Back to Journals » Cancer Management and Research » Volume 13
Myelin and Lymphocyte Protein (MAL): A Novel Biomarker for Uterine Corpus Endometrial Carcinoma
Authors Li D, Zhang J , Wu L, Yang X, Chen Z, Yuan J
Received 4 May 2021
Accepted for publication 30 August 2021
Published 21 September 2021 Volume 2021:13 Pages 7311—7323
DOI https://doi.org/10.2147/CMAR.S317319
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Dong Li,1,* Juan Zhang,2,* Lilong Wu,2 Xiaoming Yang,1 Zheng Chen,1 Jiangjing Yuan1
1Department of Obstetrics and Gynecology, International Peace Maternity and Child Health Hospital, Shanghai Key Laboratory of Embryo Original Diseases, Shanghai Municipal Key Clinical Speciality, School of Medicine, Shanghai Jiao Tong University, Shanghai, People’s Republic of China; 2Department of Obstetrics and Gynecology, Jinan Maternal and Child Health Care Hospital, Jinan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jiangjing Yuan Email [email protected]
Purpose: Myelin and lymphocyte protein (MAL) plays an essential role in esophageal cancer, classic Hodgkin’s lymphoma and breast cancer. However, its role in uterine corpus endometrial carcinoma (UCEC) has not been explored. Therefore, the current study sought to explore the role of MAL in UCEC.
Patients and Methods: Differentially expressed genes (DEGs) were identified by using Limma package in R based on TCGA-UCEC data. Kaplan–Meier plotter analysis was performed to explore the prognostic value of MAL. Function enrichment analyses were performed using GSVA. Further, roles of MAL in UCEC were validated using clinical cohort, which included 120 tumor and adjacent tissues. qRT-PCR and immunohistochemistry analyze the samples. Chi-square tests were performed to explore the associations between MAL expressions and clinicopathological features.
Results: The findings showed that overexpression level of MAL in tumor was correlated with worse survival (p = 0.000424). MAL exhibited predictive power for survival time of UCEC patients (3 years: AUC = 0.635; 5 years: AUC = 0.635). Notably, high expression level of MAL was correlated with advanced stage of UCEC. MAL overexpression was significant in UCEC with microsatellite instability (MSI). Enrichment analysis showed that MAL was enriched mainly in MYC targets, epithelial mesenchymal transition and KRAS signaling. Furthermore, MAL was associated with infiltration of immune cells in the tumor micro-environment and immune checkpoint. Analysis showed a positive association between MAL and T cell (CD4+ memory resting). Correlation analysis showed that MAL was significantly positively correlated with several immune checkpoint, including CD274 (R = 0.3389, p = 0.0081), LAG3 (R = 0.2913, p = 0.0229), PDCD1LG2 (R = 0.5345, p < 0.0001). The prognosis value of MAL was confirmed through the experiment.
Conclusion: The findings of the current study indicated that MAL is an effective prognostic biomarker and potential therapeutic target for UCEC patients. These results indicated that MAL functions as a diagnosis and therapeutic marker in UCEC treatment.
Keywords: MAL, UCEC, prognosis, immune micro environment, biomarker
Introduction
UCEC is a commonly diagnosed gynecological malignancy of the genital tract. The current global cancer statistics report that the incidence of UCEC is approximately 4.4%.1 The conventional therapy for UCEC is surgery treatment followed by adjuvant radio- and/or chemotherapy depending on the tumor characteristics.2,3 Hormone therapy is an alternative treatment for patients with metastatic or recurrent disease, with limited treatment options and those who wish to maintain their fertility.4 Alternative therapeutic approaches for UCEC, such as molecular targeted therapy, have attracted increasing attention. Yen et al summarized several potential targeted approaches for the treatment of UCEC, including endocrine therapy, targeting HER2/Neu, targeting PI3K-AKT-mTOR pathway and immune checkpoint inhibitors.5 Tucci et al reported immunotherapeutic options currently available for UCEC. IGF, EGF, genomic instability and angiogenesis represent the hallmarks of UCEC, most investigated in the last decade with significant findings. In addition, adoptive T cell therapies are presented as a promising strategy for the treatment of UCEC.6 Tumorigenesis is highly correlated with abnormal activation of multiple signaling pathways, and molecular drugs can specifically target these abnormally activated signaling pathways, regulate the signaling pathways, reduce damage to normal cells, thus achieving effective and specific tumor killing. Temsirolimus is a water-soluble rapamycin ester and a highly specific molecular drug. Approximately 26% partial response rate was documented in a Phase II trial using this agent to treat 31 women with recurrent or metastatic UCEC.7 Currently, effectiveness of molecular therapy is unsatisfactory owing to lack of accurate biomarkers for molecular therapy. Therefore, there is a need to identify effective biomarkers for molecular therapy.
