Back to Journals » Drug Design, Development and Therapy » Volume 15
Network Pharmacology and Experimental Evidence: PI3K/AKT Signaling Pathway is Involved in the Antidepressive Roles of Chaihu Shugan San
Authors Zhang S , Lu Y, Chen W, Shi W, Zhao Q, Zhao J, Li L
Received 29 April 2021
Accepted for publication 21 July 2021
Published 5 August 2021 Volume 2021:15 Pages 3425—3441
DOI https://doi.org/10.2147/DDDT.S315060
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tuo Deng
Shan Zhang,1 Yujia Lu,1 Wei Chen,2 Wei Shi,1 Qian Zhao,1 Jingjie Zhao,1,3 Li Li1,3
1Department of Traditional Chinese Medicine, Beijing Friendship Hospital, Capital Medical University, Beijing, 100050, People’s Republic of China; 2Experimental and Translational Research Center, Beijing Friendship Hospital, Capital Medical University, Beijing, 100050, People’s Republic of China; 3Department of Integrated Traditional and Western Medicine, Capital Medical University, Beijing, 100050, People’s Republic of China
Correspondence: Jingjie Zhao; Li Li
Department of Traditional Chinese Medicine, Beijing Friendship Hospital, Capital Medical University, NO. 95 Yong-an Road, Beijing, 100050, People’s Republic of China
Tel/Fax +86 10-63139096
Email [email protected]; [email protected]
Objective: Chaihu Shugan San (CSS) is a common antidepressant prescription in traditional Chinese medicines. However, its active ingredients and mechanisms are unknown. The aim of this study was to explore the potential active ingredients and pharmacological mechanisms of CSS for the treatment of major depressive disorder (MDD).
Methods: Active compounds in CSS were screened using the Traditional Chinese Medicine Systems Pharmacology database. Compound-related targets were retrieved using the SwissTargetPrediction database. MDD-related targets were determined using DisGeNET, Therapeutic Target Database and DrugBank databases. The common targets of active compounds in CSS and MDD were retained to construct a compound-MDD target network. Then, functional enrichment analysis and protein–protein interaction analysis were performed to identify hub targets and explore the underlying molecular mechanisms. Finally, hub-targeted genes and pathways were validated by Western blotting and immunofluorescence using chronic unpredictable mild stress (CUMS) mice with or without CSS treatment. The affinities between the active compounds in CSS and hub-targeted genes were evaluated by molecular docking.
Results: Network pharmacology analysis revealed 24 potential targets for treatment of MDD by CSS. Functional enrichment analysis showed that PI3K/AKT signaling pathway was likely to be evidently affected by CSS in the treatment of MDD. In vivo experiments showed that CSS could improve depressive-like behaviors and promote neurogenesis in CUMS mice. Furthermore, CSS could increase phosphorylated (p) PI3K/PI3K and pAKT/AKT levels and decrease the pGSK3β/GSK3β level in the hippocampus of CUMS mice. The active compounds mainly included quercetin and luteolin, which showed good docking scores targeting the PI3K protein.
Conclusion: This network pharmacological and experimental study highlights that the PI3K/AKT pathway is the potential mechanism by which CSS is involved in MDD treatment. Quercetin, luteolin, and kaempferol are probable active compounds in CSS, and these results might provide valuable guidance for further studies of MDD treatment.
Keywords: Chaihu Shugan San, major depressive disorder, network pharmacology, PI3K/AKT signaling pathway
Graphical Abstract:
Introduction
Major depressive disorder (MDD) is a prevalent mental disease characterized by altered emotional, cognitive, and behavioral functions. Increasing attention has been focused on MDD because of its high incidence, low recovery rate, and long duration.1,2 Huang et al reported that the prevalence of MDD in China was 3.6% during a 12-month period and 6.8% during the participants’ entire lifetime.3 Selective serotonin reuptake inhibitors are the most commonly prescribed drugs for MDD treatment but have side effects and high failure rates.4–7 Therefore, additional efforts should be made to identify alternative therapeutic options for depression with increased effectiveness and safety. In recent years, the development of new antidepressants from natural herbs has become an important area of research.
In Chinese medicine theory, MDD patients show long-term depressive symptoms because of the stagnation of “Qi”. Chaihu Shugan San (CSS) is a representative type of the “Li qi” prescription, which is used to smooth the “liver” and promote “Qi” flow. It consists of Bupleurum falcatum L. (Chai hu), Cyperus rotundus L. (Xiang fu), Ligusticum chuanxiong Hort. (Chuan xiong), Citrus reticulata Blanco (Chen pi), Citrus × aurantium L. (Zhi Qiao), Paeonia lactiflora Pall. (Bai shao), and Glycyrrhiza uralensis Fisch. (Gan cao). CSS has been used to clinically and pre-clinically treat depression and has shown good safety and effectiveness.8,9 Previous pharmacological studies have shown that CSS can alleviate depressive symptoms in the chronic unpredictable mild stress (CUMS) mouse model, to some extent, by regulating neurotrophic signaling10 and reducing neuron apoptosis.11,12 However, because the ingredients of traditional Chinese medicine (TCM) are diverse and the interactions of TCM with the human body are complex, the specific molecular mechanisms involved in MDD treatment with CSS remain unclear, which greatly impedes the clinical application of CSS.
The network pharmacology approach proposed is novel and integrates information from bioinformatics, systems biology, and polypharmacology.13 Using a “disease-target-herb” network model, it explores the molecular mechanism of TCM from a multidimensional perspective.14–16 A recent network pharmacology study found that the antidepressant mechanism of CSS was closely related to the neural plasticity, growth, transfer conditions, and gene expression of neuronal cells.17 However, the candidate CSS targets in this study were limited to published papers, and therefore novel targets could not be identified; additionally, the active compounds of CSS were not mentioned.
Our current study aims to screen hub targets from a protein interaction map, which may have a greater potential to identify novel biomarkers.18 The compound-MDD target network was used to predict the active ingredients of CSS involved in MDD treatment. Based on the network pharmacology analysis results, molecular docking and in vivo experiments were performed to further verify the findings.
