and Experimental Groups
Fifty SPF C57BL/6 mice (50 males) weighing between 18 g and 22 g were purchased from the SPF (Beijing) Biotechnology Co., Ltd. (Beijing China, License No. SYXK 2019–0010). The animal experiment was approved by the Animal Ethics Committee of the SPF (Beijing) Biotechnology Co., Ltd. on November 15, 2020 (No. AW2020111501), as showed in Figure S1. All laboratory procedures were performed following the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People’s Republic of China. During the experiment, the operating procedures of experimental animals were strictly followed and the welfare of animals was guaranteed, and the name of the guidelines followed for the welfare and treatment of the laboratory animals was the Laboratory Animals Guidelines for Ethical Review of Welfare (GB/T 35892–2018, General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China, Standardization Administration of the People’s Republic of China).
The animals were randomly divided into five groups (n = 10/group): model (Mod), Cisplatin (Cis), YHT low-dose (YHT-L), YHT medium-dose (YHT-M) and YHT high-dose (YHT-H). The high dose is the dose documented in “Bian Zheng Qi Wen”. Because the dose recorded in the book was already higher than the doses of other Chinese herbal medicine prescriptions, the ratio of the three doses of high, medium and low was 4:2:1.
Lewis lung cancer cells (2 × 106 in 200 μL serum-free DMEM) were injected s.c. into the right armpit of mice. The subcutaneous tumors were removed 14 days after Lewis lung cancer cells injection. Then, all the mice were given the drug treatment for 14 days. The Mod group and Cis group received normal saline (10 mL/kg/d, intragastric). The Cis group was given cisplatin (3mg/kg/d, intraperitoneal injection) once a week, all the other groups were given the same volume of saline (intraperitoneal injection). Group YHT-L, YHT-M and YHT-H received YHT (5.5575g/kg/d, 11.115g/kg/d, and 22.23g/kg/d, respectively, intragastric, calculated by dose of herbal medicines). On day 28 after the injection of tumor cells, all animals were sacrificed to collect relevant samples.
Immunochemistry
Tumor tissues were completely stripped after all the animals had been sacrificed. One side of the tumor tissues was used for immunochemistry staining, according to methods described in a previous study.38 Primary antibody used was anti-Ki-67 (Abcam, USA). Other tumor tissues were prepared for further analysis.
RNA Sequencing
RNA sequencing was based on the Illumina Novaseq 6000 platform. The sequencing experiment used the Illumina TruseqTM RNA sample prep kit method for library construction. The Differential Expression Genes (DEGs) between the tumor samples of Mod group and YHT-H group were screened by EdgeR. R software was used to perform KEGG pathway enrichment analysis. Three tumor tissues were randomly selected from each group for RNA sequencing experiment.
Real-Time Quantitative Polymerase Chain Reaction (PCR)
Three tumor tissues were randomly selected from each group for real-time quantitative PCR, according to the methods described in our previous study.39 The primer sequences of relative genes are listed in Table 2.
 |
Table 2 The Primer Sequences of Relative Genes |
Western Blot
Three tumor tissues were randomly selected from each group for Western blot analysis, according to the methods described in our previous study.39 Primary antibodies used were as follows: S1P (Bioss, Beijing, China); S1PR5 (Proteintech, Wuhan, China); GNAI1 (Proteintech, Wuhan, China); Rac1 (Proteintech, Wuhan, China); KRAS (Proteintech, Wuhan, China); ERK1/2 (Proteintech, Wuhan, China); RAF1 (Proteintech, Wuhan, China); Phospho-ERK1/2 (Proteintech, Wuhan, China); MEK1 (Santa, USA); Phospho-Raf1 (Abcam, USA); Phospho-MEK1 (Abcam USA); PI3K p85 alpha (Proteintech, Wuhan, China), Phospho-PI3K p85 alpha (Abcam USA); RAC1 (Proteintech, Wuhan, China); GAPDH (Proteintech, Wuhan, China).
Molecular Docking
The physicochemical and absorption, distribution, metabolism, and excretion (ADMET) parameters of pivotal compounds were predicted using an online webserver ADMETlab 2.0 (https://admetmesh.scbdd.com/).40 Drug-like properties were assessed based on the prediction of Lipinski’s “rule of 5”, including molecular weight, logP value, number of hydrogen bonds donor, and acceptor.
