Back to Journals » Infection and Drug Resistance » Volume 14
Mycobacterium colombiense and Mycobacterium avium Complex Causing Severe Pneumonia in a Patient with HIV Identified by a Novel Molecular-Based Method
Received 15 September 2020
Accepted for publication 18 November 2020
Published 6 January 2021 Volume 2021:14 Pages 11—16
DOI https://doi.org/10.2147/IDR.S282190
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Xiaoling Yu, Wenqian Jiang
Department of Infectious Diseases, Mengchao Hepatobiliary Hospital of Fujian Medical University, Fuzhou, People’s Republic of China
Correspondence: Xiaoling Yu
Department of Infectious Diseases, Mengchao Hepatobiliary Hospital of Fujian Medical University, Fuzhou 350025, Fujian, People’s Republic of China
Tel/Fax +86 591 88116019
Email [email protected]
Abstract: Non-tuberculous mycobacteria are conditional pathogens that can cause many diseases, among which pulmonary infections are the most common (65– 90%). Mycobacterium avium and Mycobacterium abscessus are non-tuberculous mycobacteria most often associated with lung diseases. Mass spectrometry diagnostic techniques were not effective in Mycobacterium avium complex infection. We report a case of Mycobacterium colombiense and Mycobacterium avium complex causing severe pneumonia in an adult with HIV. Our group developed a novel molecular-based method to identify Mycobacterium species. Novel techniques such as molecular cloning which we have described here can make up for the inability of matrix-assisted laser desorption ionization-time of flight mass spectrometry to distinguish the multiple microorganisms present, and may add to the diagnostic toolkit and increase the accuracy and rapidity of diagnosis in the future.
Keywords: non-tuberculous mycobacteria, Mycobacterium avium complex, Mycobacterium colombiense, human immunodeficiency virus infection, pneumonia
Introduction
Non-tuberculous mycobacteria (NTM) are mycobacterial species other than Mycobacterium tuberculosis and Mycobacterium leprae that cause tuberculosis and leprosy, respectively.1 NTM, which are conditional pathogens, can cause chronic skin infection and lymphadenitis, and may further develop into a disseminated disease, of which pulmonary infections are the most common (65–90%). The Mycobacterium avium (M. avium) complex and Mycobacterium abscessus complex is the NTM most often associated with lung diseases.1 NTM are highly prevalent in the environment; however, non-immunocompromised hosts can withstand infection. Conversely, immunocompromised patients such as organ transplant recipients and people living with human immunodeficiency virus (HIV) are at greater risk of NTM infection.2 Herein, we report a case of severe pneumonia caused by a Mycobacterium colombiense (M. colombiense) and M. avium complex (MAC) in a patient with HIV. The blood culture of the patient was positive, but the pathetic cultures could not be identified through matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS). Sanger sequencing results of 16S rDNA polymerase chain reaction (PCR) of the cultured pathogenic microorganisms showed double peaks which indicated the existence of more than one pathogenic microorganism. MAC are difficult to decipher by re-scribing. Using molecular cloning and 16S rDNA sequencing, multiple microorganisms can be distinguished and identified separately.
Case Description
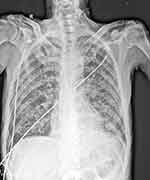
A 62-year-old man presented with anhelation, fatigue and anorexia for two months before hospitalization. The patient was initially seen in a medical centre in Fuzhou, Fujian, China with no medical history and was a smoker for 35 years. Due to the patient’s history of multiple sexual encounters with various female partners as well as his severe condition at presentation, he was tested for chronic viral infections including HIV. The patient’s examination results were as follows: body weight, 40 kg; malnourished, emaciated, weight loss in the last 2 months, 10 kg; body temperature, 37 °C; respiratory rate, 23 breaths/minute; heart rate, 96 beats/min; blood pressure, 116/61 mmHg; T lymphocyte subgroup detection, CD4+ T-cell (absolute value) count of 15 cells/μL [reference range: 410–1590 cells/μL]; high-sensitivity quantitative HIV nucleic acid detection, 2.48×105 copies/mL. The initial complete blood count (CBC) showed the following: white blood cell count, 11.63×109 cells/L (90.8% neutrophils, 5.0% lymphocytes, 2.7% monocytes, 1.0% eosinophils and 0.5% basophil); haemoglobin, 97 g/dL [reference range: 130–175 g/dL]; and platelets, 286×109 cells/L. The erythrocyte sedimentation rate was 79 mm/hr [reference range: 0–15 mm/hr], and the C-reactive protein level was 119.39 mg/L [reference range: 0–8 mg/L]. Initial chest digital radiography (DR) showed extensive bilateral lung lesions involving almost all pulmonary lobules (Figure 1). Blood gas analysis (FiO2: 63%) showed the following: pH, 7.46; PCO2, 23 mmHg; PO2, 161 mmHg; and oxygenation index, 256. The β-d-glucan (βDG) test was 159.2 pg/mL [reference range: negative < 70 pg/mL; grey area = 70–95 pg/mL; positive > 95 pg/mL]. Due to the low CD4+ T-cell count, the patient was subsequently diagnosed with HIV infection via Western blot testing. Additionally, lung CT showed an interstitial pattern at the inferior lobe of the left lung with diffuse pulmonary bullae indicating emphysema. The patient was then treated speculatively with fluconazole and ceftazidime for possible fungal and bacterial infections, respectively. As the patient did not improve after ten days, he was referred to the infectious service of Mengchao Hepatobiliary Hospital of Fujian Medical University.