Materials and Methods
Screening of Compounds in CSS
The Traditional Chinese Medicine System Pharmacology Database and Analysis Platform (TCMSP,http://tcmspnw.com/, updated on May 31, 2014) is a unique pharmacology database of Chinese herbal medicines, cataloging a variety of herbs, components, targets, and relationships with various diseases.19,20 We searched this online database to identify potential compounds in CSS. The pharmacokinetic properties of compounds, including absorption, distribution, metabolism, and excretion (ADME), are important contributors that affect bioactivity. As suggested by the TCMSP, the two ADME-related parameters of oral bioavailability (OB)≥30% and drug-likeness (DL)≥0.18 were used as the inclusion criteria for bioactive chemical compounds in CSS.21
Prediction of CSS-Related and MDD-Related Targets
The SwissTargetPrediction database (http://www.swisstargetprediction.ch/, updated in 2019) provides a probability value derived from cross-validation analyses that can be used to rank targets and estimate the accuracy of predictions.22,23 In this study, we limited target selection to the species “Homo sapiens” and used a probability value≥0.4 to obtain predicted targets of CSS. The official gene symbol and UniProt ID were acquired from the UniProtKB database (https://www.uniprot.org/, updated on October 15, 2019). The Therapeutic Target Database (TTD) (https://db.idrblab.org/ttd/, updated on June 1, 2020),24 DisGeNET database (http://www.disgenet.org/web/DisGeNET/, updated in May, 2020),25 and DrugBank database (http://www.drugbank.ca/, released on April 22, 2020)26 were used to search MDD-related gene targets, using the keywords “major depressive disorder”, “depression”, or “unipolar depression”. The overlapping genes were retained for further analyses.
Screening for Hub Targets and Potential Active Compounds
Overlapping gene targets between CSS and MDD were identified using the Venny web tool 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/). Interactive relationships among the overlapping targets were retrieved from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/, updated on January 19, 2019),27 with the species limited to “Homo sapiens” and a confidence score≥0.4 (medium confidence). The potential interactions were identified based on known interactions from curated databases and experiments. A protein-protein interaction (PPI) network of overlapping targets was visualized using Cytoscape3.7.1 software (https://cytoscape.org/). The NetworkAnalyzer tool in Cytoscape was applied to mine the hub targets by calculating the node degree (number of adjacent neighbors). Nodes with greater degrees were generally regarded as hub targets. Then, a compound-MDD target network was constructed in the same manner, and nodes with greater degrees were identified as the potential active compounds in CSS that might alleviate MDD.
Functional Enrichment Analysis
Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of the overlapping genes were analyzed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/, updated in May 2016), which is an online tool that can be used to identify major biological functions and pathways.28 A value of p<0.05 was considered significant.
Molecular Docking Analysis
SYBYL x-2.1 software (https://sybyl-x.software.informer.com/2.1/) was used to validate the network pharmacology screening results by docking the active compounds with hub proteins. The procedures were based on those in the relevant literature.29,30 The crystal structures of protein kinase B (AKT) (4gv1.pdb), phosphoinositide 3-kinase (PI3K) (1e8w.pdb), glycogen synthase kinase 3 beta (GSK3β) (1j1b.pdb), quercetin (1h1i.pdb), kaempferol (4rel.pdb), and luteolin (4qxv.pdb) were downloaded from the Protein Data Bank (http://www.rcsb.org, updated in 2020).31,32 The protein structure was prepared by removing water molecules and adding hydrogen atoms and charges. The structure of the compounds was constructed using Sybyl sketch and energy was minimized using MMFF94 force field and MMFF94 charges (conjugate gradient, termination: gradient 0.005 kcal/mol Angstrom°, and maximum iterations: 1000). After the compounds were prepared, the surflex-Dock docking mode was used. The default parameters were as follows: maximum conformations per fragment: 20, maximum number of rotatable bonds per molecule: 100, number of spins per alignment: 12, minimum root-mean-square deviation between final poses: 0.5. The top docked poses were selected based on the orientation of the test ligand with respect to the reference ligand and its score. Molecular docking simulations with high scores were regarded as having a high target affinity for each tested compound.
Drugs, Reagents, and Instruments
CSS granules were purchased from Xuanwu Traditional Chinese Medicine Hospital (Beijing, China). Buspirone hydrochloride (No. 6-EOD-111-1) was purchased from Toronto Research Chemicals (Toronto, ON, Canada). Phenytoin sodium (No. BNV229) was purchased from Bidepharm (Shanghai, China). Bromodeoxyuridine (BrdU, No. B5002-250MG) was purchased from Sigma (St. Louis, MO, USA). The chemiluminescence ECL assay kit (P90719) was purchased from Millipore (Burlington, MA, USA)
Phosphorylated (p) PI3K (#4228S), pAKT (#4060S), pGSK3β (#9323S), PI3K (#4257), and AKT (#4691) were purchased from Cell Signaling Technologies (Danvers, MA, USA). GSK3β (ab93926), brain-derived neurotrophic factor (BDNF, ab108319), neuronal nuclei (NeuN) (ab209898), BrdU (ab6326), Alexa Fluor 488 anti-rabbit antibody (ab150077), Cy3-anti-rat antibody (ab97035), 4′,6-diamidino-2-phenylindole (DAPI, ab104039), and β-actin (ab8226) were purchased from Abcam (Cambridge, MA, USA).
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed on a Shimadzu 20A series LC system (Shimadzu, Kyoto, Japan) coupled with an Applied Biosystems Qtrap 4000 triple-quadrupole/linear ion trap mass spectrometer (Applied Biosystems, Foster City, CA, USA). A Zorbax XDB C18 phase column (50 mm×2.1 mm, 3.5 μm) was purchased from Agilent (Santa Clara, CA, USA). A ChemiDoc MP Imaging system (BIO-RAD, Hercules, CA, USA) was used to image Western blots. An Olympus DP70 fluorescence microscope (Olympus, Tokyo, Japan) was used for Immunofluorescence imaging.
Animals
C57BL/6 mice (male, 7–8 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Company (Beijing, China). The animals were housed under standard light (12h light/dark cycle), temperature (25±1°C), and relative humidity (55±5%) conditions. Animals had free access to food and drinking water. All experiments were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Experimental Animal Ethics Committee of Beijing Friendship Hospital (Approval No. 19–1006).
Quality Control of CSS
The plant names, parts used, and clinical dosages are presented in Table 1. Quality control of CSS was performed by LC-MS/MS as previously described.33 The criteria for the quality of herbs in CSS were in accordance with the 2020 Chinese Pharmacopeia (the purity [%] and information regarding reference standards are provided in Supplementary Table S1). Stock solutions (1mg/mL) of 12 reference standards were separately prepared in methanol. Mixed stock solutions with different concentrations were then diluted with methanol into a series of mixed standard solutions for quantitation. Stock solutions of buspirone hydrochloride (1mg/mL) and phenytoin sodium (1mg/mL) were also prepared in dimethyl sulfoxide. CSS (0.1g/mL) was immersed in water and ultrasonically extracted for 0.5h. Then, 0.2mL aliquots of the extracted solutions were diluted to 1mL and prepared for quantitation. The samples were analyzed in positive or negative electrospray ionization mode, and monitored in the multiple reactions monitoring mode. High purity nitrogen was used as the curtain gas, ion source gas 1, and ion source gas 2, with pressures of 10psi, 55psi, and 55psi, respectively. The spray voltage was ±4.5kV, and the capillary temperature was 500°C. Other parameters are presented in Supplementary Table S2. The samples were separated on an Agilent Zorbax XDB C18 phase column. Mobile phase A and mobile phase B consisted of 0.1% formic acid in water and acetonitrile, respectively. The gradient elution program was as follows: 0–0.5 min 20% B; 0.5–4 min 20–80% B; 4-5 min 80% B; 5–5.01 min 80–20% B; 5.01–6 min 20% B. The flow rate was 0.5mL/min. The column temperature was 30°C, and a 1μL aliquot of the solution was injected. The quantitative method was used for simultaneous quantitation of 12 compounds. The chromatogram of these 12 compounds in CSS is shown in Supplementary Figure S1, and the quantitation of these compounds is presented in Supplementary Table S3.