The structures of the pivotal compounds were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/). The crystal structure of the protein was obtained from RCSB PDB (https://www.rcsb.org/). The compounds were docked to the active pocket of protein by AutoDock Vina 1.1.2 program. The best docking score poses were used to perform molecular dynamics (MD). MD simulations were performed using AMBER18 software package.41 Before MD, the topology and coordinate files required for the simulation were prepared by CHARMM-GUI (http://www.charm-gui.org) web. The process includes: complex embedded in a 2:2:1 POPC:POPE:Cholesterol Lipid Bilayer; describing acceptor AMBER force field ff14SB;42 topology and parameter files for the ligands were generated using the latest CGenFF via CHARMM-GUI.43,44 Immersed the complex system into a rectangular periodic box of pre-equilibrated TIP3P water with at least 12 Å distance around the complexes; Added appropriate numbers of NA+/CL- ions to maintain the electroneutrality of the simulation system.
During the simulation, the following sophisticated protocol was followed. Initially, each system was minimized through 2500 steps of steepest descent followed by 2500 steps of conjugate gradient method. After energy minimization, each system was gradually heated at constant volume from 0 K to 298.15 K over a coupling time of 100 ps with position restraints. To accommodate the solvent density, the whole system was equilibrated over 100 ps at a constant pressure of 1 bar. Subsequently, another 320 ps pre-equilibration was performed for pressure relaxation with a weak restraint on the protein backbone. After that, 100 ns MD simulation was conducted for each system to produce trajectories. In these simulations, periodic boundary conditions were employed, and the direct space interaction was calculated by using the particle mesh Ewald method with a long-range electrostatic interaction.45 All bonds involving hydrogen atoms were constrained with the SHAKE algorithm46 allowing an integration time step of 2 fs. The protein dynamic trajectories obtained from the simulations will be used for further studies. In addition, we performed a molecular dynamics simulation of 4 ns at the same parameters but without Lipid Bilayer, which was used for MM-GBSA calculations.
Statistical Analysis
The calculated results were mean ± standard deviation of at least 3 independent experiments. All the statistical analysis was done using SPSS20.0 (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered statistically significant. The differences between the groups were examined using ANOVA, chi-square test and two-tailed unpaired Student’s t-test.
Results
Compound Information
A total of 149 candidate compounds were selected through TCMSP and BATMAN-TCM databases, of which 23 only from Ba ji tian, 12 only from Fuling, 16 only from Maidong, 78 only from WuWeizi, 1 common from Bajitian and Shudi, 17 common from Fuling and Wuweizi, 2 common from Shudi and Maidong, as listed in Table S1.
Targets in YHT Active Against LUAD Recurrence
A total of 111,170 targets directly and indirectly related to LUAD relapse were obtained, and 487 YHT-related objectives were obtained, as listed in Table S2. A total of 419 shared targets were obtained, as shown in Figure 1A and listed in Table S2.
There were 128 active ingredients correlated with the 419 common targets. Active ingredients-gene symbols network was shown in Figure S2 and Table S3. And four pivotal active ingredients were identified with degree ≥46, namely stigmasterol, epiguaipyridine, nootkatone and ergotamine.
Construction of PPI Network and Identification of Hub Genes
The PPI relationship of 419 target genes was obtained with STRING tool, and the minimum interactive score was 0.9 (Figure 1B). The genes evaluated by maximal clique centrality (>87178291119, Figure 1C) and the degree (>22, Figure 1D) were identified: S1PR5, PTGER3, OPRM1, OPRK1, MTNR1B, MTNR1A, HTR1D, HTR1B, HTR1A, DRD4, DRD3, DRD2, CXCR4, CX3CR1, CNR2, CNR1, CHRM2, ANXA1, ADRA2C, ADRA2B, ADRA2A, ADORA3, ADORA1, ADCY1, ADRB2, F2, PIK3CA, PIK3R1.
Drug-Active Ingredients-Hub Genes-Disease Network Analysis
Drug-active ingredients-hub genes-disease network is shown in Figure 2, and the more details were listed in Table S4. There were 5 Chinese herbs, 48 active ingredients and 28 hub genes in the network.