MAL8 is a prototypical member of the MAL proteolipids, which belongs to a family including MAL-2, BENE, and plasmolipin.9–11 MAL is implicated in apical transport of proteins in polarized epithelial cells.9 Notably, MAL is highly expressed in trans-Golgi network (TGN) and vesicles derived from TGN, the apical region of plasma membrane, and early endosome membranes.12 MAL is a 17 kDa protein characterized by glycosphingolipid and cholesterol-enriched microdomains (GEMs) or membrane rafts (MR). MAL can be self-associated, and induces molecules to be transported from the extracellular space into the cytoplasm. MAL-containing MR can perform similar functions as Clostridium perfringens ε-toxin (CPET).13–15 MAL can modulate cellular processes including cell motility, proliferation and survival through specific pathways. MAL modulates tumor micro-environment thus promoting tumor development, in a similar mechanism as, Epstein–Barr virus oncoprotein LMP-1, which activates MEK-ERK and PIk3-Akt pathways and self-associates in GEMs.16 MAL is implicated in cell transformation. In addition, MAL is important in epithelial differentiation along with glycosphingolipids17–19 and axonal segregation. A previous study reported MAL protein in myelin synthesizing cells.20,21 Expression of MAL in T-lymphocytes was initially reported only in late stages of development, implying that it can be detected in mature T-cells; however, MAL-mRNA was not detected in proliferating T-cells.8 Notably, the role of MAL in UCEC has not been fully elucidated.
The Cancer Genome Atlas (TCGA) is a publicly funded project that aims at cataloguing and discovering major cancer-causing genomic alterations. High-throughput RNA-sequencing data from TCGA was used in the current study. The findings showed that MAL plays a role in UCEC and is a potential immunotherapy biomarker. The role of high expression of MAL on clinical prognosis was confirmed using clinical data from TCGA database. In addition, the effect of MAL on tumor prognosis was verified through tissue samples.
Patients and Methods
Data Retrieval and Identification of Differentially Expressed mRNAs
Transcriptome data of UCEC used in this study were retrieved from TCGA-UCEC database. The data included 552 UCEC tissues and 35 adjacent normal tissues. This dataset comprised 23 paired samples. Clinical information including age, gender, tumor stage, pathology information, and overall survival (OS) were obtained. Differential gene expression between the two groups was analyzed using limma R package. P-value < 0.05 and |log 2 (fold change [FC])| >1 were used as the threshold criteria.
Kaplan–Meier Plotter Database Analysis
KM-Plotter database (http://kmplot.com/analysis/) comprises survival data of UCEC patients. Relationship between mRNA expression levels of MAL and OS in UCEC was analyzed using KM-Plotter database. The median of gene expression was used as the cutoff value to divide patient samples into two groups and plots were generated. Hazard ratio (HR), 95% confidence interval (CI), and log-rank p were determined.
Gene Set Variation Analysis (GSVA)
Gene set variation analysis was performed using the GSVA R package. This analysis was performed to obtain an enrichment score and pathway by transforming a classical gene matrix (gene-by-sample) into a gene set by sample matrix (pathway score-by sample). Patients were divided into control group and test group based on the expression level of MAL. The most significant hallmark gene-set in the enrichment analysis was selected as the reference gene-set. Gene ranking was performed using Signal2Noise metric. The threshold was set at DR < 0.25 and P-value < 0.05.
