Back to Journals » International Journal of Nanomedicine » Volume 15
MXene Surface on Multiple Junction Triangles for Determining Osteosarcoma Cancer Biomarker by Dielectrode Microgap Sensor
Authors Zhou D, Gopinath SCB , Mohamed Saheed MS, Siva Sangu S, Lakshmipriya T
Received 30 September 2020
Accepted for publication 11 November 2020
Published 14 December 2020 Volume 2020:15 Pages 10171—10181
DOI https://doi.org/10.2147/IJN.S284752
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Israel (Rudi) Rubinstein
Dakai Zhou,1 Subash CB Gopinath,2,3 Mohamed Shuaib Mohamed Saheed,4,5 Sangeetha Siva Sangu,5,6 Thangavel Lakshmipriya3
1Department of Spinal Surgery, Xinxiang Central Hospital, Xinxiang City, Henan Province 453000, People’s Republic of China; 2Faculty of Chemical Engineering Technology, Universiti Malaysia Perlis (UniMAP), Arau, Perlis 02600, Malaysia; 3Institute of Nano Electronic Engineering, Universiti Malaysia Perlis (UniMAP), Kangar, Perlis 01000, Malaysia; 4Department of Mechanical Engineering, Universiti Teknologi PETRONAS, Seri Iskandar 32610, Perak Darul Ridzuan, Malaysia; 5Centre of Innovative Nanostructures & Nanodevices (COINN), Universiti Teknologi PETRONAS, Seri Iskandar 32610, Perak Darul Ridzuan, Malaysia; 6Department of Fundamental & Applied Sciences, Universiti Teknologi PETRONAS, Seri Iskandar 32610, Perak Darul Ridzuan, Malaysia
Correspondence: Subash CB Gopinath Email [email protected]
Background: In recent years, nanomaterials have justified their dissemination for biosensor application towards the sensitive and selective detections of clinical biomarkers at the lower levels. MXene is a two-dimensional layered transition metal, attractive for biosensing due to its chemical, physical and electrical properties along with the biocompatibility.
Materials and Methods: This work was focused on diagnosing osteosarcoma (OS), a common bone cancer, on MXene-modified multiple junction triangles by dielectrode sensing. Survivin protein gene is highly correlated with OS, identified on this sensing surface. Capture DNA was immobilized on MXene by using 3-glycidoxypropyltrimethoxysilane as an amine linker and duplexed by the target DNA sequence.
Results: The limitation and sensitivity of detection were found as 1 fM with the acceptable regression co-efficient value (y=1.0037⨰ + 0.525; R2=0.978) and the current enhancement was noted when increasing the target DNA concentrations. Moreover, the control sequences of single- and triple-mismatched and noncomplementary to the target DNA sequences failed to hybridize on the capture DNA, confirming the specificity. In addition, different batches were prepared with capture probe immobilized sensing surfaces and proved the efficient reproducibility.
Conclusion: This microgap device with Mxene-modified multiple junction triangles dielectrode surface is beneficial to quantify the survivin gene at its lower level and diagnosing OS complication levels.
Keywords: MXene, biosensor, bone cancer, microdevice, biomarker, survivin gene
Introduction
In past two decades, development of biosensor and biochip to be capable of characterizing and quantifying biomolecules greatly contributed to biological and medical fields.1 With the rapid production of high-sensitivity sensor, recent many scientists have developed nanostructured biosensors with nanotube, nanowire, nanoparticle and nanosheet. These kinds of nanomaterials are promising candidates for higher biomolecular capturing on the sensor surfaces and enhance the targeted analyte detection.2–4 Nanomaterials such as silver, gold, graphene, copper, silica and iron, have been utilized in the biosensor field for surface physical functionalization to promote the detection. Recently, a new layered two-dimensional (2D) material rose as a graphene analog named “MXene”, which was produced by the exfoliation of selective phases with MAX. Due to the unique physical, chemical, and electrical properties, MXene had drawn a great attention for diverse scientific applications.5 In particular, MXene contains the prominent properties for improving the performance of biosensors, which includes biocompatibility, hydrophilicity, easier surface functionalization, and larger surface area.6 In addition, it provides improvement with the immobilization of biomolecule on the sensing surface and leads to higher sensitivity and lower detection limit.7 In this research MXene was synthesized and modified on the multiple junction triangles on sensor surface to diagnose osteosarcoma cancer.