 |
Figure 2 Drug-active ingredients-hub gene symbols-disease network of YHT in the treatment of LUAD recurrence. The detail of the code in the figure was listed in Table S4. There were 5 Chinese herbs, 48 active ingredients and 28 hub genes in the network. |
GO and KEGG Pathway Enrichment Analyses
The 419 common target genes were enriched with GO and KEGG analysis. It turned out that the main biological processes were “oxidation-reduction process”, “response to hypoxia”, “regulation of postsynaptic membrane potential”, “regulation of ion transmembrane transport”, “G-protein coupled receptor signaling pathway” and “coupled to cyclic nucleotide second messenger”; the cellular component terms were enriched in “plasma membrane”, “integral component of plasma membrane”, “voltage-gated sodium channel complex”, “GABA-A receptor complex and postsynaptic membrane”; the molecular function terms were enriched in “drug binding”, “extracellular ligand-gated ion channel activity”, “L-ascorbic acid binding”, “GABA-A receptor activity” and “voltage-gated sodium channel activity”. In terms of signaling pathway enrichment analysis, we found target genes mainly focused on inflammation, metabolism, immune responses, and apoptosis, such as “Purine metabolism (hsa00230)”, “AMPK signaling pathway (hsa04152)”, “Human cytomegalovirus infection (hsa05163)”, “Apoptosis (hsa04210)”, “Insulin signaling pathway (hsa04910)”, “Sphingolipid signaling pathway (hsa04071)”, “Human immunodeficiency virus 1 infection (hsa05170)” and “Human T-cell leukemia virus 1 infection (hsa05166)” (Figure 3). According to the enrichment analyses, the mechanism of YHT could be summed up to inflammation, metabolism, immune responses, and apoptosis.
 |
Figure 3 GO and KEGG enrichment analysis. (A) Barplot showing the BP, CC, and MF domains in the ontology; (B) Dot plot showing the top 30 of KEGG pathways. |
Identification of the Chemical Constituents in Yin-Huo-Tang
Figure 4 gives a typical total ion chromatograms (TICs) of non-volatile components extracted by YHT. Although TICs are complex, most chromatographic peaks can be well separated. Chemical compositions of YHT water extracts determined were shown in Tables S5–S7. Three of the four pivotal active ingredients obtained from the network pharmacology analysis were identified through qualitative phytochemical analysis, namely stigmasterol, nootkatone and ergotamine.
 |
Figure 4 TICs of the water extract of YHT by liquid chromatography mass spectrometry. (A) TIC of YHT in positive ion mode; (B) TIC of YHT in negative ion mode. |
In vivo Validation of the Efficacy of Yin-Huo-Tang
We established a Lewis lung cancer mouse model and evaluated the potential therapeutic effect of YHT.
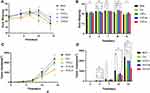
During the first 7 days of treatment, the weights of all animals gradually increased. During the last 7 days of treatment, the weights of the model and Cisplatin-treated mice decreased. During the last 3 days of treatment, the body weights of YHT-L, YHT-M and YHT-H mice decreased. On the tenth day, body weight of the Cis group was the lowest (P < 0.05, Figure 5). However, after 14 days of treatment, there was no significant difference, which was related to the increasing tumor burden. During the drug treatment, the tumor volumes of all mice gradually increased. On the fourteenth day, tumor volumes of Cisplatin-treated and YHT-H-treated mice were significantly lower than those of the model, YHT-M-treated and YHT-L-treated mice (P < 0.05, Figure 5). The number of recrudescent mice was potentially reduced in the cisplatin and YHT-H treatment groups compared to the model group after treatment for 4 days (Table 3). After treatment for 7 days, the tumor had recurred in all mice, which was related to the high invasiveness of Lewis lung carcinoma cells. As expected, cisplatin and YHT-H decreased Ki-67 (a marker of cell proliferation) in tumor tissues (Figure 6). We believed that high doses of YHT could significantly inhibit the recurrence of lung adenocarcinoma.
 |
Table 3 Tumor Recurrence Information |
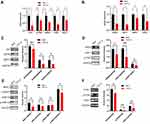
RNA sequencing was used to explore the mRNA expression differences between tumor tissues in the model mice and YHT-H-treated mice. There were 2099 differentially expressed genes, including 1041 upregulated and 1058 downregulated (Figure 7). KEGG analysis was used to analyze the functional enrichment of important modules. The top 15 potential signaling pathways are listed in Figure 7. “Sphingolipid signaling pathway” and “Non-alcoholic fatty liver disease (NAFLD) pathway” were predicted by RNA sequencing and network pharmacological analysis at the same time. Sphingolipid signaling pathway was related to cell survival proliferation, migration and cytoskeletal events (Figure 8). S1PR5 was one of the hub genes identified by network pharmacology, which contained in sphingolipid signaling pathway. According to the details of the sphingolipid signaling pathway, S1PR5 played an important role through S1P/S1PR5/GNAI1 signaling (Figure 8). Then, we verified the inhibition of YHT-H for sphingolipid signaling pathway by PCR and WB analysis. As expected, YHT-H suppressed Lewis lung cancer cells proliferation by inhibiting S1P/S1PR5/GNAI/KRAS/RAF1/MEK1/ERK1/2 pathway, and inhibited migration through S1P/S1PR5/GNAI/PI3K/RAC1 pathway (Figure 9). YHT could inhibit the recurrence of LUAD by modulating the sphingolipid signaling pathway.