Cell Type Identification by Estimating Relative Subsets of RNA Transcripts
To explore the landscape of immune infiltration in UCEC patients with high and low MAL expression level, CIBERSORT tool was used to determine the mRNA expression profile. CIBERSORT tool can effectively and specifically discriminate 22 human immune cell phenotypes, including naive B cells, memory B cells, CD8 T cells, different CD4 T cell types, Tregs, NK cells, plasma cells, monocytes, three macrophage types, and dendritic cells. The average CIBERSORT score of MAL low-expression group and MAL high-expression group for the 22 immune cells was calculated. In addition, transcriptional expression level of 8 immune checkpoints was determined between the two groups.
Tissue Samples
Cancer tissues and their corresponding adjacent normal tissues from 120 cases diagnosed with UCEC at the Jinan Maternity and Child Health Care Hospital between January 2015 and January 2016. Moreover, clinical information was obtained from electronic medical records. The tissue samples were immediately snap-frozen in liquid nitrogen and stored under −80°C for further processing. Tumor samples were fixed with formalin, and embedded using paraffin for histologic and immunohistochemical analysis. The study design and protocol were approved by the ethics committee of Jinan Maternity and Child Health Care Hospital. All patients included in this study signed informed consent forms.
Immunohistochemistry (IHC)
Immunohistochemistry was performed following standard protocols. Anti-MAL primary antibody and a secondary antibody were used to determine protein expression level of MAL in UCEC tumor samples. Patients were classified into four groups, including score 0, score 1, score 2 and score 3 four groups. Score 0 and score 1 represented low MAL expression group and score 2 and score 3 represented high MAL expression. A survival curve was plotted based on patients’ follow-up data.
RNA Extraction and Reverse Transcription and Quantitative (RT-PCR)
Total RNA was extracted from UCEC tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was reverse-transcribed into complementary DNA (cDNA) using HiScript II. Further, qRT-PCR was performed using SYBR Green I (Vazyme, Shanghai, China) on ABI 7900 system (Applied Biosystems, Carlsbad, CA, USA). Primers for MAL, CD274, CTLA4, LAG3, PDCD1LG2, GAPDH are presented in Table S1.
Western Blotting
Western blotting was performed to explore protein expression levels. Radioimmunoprecipitation assay (RIPA) containing protease and phosphatase inhibitors was used to prepare the protein extract. Lysates were then diluted with 5× loading buffer. The mixture was then boiled at 95 °C for 10 min. Proteins were isolated using gradient SDS-page (10%). After blocking with 5% bull serum albumin for 2 hours, membranes were blotted with primary antibodies overnight at 4°C. Protein bands in the membrane were visualized using ECL substrate kit.
Statistical Analysis
The relationship between MAL expression and clinicopathological factors was analyzed using the χ2 test. Survival statistics were performed using Kaplan–Meier curve and log rank test. Univariate and multivariate survival analyses were performed using Cox regression analysis. Correlation between MAL expression and expression of immune checkpoint genes was analyzed using Pearson correlation analysis. Comparisons between the two groups were carried out using Student’s t-test. P < 0.05 was considered statistically significant. R software was used for all analyses.
Results
Identification of Differentially Expressed Genes
Differential expression analysis was performed based on 552 cancerous and 35 adjacent normal tissues. The findings showed that 8887 genes were differentially expressed between UCEC cancerous and adjacent normal tissues (Figure 1A). Further, differential gene expression analysis was performed on 23 paired samples to reducing the bias owing to the unbalanced characteristics of the data retrieved from TCGA-UCEC (Figure 1B). A total of 6747 genes were selected based on the intersection of the first gene set and the second gene set (Figure 1C). Univariate and multivariate Cox proportional hazard analysis was performed using these genes to explore their prognostic value in predicting survival of UCEC patients. Univariate survival analysis showed that 35 genes were correlated with the overall survival of UCEC patients (Table S2). Moreover, 5 cancer‐promoting genes (Figure 1D), including RNF2P1 (HR:2.38 (1.13, 4.12)), AC012354.1 (HR:1.43 (1.11, 1.86)), AC005381.1 (HR:2.16 (1.53, 3.04)), PHACTR3 (HR: 1.65 (1.11, 2.44)) and MAL (HR: 1.22 (1.05, 1.41)) were identified by multivariate survival analysis.