The elected multiple junction triangles sensor is the electrical-based biosensing system to recognize various biomolecular interactions on the electrode surface through the response of current.8–10 It offers several advantages such as label-free, low-power, low-cost, portable, higher sensitivity and easy to use. Numerous devices with nanogap/microgap have been fabricated with various techniques and reported by researchers for biosensing applications.11 In this research a microgap integrated electrode was fabricated on silica wafer as the base layer and then aluminium was deposited with triangle junctions. Silica helps to hold the recognition element through the chemical modification. Since aluminium was deposited on SiO2 through thermal evaporator method, it is quite stable. Moreover, a thin layer of aluminium helps to protect the sensing surfaces from leakage. This dielectric layer separates the two electrodes and also works to hold the receptor. Current response changes upon biomolecular interaction on the electrode surfaces were noticed. The current research was developed by an MXene modified surface on the sensing surface to diagnose osteosarcoma cancer biomarker.
Osteosarcoma (OS) is the most common bone cancer, mainly occurs in children and young adults between the ages of 10 and 30 years during the bone development period with adolescence.12,13 OS has been noticed to be responsible for 3% of cancers occurring in childhood. Currently neoadjuvant chemotherapy and radical surgery are the treatments for OS, which increases the survival rate from 20 to 75%. Early identification of OS with a suitable biomarker is the key factor to ensure effective treatment. Recent studies proved that survivin gene is highly correlated with OS, so that detecting the survivin helps to identify the OS and its condition. Survivin protein inhibits the caspase activation, leads to the negative impact of programmed cell death and apoptosis. In addition, it was proved that survivin protein is highly expressed in fatal tissue and most of the human cancers, which include breast, gastric, colorectal, lung, urinary bladder cancers, and osteosarcoma, but it was completely absent in the normal cell.14 Therefore, quantifying the survivin gene helps detect OS.
In this research, a DNA based electrode sensor was used to detect the survivin gene, which is related to osteosarcoma. To immobilize the capture DNA, the sensing surface was modified with 2D-nanomaterial MXene. The oxidized MXene with H2O2 was attached on the sensing surface through the modification by 1,1ʹ-carbonyldiimidazole. Further, NH2-capture DNA was immobilized on Mxene through 3-glycidoxypropyltrimethoxysilane as the linker. This capture DNA immobilized surface was utilized to identify the target DNA sequence for survivin. Specificity experiments were conducted with control sequences instead of target sequence and the sensing surface reproducibility was evaluated for the stability of capture probe immobilization. This research with an ideal methodology can detect the survivin gene and helps to diagnose the OS cancer and its severity.
Materials and Procedures
Biomolecules and Chemicals
3-glycidoxypropyltrimethoxysilane (GOPTS) was purchased from Shin-Etsu Chemical Co., Ltd, Tokyo, Japan. Transmission electron microscope (JEOL JEM 2100F TEM), field-emission scanning electron microscope (GEMINI, FESEM), and X-ray Photoelectron spectroscopy (XPS; Thermo Scientific, K-Alpha), were used to characterize the MXene by following the procedures outlined earlier.15 1,1ʹ-Carbonyldiimidazole (CDI), bovine serum albumin (BSA) and PBS were from Sigma-Aldrich (USA). MAX powder was acquired from Luoyang Tongrun Info Technology Co., Ltd, China. DNA oligos were purchased from the local supplier, Apical Scientific Oligo (Kuala Lumpur, Malaysia). The synthesized sequences were followed as described,16 immobilized probe: 5′-NH2-TTGGAGGGCTGCGCCTGCACCC-3′; target DNA: 5′-GGGTGCAGGCGC AGCCCTCCAA-3′; single-mismatch: 5′-GGGTGCAGCCGCAGCCCTCCAA-3′; triple-mismatch: 5′-GGGTGCAGCGCCAGCCCTCCAA-3′; noncomplementary: 5′-TTGGAGGGCTGCGCCTGCACCC-3′.