 |
Table 1 The Basic Information of Components in CSS |
CUMS Procedure and Drug Administration
Mice were randomly divided into three groups: the CUMS group (subjected to CUMS), the CSS group (subjected to CUMS and CSS), and the control (CON) group (not subjected to CUMS and CSS). The CUMS procedure was performed as in previous studies with slight modification.34,35 The CUMS and CSS groups were housed in a single cage and subjected to CUMS stimulation (overnight exposure to damp sawdust; lights on overnight; 24h of food deprivation; 24h of water deprivation; 24h during which the cages were tilted at 45°; 10min of tail clamping; 10min of exposure to mice scream; and 2h of being confined in a tube) over a 6-week period. Each animal received a long-lasting stimulus and a short-lasting stimulus at arbitrary times of the day. The same stimulation was not performed for two consecutive days. During this period, CSS (dose: 2.7g/kg body weight) dissolved in water was administered intragastrically to the CSS group once a day according to previous studies.36,37 The CON and CUMS groups received the same volume of distilled water. BrdU (dose: 50mg/kg body weight) dissolved in 0.9% NaCl was injected intraperitoneally once per day for 7 days prior to the end of the experiment.38
Behavioral Testing
The sucrose preference test (SPT) reflects anhedonia, which is a core symptom of depression.39,40 Three days before the test, the mice were adapted to the presence of two water bottles placed in each cage. After adaptation, the mice were deprived of water and food for 12h. During the SPT, which lasted for 24h, mice housed in individual cages had free access to a 2% sucrose solution or plain water. The sucrose preference was calculated using the following equation: sucrose preference (%)=sucrose intake/total liquid intake.
During the tail suspension test (TST), each mouse was suspended 30cm above the floor by the tail via tape placed approximately 1cm from the tip of the tail. The test was conducted for 10min. The cumulative time during which the mouse was completely immobile was calculated.41,42
The forced swim test (FST) was performed by placing the mice in an open glass beaker (18.5cm in height and 14.5cm in diameter) containing 25°C water 10cm deep to prevent the mice from supporting themselves by touching the bottom of the beaker with their tails.43 Floating passively without struggling in the water was defined as immobility.44 The total immobility time during the 10min test was recorded.
Immunofluorescence Staining
Mice were anesthetized with 1% pentobarbital and sacrificed. Mouse brains were cut into 10-µm-thick sections using a cryostat. The slices were fixed with 10% paraformaldehyde (10min) and subjected to acid hydrolysis using 2N HCl for 30min to denature DNA. Slices were immersed in 0.2%Triton X-100 in phosphate-buffered saline (PBS) for 0.5h and blocked with 5% normal donkey serum at room temperature for 1h. The sections were then incubated overnight at 4°C with antibodies against NeuN (1:200) and BrdU (1:200), followed by incubation with Alexa Fluor 488 anti-rabbit (1:400, 488nm emission, green) and Cy3-anti-rat (1:400, 555nm emission, red) secondary antibodies. After washing with PBS, the slices were counterstained with DAPI. The slides were viewed at 10× and 60× magnification using a fluorescence microscope. The number of NeuN+/BrdU+ co-labeled cells in the dentate gyrus was determined by calculating the average number of cells in three biological replicates; six brain slices were used for each replicate.45,46
Western Blot Analysis
The mouse hippocampus was homogenized and lysed in radio immunoprecipitation assay buffer supplemented with protease and phosphatase inhibitor. The total protein concentration was determined by a bicinchoninic acid assay. A total of 30µg of protein from each sample was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose filter membrane. The membranes were blocked with 5% skim milk for 2h, followed by incubation with pPI3K (1:1000), pAKT (1:1000), pGSK3β (1:1000), PI3K (1:1000), AKT (1:1000), GSK3β (1:1000), or BDNF (1:1000) overnight at 4°C. Immunoblots were washed with Tris-buffered saline with Tween and incubated with secondary antibodies for 1h at room temperature. Protein signals were detected using a Bio-Rad imaging system, and protein band quantification was performed using ImageJ software. All protein expression levels were normalized to the β-actin level.
Statistical Analysis
Normal distribution was tested using SPSS 22.0 software. Data are presented as the mean±standard error of the mean and were analyzed by one-way analysis of variance, followed by the least significant difference test. P-values less than 0.05 were considered significant.