Association Between MAL Expression and Clinical Prognosis in UCEC
MAL was used for further analysis as differential genes. Expression analysis of cancerous tissues and adjacent normal tissues showed that the other 4 gene expressions were too low for clinical application (Figure 2A). Notably, higher MAL expression level was observed in stage III and IV, indicating that MAL may be an oncogene in UCEC (p < 0.05, Figure 2B). In addition, Kaplan–Meier plotter analysis was performed to generate survival curve for UCEC patient based on MAL expression. High expression of MAL was correlated with poorer overall survival of UCEC patients (p=0.000424, Figure 2C and D). Moreover, MAL could accurately predict 1-year (AUC=0.559), 3-years (AUC=0.635) and 5-years (AUC=0.635) survival rates of UCEC patients (Figure 2E). These findings indicated that MAL is a potential prognosis predictor of UCEC.
Relationship Between MAL and Immune Response
The protein encoded by MAL is a member of MAL family of proteolipids. The protein is a highly hydrophobic integral membrane protein. The protein is expressed predominantly in endoplasmic reticulum of T-cells and compact myelin of cells in the nervous system. It is implicated in ligation of T cell signal transduction, formation, stabilization and maintenance of glycosphingolipid-enriched membrane microdomains. Changes in expression level of MAL gene are associated with tumor progression. Multiple transcript variants of MAL arise from alternative splicing owing to the presence of exons 2 and 3 or not. These findings indicate that MAL may play a role in immune response; thus, CIBERSORT tool was used to analyze differences in micro-immune microenvironment between high and low MAL expression groups.
A total of 7 immune cells including B cell memory, T cell CD8+, T cell regulatory (Tregs), T cell gamma delta, NK cell activated, macrophage M1 and Myeloid dendritic cell activated were used in differential expression analysis between high and low MAL expression groups (Figure 3A and B). To further explore the role of MAL in immune response, the association between MAL and immune checkpoints was evaluated using 23 paired samples (Figure 4A). The findings showed a difference in CD274, CTLA4, LAG3 and PDCDILG2 scores between high and low MAL expression groups. Notably, Pearson correlation analysis showed correlations between MAL and CD274 (R=0.3389, p=0.0081), CTLA4 (R=−0.2913, p=0.0239), LAG3 (R=0.2933, p=0.0229) and PDCDILG2 (R=0.5345, p < 0.0001) (Figure 4B).
Moreover, GSVA was performed to explore the dynamics of biological processes and pathways for hallmark gene sets based on MAL expression. The findings showed that MAL is implicated in regulating various cellular processes. Significantly enriched signaling pathways between MAL high group and MAL low group were identified, including 17 upregulated pathways and 16 downregulated pathways. Notably, upregulated pathways included MYC V2 targets, KRAS signaling pathway, late estrogen response pathway, myogenesis and TNFα-signaling through NFκB. On the other hand, downregulated pathways included epithelial mesenchymal transition, inflammatory response, IL2/STAT5 signaling, protein secretion and early estrogen response (Figure 5).
 |
Figure 5 GSVA analysis for tumor and adjacent tissue. |
Expression of MAL in Clinical UCEC Cohorts and Its Prognosis Validation
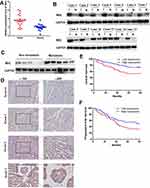
Further, Western blotting and qRT-PCR assays were performed to determine MAL expression levels in UCEC tumor and adjacent normal tissues. qRT-PCR analysis showed that transcriptional expression of MAL was higher in tumor tissues compared with the paired non-tumor controls (Figure 6A). Notably, 11 of the 13 pairs UCEC tissues showed higher levels of MAL compared with their adjacent normal tissues (Figure 6B). In addition, metastatic UCEC showed higher MAL expression level compared with non-metastatic tissues (Figure 6C).
To validate results obtained from bioinformatics analysis, IHC staining assay was performed on tissue microarrays (TMAs) of UCEC samples of 120 cases. The relationship between MAL expression and clinicopathological features of UCEC patients is summarized in Table 1. Analysis showed no significant association between MAL expression level with age and menopause. However, the findings showed that high expression of MAL was correlated with advanced histological grade (Figure 6D). To validate the prognostic value of MAL, Kaplan–Meier analysis was performed using UCEC cohort. Kaplan–Meier analysis showed that high MAL expression level was correlated with a poor OS (P < 0.05) and progression-free survival (PFS) (P < 0.05) (Figure 6E and F).