Synthesis of MXene Using HCl-LiF as Etchant
MXene powder was prepared by etching the MAX powder in a solution of HCl-LiF (hydrochloric acid + lithium fluoride). The etchant was prepared by adding desired LiF to 9 M HCl and left under continuous stirring for five minutes. MAX powder was gradually added over the course of five minutes to the prepared etchant, and the reaction was stirred for 24 h at room temperature. Then the acidic mixture was washed with deionized water by centrifugation at 7500 rpm for multiple cycles. After each cycle, the acidic supernatant was discharged as waste. These washing cycles were repeated until pH 5 was achieved. Afterwards, the obtained black Ti3C2Tx slurry was dissolved in deionized water and ultrasonicated. This solution was then vacuum-filtered to produce thick, black clay and subsequently dried in a vacuum oven at 80°C for six hours. The produced powder was stored in a tight container to prevent oxidation.
Multiple Junction Triangles Dielectrode Fabrication
Sensing microgap electrode surface was prepared by using the wet-etching traditional process. Before that the pattern of the electrode was designed by AutoCAD software.1,17 The sensing surface was arranged by twin electrodes with multiple junction triangles with the microgap distances. To fabricate the sensor, initially the basic substrate of cleaned silicon wafer was oxidized by thermal oxidation method at 500°C to grow the SiO2 layer. After that, aluminum (Al) was deposited on the SiO2 layer assisted by thermal evaporator. Next, positive photoresist with a 2000 nm layer was applied on Al substrate by using the spin coating method, followed by a soft-bake carried out at 90°C for one hour to remove the moisture on the substrate and also help to harden the layer of photoresist. Further, the pattern of the electrode was transformed on the photoresist layer by UV light exposure for 10 seconds followed by immersing the surface in a developing solution to remove the exposed area, and then hard-baked at 110°C for one minute. Next, the device was immersed in an Al-etching solution to clear the unexposed area. Finally, the sensor surface was washed with distilled water followed by acetone and kept in a dry place for further surface modification.
Construction of Capture DNA Immobilized Mxene Surface
Capture DNA was constructed on Mxene surface by the epoxylation process using GOPTS as a linker. Before that, the surface of MXene was oxidized with H2O2 solution to modify the surface into oxide. This process of carried out by mixing one gram of Mxene with 100 mL of distilled water under stirring condition and then added 10 mL of 30% H2O2. The H2O2 MXene treated was cleaned with distilled water then separated by centrifugation followed by drying under vacuum for further modification.
The sensing surface was modified into Mxene by CDI linking and then capture probe was attached. One gram of Mxene was suspended in PBS, spotted on the surface for three hours at room temperature (RT). The unbound MXene was cleared by dipping the surface in distilled water, and then GOPTS was added on the surface. For that, 2% GOPTS was diluted in toluene, dropped on Mxene-modified sensing surface and kept for three hours at 50°C followed by washing with toluene and distilled water to clear the unbound GOPTS. Then, 1 µM of NH2-capture DNA was dropped on GOPTS surface and rest for 30 min at RT followed by covered the surface with 0.5 M BSA. After washing the electrode with PBS, determination of target DNA was conducted on capture DNA constructed MXene-surface for diagnosing OS.
Detection of Target DNA on MXene-capture DNA Surface
Target DNA was detected and quantified on capture DNA modified IDE-Mxene. For that, target concentration with 100 pM was initially dropped on capture DNA and rest for 10 min at RT. The surface was rinsed with PBS buffer, and then the response of current was recorded to confirm the hybridization of target. Furthermore, to evaluate the detection performance of target DNA, dilutions of target from 1 fM with 10 orders until 10 pM were spotted independently on capture DNA probe immobilized surface. The current responses with before and after adding target were recorded for each concentration. The difference in current was plotted in an Excel spreadsheet to calculate the detection limit and sensitivity of target DNA duplex formation. All measurements were performed at room temperature using Keithley 6487 Picoammeter voltage/current supply at the voltage 0–2 V with a current linear sweep of 0.1 V. Buffering solution (10 mM PBS) at pH 7.4 was used as the electrolyte. For the measurements 5–10 µL reaction solution was utilized to cover the active sensing area. Between each of the measurements/reactions the surface was washed thoroughly using 20 volume of the above buffer. All measurements were performed in triplicates and maintained the wet condition throughout the experiments, unless otherwise stated.