Results
Prediction of Hub Targets and Potential Active Compounds of CSS Involved in MDD Treatment
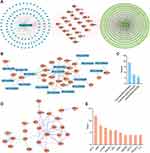
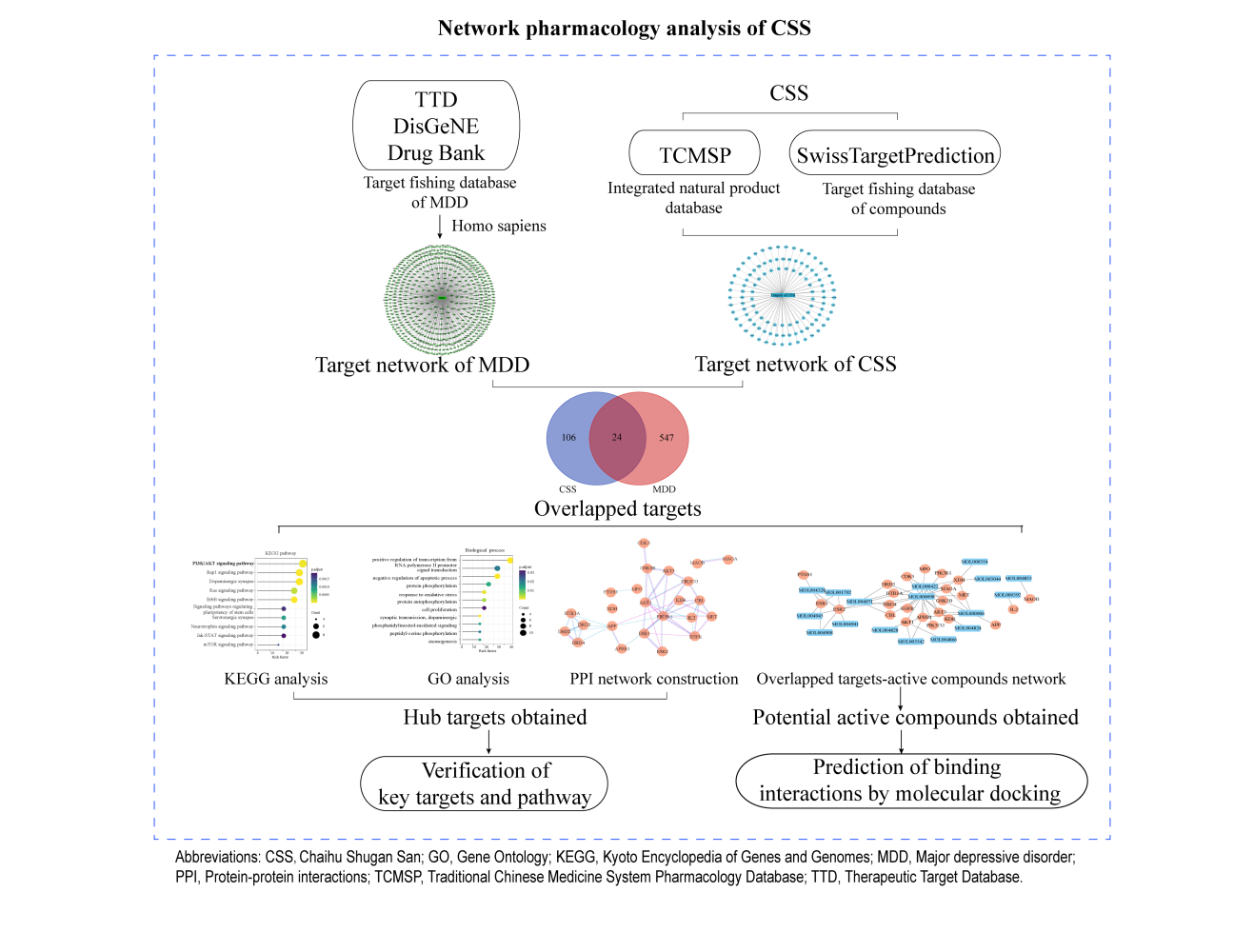
The network pharmacological flowchart in this study is summarized in Figure 1. In total, 38 active compounds in CSS were identified from the TCMSP database after eliminating duplicates. Detailed information regarding the active compounds is provided in Supplementary Table S4. According to the target prediction system using the SwissTargetPrediction database, 130 CSS targets (confidence score ≥0.4) were identified (Supplementary Table S5). Moreover, 571 MDD-related targets were retrieved from the TTD, DrugBank, and DisGeNET databases after eliminating redundant targets. All identified targets are displayed in Supplementary Table S6. As shown in Figure 2A, a network containing CSS targets and MDD targets was constructed, and 24 potential CSS targets associated with MDD were obtained. Thus, the 24 proteins were considered to be potential effective targets of CSS during MDD treatment.
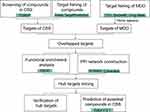 |
Figure 1 The workflow for the network pharmacology analysis. |
A compound-MDD target network was constructed to identify potential active ingredients that interact with overlapping targets of MDD and CSS. There were 17 CSS compounds included in this network. (Figure 2B). The nodes with higher degrees included quercetin (MOL000098), kaempferol (MOL000422), and luteolin (MOL000006), which indicated that these components were associated with a greater number of targets and might play important roles in mediating the antidepressant activity of CSS. The pharmacokinetic properties of the top three potential compounds are exhibited in Table 2. These candidate compounds exhibited relatively high OB (≥30%)47 and good pharmaceutical properties (≥0.18).48 For example, the OB of quercetin was 46.43%, and the DL was 0.28; the OB of kaempferol was 41.88%, and the DL was 0.24. The analysis also showed that most of the active compounds were derived from the “monarch herb”-Chai hu and the “minister herb”-Xiang fu. As shown in Figure 2C, quercetin was considered to have the highest degree of interaction among the active compounds.
 |
Table 2 The Detailed Information of Potential Active Ingredients in CSS Against MDD |
A PPI network of the shared targets was constructed based on the STRING database (Figure 2D). The resultant interactive PPI network comprised 22 nodes and 34 edges. A hub node in a network is regarded as a crucial target and can be used to measure the importance of the entire network. Among these targets, AKT1, amyloid precursor protein, endothelial growth factor receptor, PI3K regulatory subunit 1, Cbl proto-oncogene, estrogen receptor 1, GSK3β, AKT3, PI3K catalytic subunit gamma, and interleukin-2 were core targets (Top 10), with a higher “degree” (Figure 2E). It is worth noting that, these core genes were predicted mainly from potential components of CSS including quercetin, kaempferol and luteolin (Figure 2B). These bioactive compounds were postulated to exert positive antidepressant effects.
Functional Enrichment Analysis of the Common Targets of CSS and MDD
GO and KEGG enrichment analyses were performed to explore the potential functions and mechanisms of the common targets of CSS and MDD. A total of 29 terms in biological processes (BP), five terms in cell components (CC), and 12 terms in molecular functions (MF) were significantly enriched (p<0.05) (Supplementary Table S7). With respect to GO BP terms, most of the targets were significantly enriched in negative regulation of apoptotic processes, protein phosphorylation, cell proliferation, synaptic transmission, and axonogenesis (Figure 3A). The CC category involved the plasma membrane, growth cone, and membrane rafts (Figure 3B). The significantly enriched MFs were ATP binding, protein kinase activity, and protein serine/threonine kinase activity (Figure 3C). Furthermore, CSS could affect 52 pathways including the PI3K/AKT signaling pathway, dopaminergic synapses, the Rap1 signaling pathway, pluripotency of stem cells, and the neurotrophic signaling pathway, that are strongly associated with the occurrence and development of MDD (Figure 3D). It is noteworthy that most of the targeted genes were involved in the PI3K/AKT signaling pathway; additionally, these genes were hub genes in the PPI network. Therefore, we selected the PI3K/AKT pathway for further exploration to identify the underlying mechanisms through which CSS affects depression.