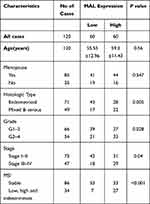 |
Table 1 Correlations Between Expression Level of MAL and Clinicopathological Features in 120 UCEC Patients |
Discussion
MAL shows different roles during development and progression of different cancer types. Previous studies report that MAL plays a role as tumor suppressor in esophageal cancer.22,23 A study by Mimor reported that promoter hypermethylation is the direct cause of low expression level of MAL.24 Similar findings have been reported in other cancer types, including colon cancer,25,26 breast cancer,27,28 stomach cancer,29,30 salivary gland cancer,31 head and neck carcinomas,32,33 non-small cell lung cancer,34 and bladder cancer.35 However, overexpression of MAL has been reported in clear cell-ovarian carcinoma.36 Notably, in the invasive serous ovarian cancer patients, MAL was associated with short-term survival.37 Moreover, overexpression of MAL has been reported in primary mediastinal large B-cell lymphoma,38,39 acute adult T-cell leukemia/lymphoma40 and classic Hodgkin’s lymphoma.41 Currently, studies have not fully explored the role and mechanism for MAL in UCEC oncogenesis.
In the current study, five genes related to cancer development and prognosis were screened based on TCGA data. Notably, MAL gene was used for subsequent analysis owing to the usefulness of the biomarker. Millan et al42 reported that MAL is involved in the formation of apical transporter and may be a functional component of vesicle transport and protein sorting between Golgi apparatus and the distal plasma membrane. Apical transport is transport of fat and protein components to the apical membrane based on cellular polarities. This transport mode is inherent in epithelial cells, and loss of functional polar transport is often associated with malignant transformation processes.43 Alonson et al8 reported that MAL is highly expressed in the middle and late stages of T cell differentiation. MAL gene is involved in intracellular transport, gene expression, immune regulation and other important cellular processes. MAL2 promotes endocytosis of tumor antigens through direct interaction with the MHC-I complex and endosome-associated RAB proteins. In preclinical models, depletion of MAL2 in breast tumor cells significantly increased cytotoxicity of tumor-infiltrating CD8+T cells and suppressed breast tumor growth, indicating that MAL2 is a potential therapeutic target for breast cancer immunotherapy.44,45
In the current study, MAL was overexpressed in UCEC tumor tissues compared with adjacent normal tissues, and this gene was associated with shorter survival of UCEC patients. Notably, these findings were validated using clinical cohorts. In addition, pathway enrichment analysis showed that MAL was involved in several pathways associated with cancer. For example, the MAL overexpression was associated with late estrogen response pathway. Some studies report the protective effect of increasing phytoestrogen consumption to reduce the risk of UCEC.46 In addition, MAL overexpression was correlated with several terms implicated in cell cycle signal pathways, such as mitotic spindle, G2M checkpoint and E2F target. These findings indicate that MAL is implicated in cell cycle regulation disorders that block cell differentiation and apoptosis, thus promoting cell proliferation in UCEC. Moreover, MAL is related to microenvironment factors (including Tregs and B cell memory and so on) and immune checkpoint genes (including CD274, CTLA4, LAG3, and PDCDILG2). Therefore, MAL may be a potential biomarker for immunotherapy efficacy in UCEC. This is the first study to explore the correlation between MAL and immune checkpoints.
Studies should explore cancer heterogeneity, mainly intratumor heterogeneity, to develop approaches for the treatment of cancer. Bulk RNA sequencing studies are based on a collection of cells, and individual cell-specific information is often obscured, thus important information is missed. Association between MAL and UCEC prognosis is not significant. Further studies should carry out single-cell sequencing to further explore the regulatory mechanism of MAL in UCEC.
In summary, the findings of the current study indicate that MAL is overexpression in UCEC tissues and expression level is correlated with worse survival. MAL is highly correlated with crucial immune cells in the immune microenvironment. Therefore, MAL is a potential therapeutic target for UCEC. Further studies should be conducted to elucidate the underlying mechanism of MAL in UCEC. The findings of the current study provide a basis for the development of new therapeutic agents for patients with UCEC.