Molecular Docking
Lipinski’s “rule of five” can be used to illustrate the drug-like properties for small molecules that are related to the oral bioavailability characteristic, including logP value (<5), molecular weight (<500), number of hydrogen bonds for the donor (≤5) and acceptor (≤10). If two properties are out of range, a poor absorption or permeability is possible, one is acceptable Table 4 reveals the drug-likeness parameters of the stigmasterol, nootkatone and ergotamine according to Lipinski’s “rule of five”. These compounds also showed a high absorption parameter. For distributions, stigmasterol is shown to have the best distribution properties. Nootkatone displays optimal PPB and VD properties, but BBB penetration is mediocre. Ergotamine is the same as Nootkatone.
 |
Table 4 Prediction of Physicochemical and ADMET Parameters |
Cytochrome P450 3A4 (CYP3A4), Cytochrome P450 2D6 (CYP2D6), Cytochrome P450 2C9 (CYP2C9) were common drug-metabolizing enzymes in the human. The results obtained in ADMETlab 2.0 showed that the structures of stigmasterol and ergotamine were substrates and inhibitors of CYP3A4, ergotamine also was inhibitor of CYP2C9, and stigmasterol was also substrate of CYP2D6. Additionally, nootkatone was neither an inhibitor nor a substrate of these enzymes.
Nootkatone and ergotamine could have long half-lives and good clearance levels. For stigmasterol, these two indicators were poor.
Stigmasterol, nootkatone and ergotamine had no predicted risk for the development of hERG, hepatotoxicity mutagenicity. Ergotamine alone might be rat oral acute toxicity.
The root mean square deviation of the molecular dynamic simulation can reflect the motion process of the complex, and the violent fluctuations indicate that the movement is intense, and conversely, the movement is smooth. As shown in Figure 10, stigmasterol, nootkatone and ergotamine fluctuated smoothly during the simulation, indicating that the binding was relatively stable. The fluctuation of proteins during MD to different molecules is not used. For example, in the Nootkatone-S1PR5, the protein behaves smoothly in the mid to late stage of the simulation (Figure 10C). As shown in the left figure (Figure 10D), the protein helix is a whole move toward the center of the molecule, making it possible for the protein to become more stable. For stigmasterol and ergotamine, the protein swings in during MD simulation (Figure 10A and E), meaning that these two molecules affect the protein structure. Further, we found that this effect is mainly to squeeze out the helix of the protein outward. This could be a key phenomenon that has the potential for small molecules to exert activity.
Based on MD simulations, the binding free energies for stigmasterol, nootkatone and ergotamine were calculated through the molecular mechanics with generalized born and surface area solvation (MM-GBSA) method. The stigmasterol, nootkatone and ergotamine presented binding free energy values of −64.85 ± 4.62 kJ/mol, −35.33 ± 0.95 kJ/mol, and −52.45 ± 2.06 kJ/mol, respectively (Table 5). The docking of S1PR5 and pivotal active ingredients was mainly through the following seven interaction forms, including conventional hydrogen bond, pi-alkyl, alkyl, pi-sigma and pi-donor hydrogen bond (Figure 11). For all compounds, van der Waals energy showed the highest negative contributions to the binding free energy. For nootkatones, low electrostatic energy may be responsible for the low overall binding energy.
 |
Table 5 Binding Free Energies and Energy Components Predicted by MM/GBSA (kcal/mol) |
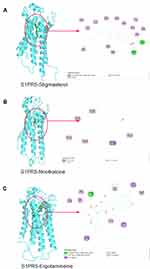 |
Figure 11 Molecular docking of three pivotal active ingredients and S1PR5. (A) S1PR5 and stigmasterol; (B) S1PR5 and nootkatone; (C) S1PR5 and ergotamine. |
Discussion
As described in the introduction, YHT is a classic traditional Chinese prescription used to prevent LUAD recurrence by “nourishing yin and clearing heat”. In this study, we firstly predicted the possible pharmacological mechanism of YHT through a network pharmacology approach, and carried out molecular docking and experiment applying Lewis lung carcinoma mice to identify it.