Meropenem, sulfamethoxazole and caspofungin were administered due to suspicion of bacterial or pneumocystis pneumonia and other fungal infections. During treatment, the patient experienced chills and had fever reaching 40 °C with low blood oxygen saturation at 80%. He was sedated and methylprednisolone was administered as an anti-inflammatory medication. After treatment, his chills improved, and blood oxygen saturation increased to 90% in room air.
On the second day after admission, the patient’s temperature was 37 °C and he again experienced chills. Intravenous immunoglobulin and linezolid were administered to enhance immune function and to further target gram-positive bacteria, respectively. Four days after admission, the patient’s condition deteriorated. The family members of the patient required automatic discharge from the hospital. The doctor informed them that the diagnosis and treatment of the condition might be delayed after discharge, resulting in unpredictable consequences. They expressed understanding, but still insisted on discharge.
Materials and Methods
The results of blood culture showed that there was microbial growth, but the microbial species could not be identified by mass spectrometry. The blood sample was then inoculated on Colombian blood agar for bacterial culture. MALDI-TOF-MS (VITEK MS, BioMerieux SA, BioMerieux Inc., France) was used to identify the cultured pathogenic microorganisms.
For the molecular identification of microorganisms, we decided to use PCR. The genome of the isolate was extracted by a one-step genome extraction kit (Weiyin, China), and 16S rDNA PCR was carried out with 16S universal primers (27F: 5ʹ AGAGTTTGATCCTGGCTCAG 3ʹ, 1492R: 5ʹ AGAGTTTGATCCTGGCTCAG 3ʹ). Then, the PCR product was sequenced using the ABI 3730xl DNA Sequencer.
The 16S rDNA PCR product was purified by a GeneJET Gel extraction kit (Thermo, USA). Clones were constructed using the pMD19-T Vector Cloning kit (Takara, Japan) and transformed into DH10B Escherichia coli. Positive clones were then screened by ampicillin and sequenced with the ABI 3730xl DNA Sequencer.
Results
We obtained a 1427 bp sequence, aligned to the NCBI nucleotide collection (nr/nt) database using BLAST. This resulted in the identification of M. avium (Accession: CP046507.1), with a query cover of 100%, an E value of 0.0, and a percent identity of 97.05%. MALDI-TOF-MS however did not successfully identify the microorganism, since the sequencing results showed two sequencing peaks (Figure 2). Therefore, we suspected that the isolate contained more than one microorganism.
 |
Figure 2 Partial Sanger sequencing peak map of the 16S rDNA PCR product of the isolate sequenced with the 1492R primer. |
We then decided to ligate the PCR product into a T vector for further identification and selected 11 positive Escherichia coli transformants for Sanger sequencing. Of these transformants, 9 were identified as M. avium and 2 as M. colombiense. These results further confirmed our suspicion that two microorganisms were present in the isolate.
Discussion
Here, we report a case of a late-stage HIV-infected adult with respiratory symptoms for two months. In accordance with other NTM-infected patients who experience relative emaciation, this patient had lost 10 kg of body weight.3 A chest CT scan indicated emphysema, while blood and sputum cultures could not identify the pathogenic culprits through MALDI-TOF-MS, and double peaks on Sanger sequencing indicated the existence of more than one pathogenic microorganism. As MAC infection was suspected but could not be distinguished, we used molecular cloning to isolate species and identify their genomes.