Abbreviations
CI, confidence interval; CPET, Clostridium perfringens ε-toxin; DEG, differentially expressed genes; GEMs, cholesterol-enriched microdomains; GSVA, Gene set variation analysis; IHC, Immunohistochemistry; MAL, Myelin and lymphocyte protein; MR, membrane rafts; MSI, microsatellite instability; OS, overall survival; PFS, progression-free survival; TMAs, tissue microarrays; Tregs, T cell regulatory; TCGA, the Cancer Genome Atlas; TGN, trans-Golgi network; RIPA, radioimmunoprecipitation assay; UCEC, uterine corpus endometrial carcinoma.
Data Sharing Statement
Publicly available datasets were analyzed in this study.
Ethics Statement
The experiments involving human participants were reviewed and approved by the Medical Ethics Committee of the Jinan Maternal and Child Health Care Hospital. Patients/participants provided written informed consent prior to participation in this study. The study complies with guidelines by the Declaration of Helsinki.
Author Contributions
Li Dong and Zhang Juan designed the study, acquired the data, and wrote the manuscript. Wu Lilong was involved in collection of samples, acquisition of data and reviewed the article. Yang Xiaoming and Chen Zheng revised the article. Yuan Jiangjing designed and supervised the study. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
Authors declare that they have no conflict of interest.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654
2. Colombo N, Creutzberg C, Amant F, et al. ESMO-ESGO-ESTRO consensus conference on endometrial cancer: diagnosis, treatment and follow-up. Ann Oncol. 2016;27(1):16–41.
3. Amant F, Mirza MR, Koskas M, Creutzberg CL. Cancer of the corpus uteri. Int J Gynaecol Obstet. 2018;143(Suppl 2):37–50. doi:10.1002/ijgo.12612
4. Carlson MJ, Thiel KW, Leslie KK. Past, present, and future of hormonal therapy in recurrent endometrial cancer. Int J Womens Health. 2014;6:429–435.
5. Yen TT, Wang TL, Fader AN, Shih IM, Gaillard S. Molecular classification and emerging targeted therapy in endometrial cancer. Int J Gynecol Pathol. 2020;39(1):26–35. doi:10.1097/PGP.0000000000000585
6. Di Tucci C, Capone C, Galati G, et al. Immunotherapy in endometrial cancer: new scenarios on the horizon. J Gynecol Oncol. 2019;30(3):e46. doi:10.3802/jgo.2019.30.e46
7. Oza AM, Elit L, Biagi J, et al. Molecular correlates associated with a phase II study of temsirolimus (CCI-779) in patients with metastatic or recurrent endometrial cancer—NCIC IND 160. J Clin Oncol. 2006;24(18_suppl):3003.
8. Alonson MA, Weissman SM. cDNA cloning and sequence of MAL, a hydrophobic protein associated with human T-cell differentiation. Proc Natl Acad Sci U S A. 1987;84(7):1997–2001. doi:10.1073/pnas.84.7.1997
9. Pérez P, Puertollano R, Alonso MA. Structural and biochemical similarities reveal a family of proteins related to the MAL proteolipid, a component of detergent-insoluble membrane microdomains. Biochem Biophys Res Commun. 1997;232(3):618–621. doi:10.1006/bbrc.1997.6338
10. Wilson SH, Bailey AM, Nourse CR, Mattei MG, Byrne JA. Identification of MAL2, a novel member of the mal proteolipid family, though interactions with TPD52-like proteins in the yeast two-hybrid system. Genomics. 2001;76(1–3):81–88. doi:10.1006/geno.2001.6610
11. de Marco MC, Martín-Belmonte F, Kremer L, et al. MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J Cell Biol. 2002;159(1):37–44. doi:10.1083/jcb.200206033
12. Puertollano R, Alonso MA. MAL, an integral element of the apical sorting machinery, is an itinerant protein that cycles between the trans-Golgi network and the plasma membrane. Mol Biol Cell. 1999;10(10):3435–3447. doi:10.1091/mbc.10.10.3435