Qualitative phytochemical analysis of the water extract of YHT confirmed stigmasterol, nootkatone and ergotamine were the pivotal active ingredients of YHT against LUAD recurrence. It has been reported that stigmasterol could cause several cancer cell apoptosis, such as ovarian cancer,47 gastric cancer48 and cholangiocarcinoma.49 However, the role of stigmasterol in lung cancer has rarely been reported. Nootkatone inhibited the growth of A549 cells and induced G1 cell cessation by activating AMPK via LKB1-independent and CAMKK2-dependent pathways.50 Ergotamine was a vasoconstricting agent, which could cause toxicity and inhibit tumor growth.51 The anti-tumor mechanism of ergotamine is still unclear, and it is worthy of further investigation. The previous literature reports support our findings. Our research also provides a direction for the development of new anti-lung cancer drugs.
In vivo experiments confirmed YHT-H reduced the postoperative recurrence of Lewis lung carcinoma and inhibited tumor cell proliferation (Figures 5 and 6). This dose was the dose documented in “Bian Zheng Qi Wen”. This provided evidence for the scientificity of the dosage of TCM prescriptions recorded in ancient medical books.
RNA sequencing, mRNA and protein expression of tumor tissues confirmed that sphingolipid signaling pathway was one of the most important mechanisms of YHT treating LUAD recurrence (Figures 7 and 9). YHT-H suppressed Lewis lung cancer cells proliferation by inhibiting S1P/S1PR5/GNAI1/KRAS/RAF1/MEK1/ERK1/2 pathway, and inhibited migration through S1P/S1PR5/GNAI1/PI3K/RAC1 pathway (Figure 9). The hub gene related to the sphingolipid signaling pathway was S1PR5. S1P can act on S1P receptors to promote tumorigenesis, and operate between cancer cells and fibroblasts to promote metastasis of cancer cells. And it had been reported that S1PR5 had the highest expression level among all the S1P receptors in malignant human tissues.52 The mitogen-activated protein kinase (MAPK) pathway is a key cell signaling pathway involved in regulating cellular growth, proliferation, and survival. RAC1 is critical for cellular adhesion, migration, motility and cell proliferation.53 PI3K/AKT signaling pathway induced RAC1 expression and activity, and then enhanced cell proliferation, survival, migration and metastasis.54
Molecular Docking showed S1PR5 could docking well with the 3 pivotal active ingredients. This further proved that YHT suppressed lung cancer recurrence by inhibiting the sphingolipid signaling pathway. Combined with the results of qualitative phytochemical analysis, we believe stigmasterol, nootkatone and ergotamine play significant roles. The results of the PCR and WB analysis confirmed the scientificity of the prediction of molecular docking. The details of the molecular docking provide directions for the study of the detailed mechanism. The application of the docking approach will assist in developing the drug in the most cost-effective and time-saving manner.55
The findings of the study confirmed the rationality of the application of multidisciplinary research methods in the field of Chinese medicine.
Conclusions
This study combined the methods of network pharmacology, RNA sequencing, liquid chromatography mass spectrometry, molecular docking, and experimental verification to explore the mechanism of YHT against LUAD recurrence. Stigmasterol, nootkatone and ergotamine were the most important pivotal active ingredients contained in YHT against LUAD recurrence. The mechanism of YHT could be summed up to inflammation, metabolism, immune responses, and apoptosis. Notably, sphingolipid signaling pathway was found to be the most important pathway, S1PR5 was the most important hub gene. And Stigmasterol, nootkatone and ergotamine could dock well with S1PR5. These findings provided a new option for the treatment of LUAD recurrence and explained the mechanism of YHT anti-recurrence of LUAD, which could help the usage of YHT in clinical.
However, our study was based on network pharmacology research and limited animal experimental research. Cell model and clinical samples were not used to further verify its authenticity. The pivotal active ingredients were verified by qualitative phytochemical analysis and molecular docking, lacking quantitative analysis and experimental verification. Our discovery should be further validated in future studies.
Abbreviations
LUAD, Lung Adenocarcinoma; YHT, Yin-Huo-Tang; TCM, Traditional Chinese Medicine; EMT, Epithelial to Mesenchymal Transition; PPI, Protein–Protein Interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, The Differential Expression Genes; TCMSP, The Traditional Chinese Medicine Systems Pharmacology database and Analysis Platform; BATMAN-TCM, Bioinformatics Analysis Tool for Molecular Mechanism of Traditional Chinese Medicine; STRING, The Search Tool for the Retrieval of Interacting Genes; TICs, Typical total ion chromatograms; ADMET, Absorption, Distribution, Metabolism, and Excretion; MD, Molecular Dynamics.