Specific Target Identification
Specific detection performance of target DNA was evaluated with the experiments by single-, triple-mismatched, and complementary of target DNA sequences. These three sequences (100 pM) were dropped separately on capture probe immobilized surface. The current response before and after adding DNAs were noted for finding the specific target identification. Further, to identify the selective detection of target DNA, 1:100 dilution of human serum was mixed with different target DNA concentrations and detected by the immobilized capture DNA.
Reproducibility of Sensing Strategies for Target DNA Identification
Reproducibility on the capture probe immobilization and target DNA identification processes was confirmed by analyzing the current responses from five different sensor surfaces. For that, five different sensing devices were tested for the immobilization steps with Mxene, GOPTS, capture DNA, BSA and target DNA. Recorded current responses with each immobilization step to find the reproducibility and validation.
Results and Discussion
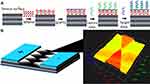
Figure 1 displays the basic strategy followed in this study for diagnosing OS on multiple junction triangles formed by dielectrode sensing system. Figure 2A shows the schematic illustration of survivin gene target sequence detection on MXene modified sensing surface sensor. As shown in Figure 2A, the electrode was initially modified by oxidized Mxene and then amine-capture DNA was attached on the surface through the GOPTS linker. The epoxy silane in GOPTS forms a covalent link to the amine-ended capture probe. Epoxy activated surface was proved to have high efficient immobilization of electrophilic and nucleophilic groups. GOPTS was dissolved in toluene to form an epoxylated surface, and it creates the secondary amine bond upon reaction. Moreover, GOPTS is highly stable at neutral pH, reactive and wettable with various nucleophilic groups to form the strong interaction with protein. In this study, on this GOPTS modified surface an amine-ended capture probe was added. This interaction happens in two ways, one is the negative charge of phosphate backbone in DNA creates the ionic bonding with the positive charged GOPTS-modified surface. In another way, the epoxy functional group in GOPTS interacted with the amine-ended capture probe.18 The excess sensing surface was blocked by BSA as a blocking agent and then target DNA was hybridized on capture DNA modified surface.
 |
Figure 1 Diagrammatic representation for the proposed sensor. Formation of bone cancer at knee joints for the detection is shown. |
Surface and Chemical Characterizations
Before performing the surface functionalization, the surface was confirmed intact by 3D-nanoprofiler imaging system and confirmed (Figure 2B). Furthermore, morphology of the synthesized MXene was analyzed by FESEM observation. Figure 3A and B shows the FESEM image of the synthesized MXene and it was clearly displayed with multilayered flake-like structure with a broad distribution.19 Size of the sheet is in the micrometer range and gap of the layers is in the nanometer range. The EDX spectra confirm the presence of Ti, Al, F, O, Cl, C on the synthesized MXene (Figure 3C).
The structural analysis of synthesized MXene was studied by XRD. The range of scale was fixed at 2θ≈0–80° using radiation operating with the current of 20 mA and the voltage at 40 kV. MXene shows the similar diffraction peak as reported earlier.20,21 The most intensive peak was observed at 39.1 indicating the Al-etching from the structure. In addition, (002) peak at 9° is indicating the lattice plane (004) peak at 19.4° belongs to Ti3Alc2.22,23 (Figure 3D).
The chemical composition on the surface of MXene was analyzed by XPS (Figure 4). The survey spectrum confirms the presence of components of titanium (T), carbon (C), oxygen (O) and fluorine (F) atoms in the synthesized MXene (Figure 4A). In the XPS spectra of Ti2p, O and C are mainly contributed from the TiC and TiOx, respectively.24 Further the deconvolutions of Ti2p, O1s, C1s were carried out to find the functional groups and their contributions. The values of binding energy components were confirmed with previous XPS analyses.25–27 As shown in figure, the peaks at 455 eV and 460.78 eV corresponds to the bond Ti-C and the peaks at 459.2 and 462. 68 eV responds for Ti-O bonds (Figure 4B). In O1s analysis, the peaks at 530.04 and 532.5 eV represent the components TiO2 and Al2O3. Peaks at 531.93 and 533.54 eV corresponds to the O atoms in C-Ti-(OH)x and C-O bonds, respectively (Figure 4C). The peak at 284.88 eV represents the C–C peak and 288.86 indicate the O–C=O peak (Figure 4D). In conclusion, XPS results reveal the presence of Ti2p, F1s, O1S, C1S, and Al2p on the surface of MXene and it was functionalized with OH groups, which are contributed from the graphitized carbides and carbons.28 Binding energies for bonds associated with deconvoluted peaks are listed in Table 1.