13. Rumah KR, Ma Y, Linden JR, et al. The myelin and lymphocyte protein mal is required for binding and activity of clostridium perfringens ε-toxin. PLoS Pathog. 2015;11(5):e1004896. doi:10.1371/journal.ppat.1004896
14. Linden JR, Ma Y, Zhao B, et al. Clostridium perfringens epsilon toxin causes selective death of mature oligodendrocytes and central nervous system demyelination. mBio. 2015;6(3):e02513. doi:10.1128/mBio.02513-14
15. Khalili S, Jahangiri A, Hashemi ZS, Khalesi B, Mard-Soltani M, Amani J. Structural pierce into molecular mechanism underlying clostridium perfringens epsilon toxin function. Toxicon. 2017;127:90–99. doi:10.1016/j.toxicon.2017.01.010
16. Meckes DG, Menaker NF, Raab-Traub N. Epstein-Barr virus LMP1 modulates lipid raft microdomains and the vimentin cytoskeleton for signal transduction and transformation. J Virol. 2013;87(3):1301–1311. doi:10.1128/JVI.02519-12
17. Frank M, van der Haar ME, Schaeren-Wiemers N, Schwab ME. rMAL is a glycosphingolipid-associated protein of myelin and apical membranes of epithelial cells in kidney and stomach. J Neurosci. 1998;18(13):4901–4913. doi:10.1523/JNEUROSCI.18-13-04901.1998
18. Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20(3):274–281. doi:10.1038/nsmb.2518
19. Frank M, Schaeren-Wiemers N, Schneider R, Schwab ME. Developmental expression pattern of the myelin proteolipid MAL indicates different functions of MAL for immature Schwann cells and in a late step of CNS myelinogenesis. J Neurochem. 1999;73(2):587–597. doi:10.1046/j.1471-4159.1999.0730587.x
20. Kim T, Fiedler K, Madison DL, Krueger WH, Pfeiffer SE. Cloning and characterization of MVP17: a developmentally regulated myelin protein in oligodendrocytes. J Neurosci Res. 1995;42(3):413–422. doi:10.1002/jnr.490420316
21. Schaeren-Wiemers N, Valenzuela DM, Frank M, Schwab ME. Characterization of a rat gene, rMAL, encoding a protein with four hydrophobic domains in central and peripheral myelin. J Neurosci. 1995;15(8):5753–5764. doi:10.1523/JNEUROSCI.15-08-05753.1995
22. Wang Z, Wang M, Xu X, et al. Studies of MAL gene in human esophageal cancer by RNA in situ hybridization. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2000;17(5):329–331.
23. Xu Z, Wang M, Cai Y. [MAL gene is down-regulated substantially in human esophageal cancer]. Zhonghua Zhong Liu Za Zhi. 1999;21(4):250–252. Chinese.
24. Mimori K, Shiraishi T, Mashino K, et al. MAL gene expression in esophageal cancer suppresses motility, invasion and tumorigenicity and enhances apoptosis through the Fas pathway. Oncogene. 2003;22(22):3463–3471. doi:10.1038/sj.onc.1206378
25. Lind GE, Ahlquist T, Kolberg M, et al. Hypermethylated MAL gene - a silent marker of early colon tumorigenesis. J Transl Med. 2008;6(1):13. doi:10.1186/1479-5876-6-13
26. Kalmár A, Péterfia B, Hollósi P, et al. DNA hypermethylation and decreased mRNA expression of MAL, PRIMA1, PTGDR and SFRP1 in colorectal adenoma and cancer. BMC Cancer. 2015;15(1):736. doi:10.1186/s12885-015-1687-x
27. Horne HN, Lee PS, Murphy SK, Alonso MA, Olson JA, Marks JR. Inactivation of the MAL gene in breast cancer is a common event that predicts benefit from adjuvant chemotherapy. Mol Cancer Res. 2009;7(2):199–209. doi:10.1158/1541-7786.MCR-08-0314
28. Guerrero-Preston R, Hadar T, Ostrow KL, et al. Differential promoter methylation of kinesin family member 1a in plasma is associated with breast cancer and DNA repair capacity. Oncol Rep. 2014;32(2):505–512. doi:10.3892/or.2014.3262