CSS Attenuated Depressive Behaviors in a CUMS-Induced Mouse Model
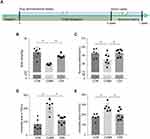
Behavioral experiments were performed to examine the antidepressant activity of CSS (Figure 4A). CSS administration increased the body weight of CUMS mice (F (2,15) =48.077, p=0.001, Figure 4B). Compared with the CON mice, the consumption of sucrose was significantly reduced in CUMS mice (F (2,17) =4.163, p=0.032, Figure 4C). However, CSS treatment restored the sucrose preference (F (2,17) =4.163, p=0.018), indicating decreased anhedonic behavior. The depressive behaviors of mice were measured by recording the immobility time in the TST (Figure 4D) and FST (Figure 4E). The duration of immobility in the TST (F (2,16) =21.194, p=0.001) and FST (F (2,16) =7.946, p=0.001) was clearly prolonged in the CUMS mice compared with CON mice. Conversely, treatment with CSS reversed the effects on immobility time in the TST (F (2,16) =21.194, p=0.004) and FST (F (2,16) =7.946, p=0.010) in CUMS mice. These results indicated that CSS has a potential antidepressant effect, which is consistent with a previous report.49
CSS Promoted Neurogenesis in CUMS-Induced Mice
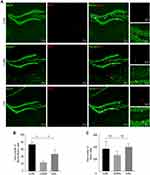
Double immunofluorescence staining for BrdU (newborn cell marker) and NeuN (neuronal marker) was used to assess neurogenesis (Figure 5A). As shown in Figure 5B, stress reduced the number of NeuN+BrdU+ cells in the CUMS group compared with the CON group (F (2,6) =26.027, p=0.001). However, administration of CSS to CUMS mice significantly increased the number of NeuN+BrdU+ cells (F (2,6) =26.027, p=0.001). Increased BrdU+ cell numbers were also observed in the CSS-treatment group, although this difference did not reach significance when compared with the CUMS mice (F (2,6) =1.324, p=0.325, Figure 5C). Thus, these results suggested that CSS ameliorated CUMS-induced impairment of hippocampal neurogenesis.
CSS Regulated the PI3K/AKT Signaling Pathway in CUMS-Induced Mice
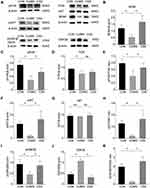
Next, we detected the expression of PI3K/AKT signaling pathway-related proteins by Western blotting analysis (Figure 6A). The pPI3K (F (2,6) =10.926, p=0.003, Figure 6C) and pAKT (F (2,6) =8.277, p=0.030, Figure 6F) protein expression in the hippocampus of CUMS mice was significantly decreased compared with unstressed CON mice. CSS treatment significantly reversed the CUMS-induced alterations in pPI3K (F (2,6) =10.926, p=0.003) and pAKT expression (F (2,6) =8.277, p=0.048). In addition, we calculated the pPI3K/PI3K and pAKT/AKT ratios in the three groups. The pPI3K/PI3K ratio (F (2,6) =5.552, p=0.017, Figure 6E) and pAKT/AKT ratio (F (2,6) =8.329, p=0.027, Figure 6H) were significantly reduced in CUMS mice, whereas CSS reversed their downregulated expression (F (2,6) =5.552, p=0.043; F (2,6) =8.329, p=0.008). However, the total PI3K (F (2,6) =4.281, p=0.070, Figure 6D) and total AKT levels (F (2,6) =0.279, p=0.766, Figure 6G) were not significantly altered in either the CUMS-induced depressive mice (compared with CON mice) or CSS-treated mice (compared with CUMS-induced depressive mice). Phosphorylation at Ser9 can inhibit GSK3β activity.50 Compared with the CON group, CUMS markedly increased GSK3β expression (F (2,6) =51.947, p=0.001, Figure 6J) and decreased the pGSK3β-Ser9 level (F (2,6) =8.750, p=0.007, Figure 6I); however, CSS supplementation reversed these expression trends (F (2,6) =51.947, p=0.001, Figure 6J; F (2,6) =8.750, p=0.024, Figure 6I). The pGSK3β/GSK3β ratio (F (2,6) =41.909, p=0.001, Figure 6K) was significantly reduced in CUMS mice, while the ratio was increased by CSS treatment (F (2,6) =41.909, p=0.001). BDNF expression was also inhibited compared with that in CUMS mice (F (2,6) =8.834, p=0.049, Figure 6B), and this effect was ameliorated by CSS (F (2,6) =8.834, p=0.006). These findings indicated that the PI3K/AKT/GSK3β pathway is involved in the antidepressant effects of CSS.
The Active Compounds in CSS Showed Good Docking Scores When Targeting PI3K/AKT Pathway- Related Proteins
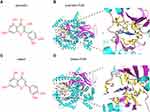
The affinities between PI3K/AKT pathway-related hub targets and active compounds were evaluated by molecular docking. A docking score greater than 5.0 indicates that the active ingredient has good binding activity with the target protein.51 The docking scores of potential compounds with their putative targets are shown in Table 3. The results revealed that the potential active compounds of CSS docked well with the PI3K/AKT signaling pathway target proteins. Interestingly, these compounds exhibited strong affinities for the PI3K protein. Figure 7 presents the docking complex of the PI3K protein targets with their strong binding components. The reference ligand, quercetin, interacted with the PI3K ligand-binding pocket via seven hydrogen bond interactions with the amino acid residues Asp964, Lys833, Tyr867, Ala885, Glu880, and Val882 (Figure 7A and B). Apart from hydrogen-bond interactions, quercetin also exhibited several hydrophobic interactions with Trp812, Met953, Ile963, le881, Val882, Ile879, and Ala885 (Figure 7B). Figure 7C and D show that luteolin, similar to quercetin, bound to the Asp964, Lys833, Val882, Glu880, and Asp950 residues, but luteolin had an obviously different docking orientation.
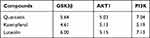 |
Table 3 The Scores of the Molecular Docking with Potential Compounds and Key Targets Proteins in PI3K/AKT Pathway |
Discussion
CSS has been reported to be an important antidepressant prescription. Its active compounds and antidepressant mechanisms have been extensively discussed. The network pharmacology approach is an effective method to explore the relationships of drugs, targets, and diseases, and is particularly suitable for TCM because of its multiple and complex ingredients. A previous study used this method to predict the targets for the antidepressant effects of CSS.17 However, these key targets were obtained from reported studies, which might partly limit the predicted key targets. The key genes in our study were screened from the protein interaction map and showed high topological importance, resulting in a higher potential for identification of novel biomarkers.18 Furthermore, our study provided evidence that CSS ameliorated depressive-like behaviors in CUMS-induced mice, which could be mediated by activation of the PI3K/AKT pathway. Our study also identified potential active compounds with antidepressive effects of CSS, which is worth further exploration for drug discovery and development.