 |
Table 1 XPS Peak Fitting of MXene |
Current Response with Capture Probe Immobilization
Capture DNA immobilization process on MXene-modified electrode surface was confirmed by observing the current response after each immobilization process. As shown in Figure 5A, bare electrode surface shows the current as 1.9E-09 A, after the surface modified into MXene, it was increased to 1.99E-09 A. This current change confirms the attachment of MXene on the sensing surface. After GOPTS was linked with MXene and current was decreased to 1.24E-09 A. This current change happened due to the chemical interaction of GOPTS and MXene modified surface. MXene and GOPTS connect strongly through the covalent bond and also MXene hybridization with GOPTS increases the film adhesiveness to the electrode surface.28 The originally prepared MXene has the surface terminations of Ti, C, F, O and to facilitate the chemical surface functionalization, we oxidized the surface using H2O2. Due to the subsequent chemical and biological functionalization layers, the further oxidation may not be severe. Based on our preliminary study the oxidized surface is quite stable for the biomolecular interaction analysis performed. On this GOPTS-modified surface, NH2-capture DNA was introduced; the current response was further changed to 8.06E-10 A. This current change was due to the reaction of amine in the capture DNA with GOPTS surface. This result confirms the binding of capture DNA on MXene modified electrode surface. Capture molecule immobilization on the sensing surface plays a crucial role for improving the detection limit. Higher and uniform distribution of capture molecules on the sensing surface improves the binding of capture and target on the sensing surface. In this research, GOPTS modified MXene is more stable and uniformly distributed on the electrode surface and attracts more capture molecules. Figure 5B shows the current differences for each biomolecular immobilization before and after adding to the surface. It was clearly seen that MXene followed by capture DNA attachments show higher changes in current after reacted GOPTS. This capture DNA modified surface was utilized to determine the target DNA duplex formation.
Survivin Target DNA Identification
Survivin target DNA was detected on capture DNA modified microgap electrode surface. To reduce the nonspecific binding and signal-to-noise ratio, before the detection of target DNA, the capture DNA modified surface was blocked by BSA. BSA can cover the excess GOPTS surface and only allows binding of the capture with target DNA sequences. As shown in Figure 5C, BSA shows the current response as 1.77E-07 A, after adding 100 pM of target DNA, the current was drastically increased to 6.47E-07 A. There are few reasons behind these changes, usually with the blocking step higher current change is expected due to its occupation on large free spaces after capture probe attachment. In addition, there are size and charge differences between DNA and BSA, which displays a huge difference. The difference in current was 4.7E-07 A and this higher change in current is responsible for the hybridization of target and capture DNA. Further, to estimate the detection limit, different concentrations of target DNA hybridization with constant capture DNA was interacted. As shown in Figure 5D, 1 fM of target DNA hybridization with capture DNA increases the current response from 1.77E-07 to 1.77E-06 A. And then with increasing the target DNA to 10 fM, 100 fM, 1 pM and 10 pM, the current was further increased to 2.6E-07, 3.74E-07, 4.48E-07 and 6.23E-07 A, respectively. This current increment was indicated a clear hybridization of each target DNA with the immobilized capture DNA. By increasing the target DNA, the current responses were gradually increased. Further, the similar concentrations of target DNA were mixed with diluted serum did not interfere the target DNA identification (Figure 6A). The differences in current were plotted and calculated the detection limit of target DNA as 1 fM. Further, the sensitivity also falls in the similar level of interaction (Figure 6B). The present sensor is operating based on the dipole moment on the sensing surface with the molecular attachment/interaction. In the case of capture DNA immobilization, there is higher negative charge due to the exposure of phosphate (negatively charged) backbone from the capture DNA. The exposure of phosphate groups has been masked upon duplex formation by the target DNA, which influences the increment in current with dose-dependent attachments of target DNA.