Data Sharing Statement
The datasets of RNA sequencing analyzed during the current study are available in the sequence read archive (SRA) repository, named PRJNA761925 (https://www.ncbi.nlm.nih.gov/sra/PRJNA761925).
The other original contributions presented in the study are included in the article/Supplementary Information, further inquiries can be directed to the corresponding authors.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (No. 2020-JYB-ZDGG-127), the National Key R&D Program of China (No. 2018YFC1705102) and Capital’s Funds for Health Improvement and Research (No. CFH 2018-1-4201).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Osarogiagbon RU, Veronesi G, Fang W, et al. Early-stage NSCLC: advances in thoracic oncology 2018. J Thorac Oncol. 2019;14(6):968–978. doi:10.1016/j.jtho.2019.02.029
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
3. Takanashi Y, Funai K, Sato S, et al. Sphingomyelin (d35:1) as a novel predictor for lung adenocarcinoma recurrence after a radical surgery: a case-control study. BMC Cancer. 2020;20(1):800. doi:10.1186/s12885-020-07306-1
4. Liu J, Yang X, Lu X, et al. Impact of T-cell receptor and B-cell receptor repertoire on the recurrence of early stage lung adenocarcinoma. Exp Cell Res. 2020;394(2):112134. doi:10.1016/j.yexcr.2020.112134
5. Yao J, Xue X, Qu D, et al. Reverse engineering a predictive signature characterized by proliferation, DNA damage, and immune escape from stage I lung adenocarcinoma recurrence. Acta Biochim Biophys Sin (Shanghai). 2020;52(6):638–653. doi:10.1093/abbs/gmaa036
6. Park IK, Hyun K, Kim ER, Park S, Kang CH, Kim YT. The prognostic effect of the epidermal growth factor receptor gene mutation on recurrence dynamics of lung adenocarcinoma. Eur J Cardiothorac Surg. 2018;54(6):1022–1027. doi:10.1093/ejcts/ezy220
7. Schwalk AJ, Ost DE, Saltijeral SN, et al. Risk factors for and time to recurrence of symptomatic malignant pleural effusion in patients with metastatic non-small cell lung cancer with EGFR or ALK mutations. Chest. 2021;159(3):1256–1264. doi:10.1016/j.chest.2020.10.081
8. Patnaik SK, Cortes EG, Kannisto ED, et al. Lower airway bacterial microbiome may influence recurrence after resection of early-stage non-small cell lung cancer. J Thorac Cardiovasc Surg. 2021;161(2):419–429.e416. doi:10.1016/j.jtcvs.2020.01.104
9. Cho J, Min HY, Pei H, et al. The ATF6-EGF pathway mediates the awakening of slow-cycling chemoresistant cells and tumor recurrence by stimulating tumor angiogenesis. Cancers. 2020;12(7):1772. doi:10.3390/cancers12071772
10. Gong FL, Wang L, Yu LG, et al. DHPAC, a novel microtubule depolymerizing agent, suppresses angiogenesis and vasculogenic mimicry formation of human non-small cell lung cancer. J Cell Biochem. 2020;121:4756–4771. doi:10.1002/jcb.29690
11. Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18(1):33–50. doi:10.1038/nrc.2017.96
12. Quinville BM, Deschenes NM, Ryckman AE, Walia JS. A comprehensive review: sphingolipid metabolism and implications of disruption in sphingolipid homeostasis. Int J Mol Sci. 2021;22(11):5793. doi:10.3390/ijms22115793
13. Piazzesi A, Afsar SY, van Echten-deckert G. Sphingolipid metabolism in the development and progression of cancer: one cancer’s help is another’s hindrance. Mol Oncol. 2021;15(12):3256–3279. doi:10.1002/1878-0261.13063
14. Ni L, Li Z, Shi X, et al. Rosthorin A inhibits non-small cell lung cancer cell growth and metastasis through repressing epithelial-mesenchymal transition via downregulating Slug. Anticancer Drugs. 2020;31(10):997–1003. doi:10.1097/CAD.0000000000000973
15. Reno TA, Kim JY, Raz DJ. Triptolide inhibits lung cancer cell migration, invasion, and metastasis. Ann Thorac Surg. 2015;100(5):