29. Buffart TE, Overmeer RM, Steenbergen RDM, et al. MAL promoter hypermethylation as a novel prognostic marker in gastric cancer. Br J Cancer. 2008;99(11):1802–1807. doi:10.1038/sj.bjc.6604777
30. Choi B, Han T-S, Min J, et al. MAL and TMEM220 are novel DNA methylation markers in human gastric cancer. Biomarkers. 2017;22(1):35–44. doi:10.1080/1354750X.2016.1201542
31. Maruya S-I, Kim H-W, Weber RS, et al. Gene expression screening of salivary gland neoplasms: molecular markers of potential histogenetic and clinical significance. J Mol Diagn. 2004;6(3):180–190. doi:10.1016/S1525-1578(10)60508-9
32. Beder LB, Gunduz M, Hotomi M, et al. T-lymphocyte maturation-associated protein gene as a candidate metastasis suppressor for head and neck squamous cell carcinomas. Cancer Sci. 2009;100(5):873–880. doi:10.1111/j.1349-7006.2009.01132.x
33. Hutajulu SH, Indrasari SR, Indrawati LPL, et al. Epigenetic markers for early detection of nasopharyngeal carcinoma in a high risk population. Mol Cancer. 2011;10(1):48. doi:10.1186/1476-4598-10-48
34. Suzuki M, Shiraishi K, Eguchi A, et al. Aberrant methylation of LINE-1, SLIT2, MAL and IGFBP7 in non-small cell lung cancer. Oncol Rep. 2013;29(4):1308–1314. doi:10.3892/or.2013.2266
35. Blaveri E, Simko JP, Korkola JE, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11(11):4044–4055. doi:10.1158/1078-0432.CCR-04-2409
36. Schwartz DR, Kardia SLR, Shedden KA, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62(16):4722–4729.
37. Berchuck A, Iversen ES, Lancaster JM, et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 2005;11(10):3686–3696. doi:10.1158/1078-0432.CCR-04-2398
38. Copie-Bergman C, Gaulard P, Maouche-Chrétien L, et al. The MAL gene is expressed in primary mediastinal large B-cell lymphoma. Blood. 1999;94(10):3567–3575. doi:10.1182/blood.V94.10.3567.422k06_3567_3575
39. Copie-Bergman C, Plonquet A, Alonso MA, et al. MAL expression in lymphoid cells: further evidence for MAL as a distinct molecular marker of primary mediastinal large B-cell lymphomas. Mod Pathol. 2002;15(11):1172–1180. doi:10.1097/01.MP.0000032534.81894.B3
40. Kohno T, Moriuchi R, Katamine S, Yamada Y, Tomonaga M, Matsuyama T. Identification of genes associated with the progression of adult T cell leukemia (ATL). Jpn J Cancer Res. 2000;91(11):1103–1110. doi:10.1111/j.1349-7006.2000.tb00892.x
41. Hsi ED, Sup SJ, Alemany C, et al. MAL is expressed in a subset of Hodgkin lymphoma and identifies a population of patients with poor prognosis. Am J Clin Pathol. 2006;125(5):776–782. doi:10.1309/98KLHRDAM5CMDHE2
42. Millán J, Puertollano R, Fan L, Rancaño C, Alonso MA. The MAL proteolipid is a component of the detergent-insoluble membrane subdomains of human T-lymphocytes. Biochem J. 1997;321(Pt 1):247–252. doi:10.1042/bj3210247
43. Marazuela M, Alonso MA. Expression of MAL and MAL2, two elements of the protein machinery for raft-mediated transport, in normal and neoplastic human tissue. Histol Histopathol. 2004;19(3):925–933.
44. Fang Y, Wang L, Wan C, et al. MAL2 drives immune evasion in breast cancer by suppressing tumor antigen presentation. J Clin Invest. 2021;131(1). doi:10.1172/JCI140837
45. Antón OM, Andrés-Delgado L, Reglero-Real N, Batista A, Alonso MA. MAL protein controls protein sorting at the supramolecular activation cluster of human T lymphocytes. J Immunol. 2011;186(11):6345–6356. doi:10.4049/jimmunol.1003771
46. Amant F, Moerman P, Neven P, Timmerman D, Van Limbergen E, Vergote I. Endometrial cancer. Lancet. 2005;366(9484):491–505. doi:10.1016/S0140-6736(05)67063-8
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.