The MAC contains a group of heterogeneous, slow-growing mycobacteria, including M. avium, Mycobacterium intracellulare (M. intracellulare), and M. colombiense.4 MAC infections are mostly caused by M. avium and M. intracellulare, while M. colombiense is extremely rare. A handful of cases of M. colombiense have been previously reported, but concurrent M. avium and M. colombiense causing pulmonary infection has not yet been encountered in the literature (Table 1).
 |
Table 1 Comparison of Case Reports of Mycobacterium colombiense Infections in Patients with and without HIV |
M. colombiense is a slow-growing Mycobacterium that was first isolated in 2006, identified from blood and sputum samples of a person living with HIV in Colombia.5 M. colombiense may be ingested or may directly invade the body through skin injury, causing the development of local lymph node lesions.6 A recent report showed that amikacin, clarithromycin, ciprofloxacin, moxifloxacin and rifabutin are effective against M. colombiense infection.7 In our case, although the sequencing results of the pathogenic microbial culture were positive, MALDI-TOF MS could not identify the strain. Therefore, it was not possible to perform drug sensitivity tests and to administer antibiotics in a targeted manner. Antiretroviral therapy (ART) was not used due to the rapid worsening of the patient’s condition. Furthermore, before the sequencing results from the blood culture were obtained, the patient elected to be discharged home on November 15, 2019, and died a week later.
MALDI-TOF MS is a mass spectrometry-based analysis technology that was developed in the mid-1980s. This method has become the gold standard for clinical laboratory microbial identification but has the disadvantage of not being able to identify complex infections.8 Moreover, the Vitek MS IVD 2.0 database contains data for M. avium but no data for M. colombiense. In this case, the identified species were MAC consisting of M. avium and M. colombiense. MAC are difficult to decipher by re-scribing, and as such, MALDI-TOF MS cannot identify them. Thus, MALDI-TOF MS is not an effective method in such cases of complex bacterial infections, including two or more microorganisms.
In addition to molecular cloning, molecular identification of some MAC members at the species level can be performed by sequencing the genes hsp65, rpoB, sodA, dnaJ, gyrB and recA.4 Members of the MAC can also be identified by next-generation metagenomic sequencing. However, the cost is greater than that of Sanger sequencing.9 In a clinical setting, next-generation sequencing (NGS) is typically used for the identification of complex microorganisms. Furthermore, the mycolic acid composition of the cell wall of Mycobacterium is critical for identifying Mycobacterium species.4 We showed that using molecular cloning and 16S rDNA sequencing allowed the identification of more than one microorganism causing infection in this patient.
Conclusion
We reported for the first time a case of severe pneumonia caused by concurrent M. avium and M. colombiense in a newly diagnosed patient with late-stage HIV with a very low CD4+ T-cell count. MAC infection is common in such patients, however, MALDI-TOF MS was not able to distinguish the multiple microorganisms present. Thus, we resorted to molecular techniques to confirm the presence of more than one pathogenic organism. In future cases where multiple pathogenic organisms cannot be identified by mass spectrometry, novel techniques such as molecular cloning which we have described here, may add to the diagnostic toolkit and increase the accuracy and rapidity of diagnosis.
Abbreviations
NTM, non-tuberculous mycobacteria; HIV, human immunodeficiency virus; M. avium, Mycobacterium avium; M. colombiense, Mycobacterium colombiense; M. intracellulare, Mycobacterium intracellulare; MAC, Mycobacterium avium complex; CBC, complete blood count; βdg, β-d-glucan; DR, digital radiography; MALDI-TOF-MS, matrix-assisted laser desorption ionization-time of flight mass spectrometry; PCR, polymerase chain reaction; ART, antiretroviral therapy; NGS, next-generation sequencing.
Ethics Approval and Informed Consent
The work described has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. Ethical approval for this study was provided by the Institutional Ethical Committee of the Faculty of Medicine, Mengchao Hepatobiliary Hospital of Fujian Medical University, Fujian, China (No. 2019_057_01). Written informed consent was provided by the patient’s father, his consent included consent to publish the case details and any accompanying images.
Acknowledgments
The authors express their gratitude to: 1) Lijuan Liu, Shaogui Wu, Shuyan Zhou (Department of Microbiology, Mengchao Hepatobiliary Hospital of Fujian Medical University, China) and You Chen (Department of Infectious Diseases, Mengchao Hepatobiliary Hospital of Fujian Medical University, China) provided support for bacterial isolates and culture. 2) Lanlan He, Yahong Chen (Department of Infectious Diseases, Mengchao Hepatobiliary Hospital of Fujian Medical University, China) supported and revised the clinical data. 3) Jun Lin (Institute of Applied Genomics, Fuzhou University, China) provided support for method, sequencing and bioinformatics analysis. 4) John Lin (Division of Hematology and Chronic Viral Illness Service, McGill University Health Centre, Montreal, Quebec, Canada) prepared the table and reviewed the literature. 5) Jean-Pierre Routy (Division of Hematology and Chronic Viral Illness Service, McGill University Health Centre, Montreal, Quebec, Canada) reviewed and proofread the manuscript. 6) Hanhui Ye (Department of Infectious Diseases, Mengchao Hepatobiliary Hospital of Fujian Medical University, China) provided support for supervision of the study.