16. Xu Z, Zhang F, Zhu Y, et al. Traditional Chinese medicine Ze-Qi-Tang formula inhibit growth of non-small-cell lung cancer cells through the p53 pathway. J Ethnopharmacol. 2019;234:180–188. doi:10.1016/j.jep.2019.01.007
17. Liu R, Zheng H, Li W, et al. Anti-tumor enhancement of Fei-Liu-Ping ointment in combination with celecoxib via cyclooxygenase-2-mediated lung metastatic inflammatory microenvironment in Lewis lung carcinoma xenograft mouse model. J Transl Med. 2015;13:366. doi:10.1186/s12967-015-0728-1
18. Yao J, Jiao L, Yao Y, et al. The effect of comprehensive rehabilitation program plus chemotherapy on quality of life in patients with postoperative non-small-cell lung cancer: study protocol of a multicenter randomized clinical trial. Trials. 2020;21(1):309. doi:10.1186/s13063-020-4162-1
19. Wang K, Chen Q, Shao Y, et al. Anticancer activities of TCM and their active components against tumor metastasis. Biomed Pharmacother. 2021;133:111044. doi:10.1016/j.biopha.2020.111044
20. Wang XQ, Zhang Y, Hou W, et al. Association between Chinese medicine therapy and survival outcomes in postoperative patients with NSCLC: a Multicenter, Prospective, Cohort Study. Chin J Integr Med. 2019;25(11):812–819. doi:10.1007/s11655-019-3168-6
21. Zhao X, Dai X, Wang S, et al. Traditional Chinese medicine integrated with chemotherapy for Stage II-IIIA patients with non-small-cell lung cancer after radical surgery: a retrospective clinical analysis with small sample size. Evid Based Complement Alternat Med. 2018;2018:4369027. doi:10.1155/2018/4369027
22. Cheng Y, Duan S, Zuo M, Hu K. Experience in treating tumor with yinhuo decoction under the mode of green therapy of cancer. China Med Herald. 2018;37(07):839–842.
23. Wang M, Hu K, Liu C, Zhou T. Treatment thought of malignant tumor based on the kidney tonification for anti-aging. China J Tradit Chin Med Pharm. 2019;34(06):2333–2336.
24. Wang M, Zhou T, Hu K. Application of yinhuo decoction in coping with side effects of radiotherapy and chemotherapy of malignant tumor. Acta Chin Med. 2019;34(06):
25. Li L, Dai W, Li W, et al. Integrated network pharmacology and metabonomics to reveal the myocardial protection effect of Huang-Lian-Jie-Du-Tang on myocardial ischemia. Front Pharmacol. 2020;11:589175. doi:10.3389/fphar.2020.589175
26. Zhang Z, Yi P, Yang J, et al. Integrated network pharmacology analysis and serum metabolomics to reveal the cognitive improvement effect of Bushen Tiansui formula on Alzheimer’s disease. J Ethnopharmacol. 2020;249:112371. doi:10.1016/j.jep.2019.112371
27. Lu F, Wang D, Li RL, He LY, Ai L, Wu CJ. Current strategies and technologies for finding drug targets of active components from traditional Chinese medicine. Front Biosci. 2021;26(9):572–589.
28. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi:10.1186/1758-2946-6-13
29. Liu Z, Guo F, Wang Y, et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechanism of traditional Chinese medicine. Sci Rep. 2016;6:21146. doi:10.1038/srep21146
30. Chen RB, Yang YD, Sun K, et al. Potential mechanism of Ziyin Tongluo Formula in the treatment of postmenopausal osteoporosis: based on network pharmacology and ovariectomized rat model. Chin Med. 2021;16(1):88. doi:10.1186/s13020-021-00503-5
31. Rappaport N, Fishilevich S, Nudel R, et al. Rational confederation of genes and diseases: NGS interpretation via GeneCards, MalaCards and VarElect. Biomed Eng Online. 2017;16(Suppl 1):72. doi:10.1186/s12938-017-0359-2
32. Baxevanis AD. Searching Online Mendelian Inheritance in Man (OMIM) for information for genetic loci involved in human disease. Curr Protoc Bioinform. 2002;Chapter 1:
33. UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Databaseissue):D204–212.
34. Zhang H, Yao S, Zhang Z, et al. Network pharmacology and experimental validation to reveal the pharmacological mechanisms of liuwei dihuang decoction against intervertebral disc degeneration. Drug Des Devel Ther. 2021;15:4911–4924. doi:10.2147/DDDT.S338439
35. Yang WJ, Wang HB, Wang WD, et al. A network-based predictive gene expression signature for recurrence risks in stage II colorectal cancer. Cancer Med. 2020;9(1):179–193. doi:10.1002/cam4.2642
36. Li X, Tang H, Tang Q, Chen W. Decoding the mechanism of huanglian jiedu decoction in treating pneumonia based on network pharmacology and molecular docking. Front Cell Dev Biol. 2021;9:638366. doi:10.3389/fcell.2021.638366
37. Nie H, Deng Y, Zheng C, et al. A network pharmacology-based approach to explore the effects of Chaihu Shugan powder on a non-alcoholic fatty liver rat model through nuclear receptors. J Cell Mol Med. 2020;24(9):5168–5184. doi:10.1111/jcmm.15166
38. Lei G, Zhang Y, Koppula P, et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30(2):146–162. doi:10.1038/s41422-019-0263-3
39. Liu D, Chen F, Yu X, et al. Do different species of sargassum in haizao yuhu decoction cause different effects in a rat goiter model? Evid Based Complement Alternat Med. 2019;2019:5645620. doi:10.1155/2019/5645620
40. Xiong G, Wu Z, Yi J, et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49:W5–W14. doi:10.1093/nar/gkab255
41. Salomon‐Ferrer R, Case DA, Walker RC. An overview of the Amber biomolecular simulation package. Wiley Interdiscip Rev Comput Mol Sci. 2013;3(2):198–210. doi:10.1002/wcms.1121
42. Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–3713. doi:10.1021/acs.jctc.5b00255
43. Kim S, Lee J, Jo S, Brooks CL
44. Vanommeslaeghe K, Hatcher E, Acharya C, et al. CHARMM general force field: a force field for drug‐like molecules compatible with the CHARMM all‐atom additive biological force fields. J Comput Chem. 2010;31(4):671–690. doi:10.1002/jcc.21367
45. Sagui C, Darden TA. Molecular dynamics simulations of biomolecules: long-range electrostatic effects. Annu Rev Biophys Biomol Struct. 1999;28(1):155–179. doi:10.1146/annurev.biophys.28.1.155
46. Kräutler V, Van Gunsteren WF, Hünenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem. 2001;22(5):501–508. doi:10.1002/1096-987X(20010415)22:5<501::AID-JCC1021>3.0.CO;2-V
47. Bae H, Song G, Lim W. Stigmasterol causes ovarian cancer cell apoptosis by inducing endoplasmic reticulum and mitochondrial dysfunction. Pharmaceutics. 2020;12(6):488. doi:10.3390/pharmaceutics12060488
48. Li K, Yuan D, Yan R, Meng L, Zhang Y, Zhu K. Stigmasterol exhibits potent antitumor effects in human gastric cancer cells mediated via inhibition of cell migration, cell cycle arrest, mitochondrial mediated apoptosis and inhibition of JAK/STAT signalling pathway. J BUON. 2018;23(5):1420–1425.
49. Kangsamaksin T, Chaithongyot S, Wootthichairangsan C, Hanchaina R, Tangshewinsirikul C, Svasti J. Lupeol and stigmasterol suppress tumor angiogenesis and inhibit cholangiocarcinoma growth in mice via downregulation of tumor necrosis factor-α. PLoS One. 2017;12(12):e0189628. doi:10.1371/journal.pone.0189628
50. Hung LVM, Moon JY, Ryu JY, Cho SK. Nootkatone, an AMPK activator derived from grapefruit, inhibits KRAS downstream pathway and sensitizes non-small-cell lung cancer A549 cells to Adriamycin. Phytomedicine. 2019;63:153000. doi:10.1016/j.phymed.2019.153000
51. Mrusek M, Seo EJ, Greten HJ, Simon M, Efferth T. Identification of cellular and molecular factors determining the response of cancer cells to six ergot alkaloids. Invest New Drugs. 2015;33(1):32–44. doi:10.1007/s10637-014-0168-4
52. Wang C, Mao J, Redfield S, Mo Y, Lage JM, Zhou X. Systemic distribution, subcellular localization and differential expression of sphingosine-1-phosphate receptors in benign and malignant human tissues. Exp Mol Pathol. 2014;97(2):259–265. doi:10.1016/j.yexmp.2014.07.013
53. Zou T, Mao X, Yin J, et al. Emerging roles of RAC1 in treating lung cancer patients. Clin Genet. 2017;91(4):520–528. doi:10.1111/cge.12908
54. Tan S, Yi P, Wang H, et al. RAC1 involves in the radioresistance by mediating epithelial-mesenchymal transition in lung cancer. Front Oncol. 2020;10:649. doi:10.3389/fonc.2020.00649
55. Kaur T, Madgulkar A, Bhalekar M, Asgaonkar K. Molecular docking in formulation and development. Curr Drug Discov Technol. 2019;16(1):30–39. doi:10.2174/1570163815666180219112421
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.