Reduction of neurogenesis in the hippocampus may be a significant factor in precipitating episodes of depression and is sufficient to induce anxiety and depression-related behaviors in mice.52 Clinical study also reported that both NeuN+ granule neurons and Nestin+ cells in the dentate gyrus of MDD patients were decreased, indicating a reduction in neural stem cells.53 Consistent with the previous viewpoint, our study showed that neurogenesis was significantly reduced in the hippocampus of CUMS mice, and CSS could reverse this change and alleviate the depressive-like behaviors.
An increasing number of studies have reported that the PI3K/AKT signaling pathway is involved in the pathogenesis of depression. Enzymatic activity of PI3K and AKT was decreased in depressed patients,54 and decreased pPI3K/PI3K and pAKT/AKT ratios in the hippocampus were found in CUMS mice.55 Evidence has also shown that activation of AKT signaling can alleviate stress-induced depressive-like behaviors in mice.55,56 Moreover, this pathway plays an important role in hippocampal neurogenesis. AKT signaling is an important upstream regulator of GSK3β. Activated AKT can inhibit GSK3β activity by phosphorylating Ser9 of GSK3β. Studies have shown that increased GSK3β activity could directly impair hippocampal neural stem cell proliferation,57 whereas this alteration could be reversed by activating the PI3K/AKT pathway.58 Our network pharmacology analysis showed that the PI3K/AKT pathway might be the major pathway involved in the effects of CSS in MDD treatment, as it included the most predicted hub targets. However, the effects of CSS on PI3K/AKT/GSK3β signaling have been rarely reported. In this study, we found that CSS treatment could augment pPI3K/PI3K, pAKT/AKT, and pGSK3β/GSK3β ratios in the hippocampus of CUMS mice. These results indicated that the antidepressant-like action of CSS is mediated, at least in part, via modulating the PI3K/AKT pathway, thereby improving hippocampal neurogenesis.
Moreover, three active ingredients of CSS were predicted in the network pharmacology analysis. Quercetin was found to have the most functional targets. Several studies have revealed that the antidepressant effects of quercetin are associated with improvement of neurotrophic function59 and anti-inflammation effects.60 Kaempferol could ameliorate depressive symptoms and upregulate the activity of the AKT/β-catenin cascade in the prefrontal cortex of chronic social defeat stress model mice.61 Luteolin was reported to exert a significant antidepressant effect and inhibit neuronal apoptosis.62 These findings indicated that the candidate compounds are a promising source of antidepressant agents. Furthermore, the roles of the active compounds may be associated with the PI3K/AKT signaling pathway. In our study, molecular docking analysis suggested that the active compounds exhibited strong interactions with PI3K/AKT signaling proteins. This study is a preliminary attempt to explore the effects of active compounds on the PI3K/AKT pathway. It may provide novel directions for further research on the molecular mechanisms underlying active compounds that ameliorate MDD.
The main limitation of our study is that we only focused on the PI3K/AKT signaling pathway in our experimental validations. Although the PI3K/AKT signaling pathway was revealed to be important in the pharmacology network analysis, most of the other predicted pathways still need to be verified. The Rap1 and Ras signaling pathway, although not enriched in most hub targets, may also provide new insights for further research. Moreover, the effect of active compounds that act on the PI3K/AKT pathway in CUMS-induced mice should be further explored.
Conclusions
In summary, our present study identified three active ingredients of CSS, determined multiple potential therapeutic targets, and found that the PI3K/AKT signaling pathway was the most promising candidate pathway for mediating the antidepressant effects of CSS. The present study provides novel insights into the effective compounds and pharmacological mechanisms of CSS with regard to MDD, which will be beneficial for the formulation development, in-depth research, and application of the traditional medicines.
Abbreviations
ADME, absorption, distribution, metabolism, and excretion; AKT, protein kinase B; BP, biological process; BrdU, Bromodeoxyuridine; CC, cellular component; CSS, Chaihu Shugan San; CUMS, chronic unpredictable mild stress; DAVID, Database for Annotation, Visualization and Integrated Discovery; DL, drug likeness; FST, Forced swimming test; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; LC-MS/MS, Liquid chromatography-tandem mass spectrometry; MDD, major depressive disorder; MF, molecular function; OB, oral bioavailability; p, phosphorylated; PBS, phosphate-buffered saline; PI3K, phosphoinositide 3-kinase; PPI, Protein-protein interactions; SPT, Sucrose preference test; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins; TCM, traditional Chinese medicines; TCMSP, Traditional Chinese Medicine System Pharmacology Database; TTD, Therapeutic Target Database; TST, tail suspension test.
Data Sharing Statement
The data that support the finding of the present study are shown in Supplementary Materials.
Ethics Approval and Informed Consent
All experiments were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Experimental Animal Ethics Committee of Beijing Friendship Hospital (Approval No.19-1006).
Acknowledgments
This study was supported by the Beijing Natural Science Foundation (Grant No. 7204250), National Natural Science Foundation of China (Grant No. 82004109), Beijing Administration of Traditional Chinese Medicine (Grant No. JJ2018-51). We are grateful to Prof. Feng Qiu of Capital Medical University for his supports to the work of LC/MS/MS analysis. We also thank Lisa Kreiner, PhD, from Liwen Bianji, (Edanz), for editing the English text of a draft of this manuscript.
Author Contributions
All authors contributed to data analysis, draft, or revision, gave final approval of the version to be published, have agreed on the journal to which the article has been submitted, and agreed to be accountable for all aspects of the work.
Disclosure
The authors declare that the research has no competing interests.