Funding
This study is sponsored by the Clinical Medicine Center Construction Program of Fuzhou, Fujian, P.R.C. [Grant numbers 2018080306], Health Research Innovation Team Cultivation Project of Fuzhou, P.R.C. [Grant numbers 2019-S-wt4] and Key Clinical Specialty Discipline Construction Program of Fujian, P.R.C.
Disclosure
The authors report no competing interests in this work.
References
1. Porvaznik I, Solovic I, Mokry J. Non-tuberculous mycobacteria: classification, diagnostics, and therapy. Adv Exp Med Biol. 2017;944:19–25.
2. Wu U-I, Holland SM. Host susceptibility to non-tuberculous mycobacterial infections. Lancet Infect Dis. 2015;15(8):968–980. doi:10.1016/S1473-3099(15)00089-4
3. Singh RK, Kumar D. Nontuberculous mycobacterial pulmonary disease in an immunocompromised patient. Cureus. 2019. doi:10.7759/cureus.6065
4. Leguizamon J, Hernandez J, Murcia MI, Soto CY. Identification of potential biomarkers to distinguish Mycobacterium colombiense from other mycobacterial species. Mol Cell Probes. 2013;27(1):46–52. doi:10.1016/j.mcp.2012.08.009
5. Murcia MI, Tortoli E, Menendez MC, Palenque E, Garcia MJ. Mycobacterium colombiense sp. nov., a novel member of the Mycobacterium avium complex and description of MAC-X as a new ITS genetic variant. Int J Syst Evol Microbiol. 2006;56(Pt 9):2049–2054. doi:10.1099/ijs.0.64190-0
6. Inagaki Y, Ito T, Kato T, Ono Y, Sawa M. Disseminated cutaneous infection of Mycobacterium colombiense in a patient with myelodysplastic syndrome. Intern Med. 2018;57(3):423–427. doi:10.2169/internalmedicine.7890-16
7. Maurer FP, Pohle P, Kernbach M, et al. Differential drug susceptibility patterns of Mycobacterium chimaera and other members of the Mycobacterium avium-intracellulare complex. Clin Microbiol Infect. 2019;25(3):379e371–379 e377. doi:10.1016/j.cmi.2018.06.010
8. Schubert S, Kostrzewa M. MALDI-TOF MS in the microbiology laboratory: current trends. Curr Issues Mol Biol. 2017;23:17–20. doi:10.21775/cimb.023.017
9. Yu X, Jiang W, Shi Y, Ye H, Lin J. Applications of sequencing technology in clinical microbial infection. J Cell Mol Med. 2019;23(11):7143–7150. doi:10.1111/jcmm.14624
10. Vuorenmaa K, Ben Salah I, Barlogis V, Chambost H, Drancourt M. Mycobacterium colombiense and pseudotuberculous lymphadenopathy. Emerg Infect Dis. 2009;15(4):619–620. doi:10.3201/eid1504.081436
11. Poulin S, Corbeil C, Nguyen M, et al. Fatal Mycobacterium colombiense/cytomegalovirus coinfection associated with acquired immunodeficiency due to autoantibodies against interferon gamma: a case report. BMC Infect Dis. 2013;13:24. doi:10.1186/1471-2334-13-24
12. Barretto AR, Felicio JS, Sales LHM, Yamada ES, Lopes ML, da Costa ARF. A fatal case of pulmonary infection by Mycobacterium colombiense in Para State, Amazon Region, Brazil. Diagn Microbiol Infect Dis. 2016;85(3):344–346. doi:10.1016/j.diagmicrobio.2016.02.011
13. Irizarry P, Basler K, Mifsud M, et al. HIV associated Mycobacterium colombiense cervical lymphadenitis: a case report. J Microbiol Infect Dis. 2017;7:101–103.
14. Pena E, Machado D, Viveiros M, Jordao S. A case report of disseminated Mycobacterium colombiense infection in an HIV patient. Int J Mycobacteriol. 2019;8(3):295–297. doi:10.4103/ijmy.ijmy_100_19
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.