References
1. Smith K. Mental health: a world of depression. Nature. 2014;515(7526):181. doi:10.1038/515180a
2. Monteggia LM, Malenka RC, Deisseroth K. Depression: the best way forward. Nature. 2014;515(7526):200–201. doi:10.1038/515200a
3. Huang Y, Wang Y, Wang H, et al. Prevalence of mental disorders in China: a cross-sectional epidemiological study. Lancet Psychiatry. 2019;6(3):211–224. doi:10.1016/s2215-0366(18)30511-x
4. Kirsch I, Jakobsen JC. Network meta-analysis of antidepressants. Lancet (London, England). 2018;392(10152):1010. doi:10.1016/s0140-6736(18)31799-9
5. Fournier JC, DeRubeis RJ, Hollon SD, et al. Antidepressant drug effects and depression severity: a patient-level meta-analysis. JAMA. 2010;303(1):47–53. doi:10.1001/jama.2009.1943
6. Orgeta V, Tabet N, Nilforooshan R, et al. Efficacy of antidepressants for depression in alzheimer’s disease: systematic review and meta-analysis. J Alzheimers Dis. 2017;58(3):725–733. doi:10.3233/jad-161247
7. Papakostas GI, Jackson WC, Rafeyan R, et al. Inadequate response to antidepressant treatment in major depressive disorder. J Clinl Psychiatry. 2020;81(3):OT19037COM5. doi:10.4088/jcp.Ot19037com5
8. Yang L, Shergis JL, Di YM, et al. Managing depression with bupleurum chinense herbal formula: a systematic review and meta-analysis of randomized controlled trials. J Altern Compl Med. 2020;26(1):8–24. doi:10.1089/acm.2019.0105
9. Qiu J, Hu SY, Shi GQ, et al. Changes in regional cerebral blood flow with Chaihu-Shugan-San in the treatment of major depression. Pharmacogn Mag. 2014;10(40):503–508. doi:10.4103/0973-1296.141775
10. Chen XQ, Li CF, Chen SJ, et al. The antidepressant-like effects of Chaihu Shugan San: dependent on the hippocampal BDNF-TrkB-ERK/Akt signaling activation in perimenopausal depression-like rats. Biomed Pharmacother. 2018;105:45–52. doi:10.1016/j.biopha.2018.04.035
11. Sun KH, Jin Y, Mei ZG, et al. Antidepressant-like effects of chaihu shugan powder () on rats exposed to chronic unpredictable mild stress through inhibition of endoplasmic reticulum stress-induced apoptosis. Chin J Integr Med. 2021;27(5):353–360. doi:10.1007/s11655-020-3228-y
12. Liu Y, Ma S, Qu R. SCLM, total saponins extracted from Chaihu-jia-longgu-muli-tang, reduces chronic mild stress-induced apoptosis in the hippocampus in mice. Pharm Biol. 2010;48(8):840–848. doi:10.3109/13880200903296154
13. Zhang Q, Yu H, Qi J, et al. Natural formulas and the nature of formulas: exploring potential therapeutic targets based on traditional Chinese herbal formulas. PLoS One. 2017;12(2):e0171628. doi:10.1371/journal.pone.0171628
14. Kibble M, Saarinen N, Tang J, et al. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat Prod Rep. 2015;32(8):1249–1266. doi:10.1039/c5np00005j
15. Xu J, Guan Z, Wang X, et al. Network pharmacology and experimental evidence identify the mechanism of astragaloside IV in oxaliplatin neurotoxicity. Drug Des Devel Ther. 2021;15:99–110. doi:10.2147/dddt.S262818
16. Panossian AG, Efferth T, Shikov AN, et al. Evolution of the adaptogenic concept from traditional use to medical systems: pharmacology of stress- and aging-related diseases. Med Res Rev. 2021;41(1):630–703. doi:10.1002/med.21743
17. Liu YY, Hu D, Fan QQ, et al. Mechanism of chaihu shugan powder () for treating depression based on network pharmacology. Chin J Integr Med. 2020;26(12):921–928. doi:10.1007/s11655-019-3172-x
18. Jiang Y, Zhong M, Long F, et al. Network pharmacology-based prediction of active ingredients and mechanisms of lamiophlomis rotata (benth.) kudo against rheumatoid arthritis. Front Pharmacol. 2019;10:1435. doi:10.3389/fphar.2019.01435
19. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi:10.1186/1758-2946-6-13
20. Liu H, Wang J, Zhou W, et al. Systems approaches and polypharmacology for drug discovery from herbal medicines: an example using licorice. J Ethnopharmacol. 2013;146(3):773–793. doi:10.1016/j.jep.2013.02.004
21. Feng SH, Zhao B, Zhan X, et al. Danggui buxue decoction in the treatment of metastatic colon cancer: network pharmacology analysis and experimental validation. Drug Des Devel Ther. 2021;15:705–720. doi:10.2147/dddt.S293046
22. Gfeller D, Grosdidier A, Wirth M, et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(WebServer issue):W32–W38. doi:10.1093/nar/gku293
23. Gfeller D, Michielin O, Zoete V. Shaping the interaction landscape of bioactive molecules. Bioinformatics. 2013;29(23):3073–3079. doi:10.1093/bioinformatics/btt540
24. Wang Y, Zhang S, Li F, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031–D1041. doi:10.1093/nar/gkz981
25. Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2019;48(D1):D845–D855. doi:10.1093/nar/gkz1021
26. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi:10.1093/nar/gkx1037
27. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi:10.1093/nar/gkw937
28. Jiao X, Sherman BT, Huang da W, et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28(13):1805–1806. doi:10.1093/bioinformatics/bts251
29. Wang C, Chen Y, Wang Y, et al. Inhibition of COX-2, mPGES-1 and CYP4A by isoliquiritigenin blocks the angiogenic Akt signaling in glioma through ceRNA effect of miR-194-5p and lncRNA NEAT1. J Exp Clinl Cancer Res. 2019;38(1):371. doi:10.1186/s13046-019-1361-2
30. Song H, Park J, Choi K, et al. Ginsenoside Rf inhibits cyclooxygenase-2 induction via peroxisome proliferator-activated receptor gamma in A549 cells. J Ginseng Res. 2019;43(2):319–325. doi:10.1016/j.jgr.2018.11.007
31. Young JY, Berrisford J, Chen M. wwPDB biocuration: on the front line of structural biology. Nat Methods. 2021;18(5):431–432. doi:10.1038/s41592-021-01137-z
32. Burley SK, Berman HM, Bhikadiya C. Protein data bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019;47(D1):D520–D528. doi:10.1093/nar/gky949
33. Liu H, Qiu F, Bian B, et al. Integrating qualitative and quantitative assessments of Yougui pill, an effective traditional Chinese medicine, by HPLC-LTQ-Orbitrap-MSn and UPLC-QqQ-MS/MS. Anal Methods. 2017;9(23):3485–3496. doi:10.1039/c7ay00259a
34. Sun HY, Li Q, Liu YY, et al. Xiao-Yao-San, a Chinese medicine formula, ameliorates chronic unpredictable mild stress induced polycystic ovary in rat. Front Physiol. 2017;8:729. doi:10.3389/fphys.2017.00729
35. Su WJ, Zhang Y, Chen Y, et al. NLRP3 gene knockout blocks NF-κB and MAPK signaling pathway in CUMS-induced depression mouse model. Behav Brain Res. 2017;322(Pt A):1–8. doi:10.1016/j.bbr.2017.01.018
36. Zhao M, Tang X, Gong D, et al. Bungeanum improves cognitive dysfunction and neurological deficits in D-Galactose-induced aging mice via activating PI3K/Akt/Nrf2 signaling pathway. Front Pharmacol. 2020;11:71. doi:10.3389/fphar.2020.00071
37. Li L, Yu AL, Wang ZL, et al. Chaihu-Shugan-San and absorbed meranzin hydrate induce anti-atherosclerosis and behavioral improvements in high-fat diet ApoE (-/-) mice via anti-inflammatory and BDNF-TrkB pathway. Biomed Pharmacother. 2019;115:108893. doi:10.1016/j.biopha.2019.108893
38. Gao C, Du Q, Li W, et al. Baicalin modulates APPL2/Glucocorticoid receptor signaling cascade, promotes neurogenesis, and attenuates emotional and olfactory dysfunctions in chronic corticosterone-induced depression. Mol Neurobiol. 2018;55(12):9334–9348. doi:10.1007/s12035-018-1042-8
39. Liu MY, Yin CY, Zhu LJ, et al. Sucrose preference test for measurement of stress-induced anhedonia in mice. Nature Protoco. 2018;13(7):1686–1698. doi:10.1038/s41596-018-0011-z
40. Iñiguez SD, Riggs LM, Nieto SJ, et al. Social defeat stress induces a depression-like phenotype in adolescent male c57BL/6 mice. Stress. 2014;17(3):247–255. doi:10.3109/10253890.2014.910650
41. Sahu P, Mudgal J, Arora D, et al. Cannabinoid receptor 2 activation mitigates lipopolysaccharide-induced neuroinflammation and sickness behavior in mice. Psychopharmacology. 2019;236(6):1829–1838. doi:10.1007/s00213-019-5166-y
42. Steru L, Chermat R, Thierry B, et al. The tail suspension test: a new method for screening antidepressants in mice. Psychopharmacology. 1985;85(3):367–370. doi:10.1007/bf00428203
43. Walker AK, Budac DP, Bisulco S, et al. NMDA receptor blockade by ketamine abrogates lipopolysaccharide-induced depressive-like behavior in C57BL/6J mice. Neuropsychopharmacology. 2013;38(9):1609–1616. doi:10.1038/npp.2013.71
44. Liu W, Liu J, Huang Z, et al. Possible role of GLP-1 in antidepressant effects of metformin and exercise in CUMS mice. J Affect Disord. 2019;246:486–497. doi:10.1016/j.jad.2018.12.112
45. Chen D, Wei L, Liu ZR, et al. Pyruvate Kinase M2 Increases Angiogenesis, Neurogenesis, and Functional Recovery Mediated by Upregulation of STAT3 and focal adhesion kinase activities after ischemic stroke in adult mice. Neurotherapeutics. 2018;15(3):770–784. doi:10.1007/s13311-018-0635-2
46. Wei L, Fraser JL, Lu ZY, et al. Transplantation of hypoxia preconditioned bone marrow mesenchymal stem cells enhances angiogenesis and neurogenesis after cerebral ischemia in rats. Neurobiol Dis. 2012;46(3):635–645. doi:10.1016/j.nbd.2012.03.002
47. Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 2012;13(6):6964–6982. doi:10.3390/ijms13066964
48. Tao W, Xu X, Wang X, et al. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J Ethnopharmacol. 2013;145(1):1–10. doi:10.1016/j.jep.2012.09.051
49. Qiu J, Hu SY, Zhang CH, et al. The effect of Chaihu-Shugan-San and its components on the expression of ERK5 in the hippocampus of depressed rats. J Ethnopharmacol. 2014;152(2):320–326. doi:10.1016/j.jep.2014.01.004
50. Yang JL, Lin XG, Pan Y, et al. Critical roles of mTOR Complex 1 and 2 for T follicular helper cell differentiation and germinal center responses. eLife. 2016;5:e17936. doi:10.7554/eLife.17936
51. Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003;46(4):499–511. doi:10.1021/jm020406h
52. Tunc-Ozcan E, Peng CY, Zhu Y, et al. Activating newborn neurons suppresses depression and anxiety-like behaviors. Nat Commun. 2019;10(1):3768. doi:10.1038/s41467-019-11641-8
53. Boldrini M, Galfalvy H, Dwork AJ, et al. Resilience is associated with larger dentate gyrus, while suicide decedents with major depressive disorder have fewer granule neurons. Bioll Psychiatry. 2019;85(10):850–862. doi:10.1016/j.biopsych.2018.12.022
54. Karege F, Perroud N, Burkhardt S, et al. Alterations in phosphatidylinositol 3-kinase activity and PTEN phosphatase in the prefrontal cortex of depressed suicide victims. Neuropsychobiology. 2011;63(4):224–231. doi:10.1159/000322145
55. Xian YF, Ip SP, Li HQ, et al. Isorhynchophylline exerts antidepressant-like effects in mice via modulating neuroinflammation and neurotrophins: involvement of the PI3K/Akt/GSK-3β signaling pathway. FASEB J. 2019;33(9):10393–10408. doi:10.1096/fj.201802743RR
56. Guo LT, Wang SQ, Su J, et al. Baicalin ameliorates neuroinflammation-induced depressive-like behavior through inhibition of toll-like receptor 4 expression via the PI3K/AKT/FoxO1 pathway. J Neuroinflammation. 2019;16(1):95. doi:10.1186/s12974-019-1474-8
57. Pardo M, Abrial E, Jope RS, et al. GSK3β isoform-selective regulation of depression, memory and hippocampal cell proliferation. Genes Brain Behav. 2016;15(3):348–355. doi:10.1111/gbb.12283
58. Wang YH, Chern CM, Liou KT, et al. Ergostatrien-7,9(11),22-trien-3β-olfrom Antrodia camphorata ameliorates ischemic stroke brain injury via downregulation of p65NF-κ-B and caspase 3, and activation of Akt/GSK3/catenin-associated neurogenesis. Food Funct. 2019;10(8):4725–4738. doi:10.1039/c9fo00908f
59. Fang K, Li HR, Chen XX, et al. Quercetin alleviates LPS-induced depression-like behavior in rats via regulating bdnf-related imbalance of copine 6 and TREM1/2 in the hippocampus and PFC. Front Pharmacol. 2019;10:1544. doi:10.3389/fphar.2019.01544
60. Merzoug S, Toumi ML, Tahraoui A. Quercetin mitigates Adriamycin-induced anxiety- and depression-like behaviors, immune dysfunction, and brain oxidative stress in rats. Naunyn Schmiedebergs Arch Pharmacol. 2014;387(10):921–933. doi:10.1007/s00210-014-1008-y
61. Gao W, Wang W, Peng Y, Deng Z. Antidepressive effects of kaempferol mediated by reduction of oxidative stress, proinflammatory cytokines and up-regulation of AKT/β-catenin cascade. Metab Brain Dis. 2019;34(2):485–494. doi:10.1007/s11011-019-0389-5
62. Crupi R, Paterniti I, Ahmad A, et al. Effects of palmitoylethanolamide and luteolin in an animal model of anxiety/depression. CNS Neurol Disord Drug Targets. 2013;12(7):989–1001. doi:10.2174/18715273113129990084
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.