Specificity and Reproducibility with Device Performance
Specific target DNA detection was confirmed with three different control experiments. Single- and triple-mismatched and complementary of target DNA sequences interactions with immobilized capture probe was analyzed. As shown in Figure 6C, all control sequences did not display significant changes in current, while the hybridization of target with capture shows a clear current change, indicating the specific target DNA detection on MXene-modified sensing surfaces.
To check the reproducibility of the sensing system and to reflect the performance of sensor, capture probe immobilization and target identification processes were repeated. For that, five different devices were prepared separately and immobilization steps with MXene, GOPTS, capture DNA, BSA attachments were done. On these surfaces 100 pM of target and capture probe hybridization was tested. As shown in Figure 6D, with all five surfaces prepared at the same time the signal was apparently similar and obvious difference was noted. This result confirms the reproducibility of our sensing system for target DNA identification. Overall, the sensor demonstrated in the current study displayed a better performance than other currently available DNA sensors for survivin detection (Table 2).
 |
Table 2 Comparison on Current DNA Sensors for Determining Survivin |
Conclusion
Osteosarcoma is the bone cancer originating in the bone cells and commonly found in a long bone of the leg with young adults and teenagers. Chemotherapy and surgery are the current treatments for OS. Diagnosing OS with a suitable biomarker is mandatory for further treatment and reduce the symptoms caused by OS. Survivin protein gene found as the efficient biomarker and highly correlated with OS. This work focused on detecting the target sequence of survivin gene on MXene-coated multiple junction triangles dielectrode surface. Capture DNA was immobilized on MXene through the GOPTS as a linker, and the detection limit of target hybridization was reached to 1 fM. Further, control sequences of single- and triple-mismatched and complementary of target DNA sequences did not show significant current responses, indicating the specific detection of survivin DNA sequence. This research work can effectively detect the target DNA sequence as a biomarker, helps to diagnose OS.
Acknowledgment
SCBG was supported by a special short-term grant (9001-00596) by Universiti Malaysia Perlis. Authors thank the Servier Medical Art.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Letchumanan I, Gopinath SCB, Md Arshad MK, et al. Gold nano-urchin integrated label-free amperometric aptasensing human blood clotting factor IX: a prognosticative approach for “Royal disease”. Biosens Bioelectron. 2019;131:128–135. doi:10.1016/j.bios.2019.02.006
2. Chen C, Chen M, Ke J, et al. Surface effects on optical and electrical properties of ZnO nanostructures. Pure Appl Chem. 2010;82(11):2055–2073. doi:10.1351/PAC-CON-09-12-05
3. Maiti S, Shrivastava NK, Suin S, et al. Polystyrene/MWCNT/graphite nanoplate nanocomposites: efficient electromagnetic interference shielding material through graphite nanoplate–MWCNT–graphite nanoplate networking. ACS Appl Mater Interfaces. 2013;5(11):4712–4724. doi:10.1021/am400658h
4. Yang X, Zhang F, Hu Y, et al. Gold nanoparticles doping graphene sheets nanocomposites sensitized screen-printed carbon electrode as a disposable platform for voltammetric determination of guaiacol in bamboo juice. Int J Electrochem Sci. 2014;9:5061–5072.
5. Zanchetta G, Lanfranco R, Giavazzi F, et al. Emerging applications of label-free optical biosensors. Nanophotonics. 2017;6(4):627–645. doi:10.1515/nanoph-2016-0158
6. Chia HL, Mayorga-Martinez CC, Antonatos N, et al. MXene titanium carbide-based biosensor: strong dependence of exfoliation method on performance. Anal Chem. 2020;92(3):2452–2459. doi:10.1021/acs.analchem.9b03634
7. Lakshmipriya T, Horiguchi Y, Nagasaki Y. Co-immobilized poly(ethylene glycol)-block-polyamines promote sensitivity and restrict biofouling on gold sensor surface for detecting factor IX in human plasma. Analyst. 2014;139(16):3977–3985. doi:10.1039/C4AN00168K
8. Balakrishnan SR, Hashim U, Gopinath SCB, et al. A point-of-care immunosensor for human chorionic gonadotropin in clinical urine samples using a cuneated polysilicon nanogap lab-on-chip. PLoS One. 2015;10(9):e0137891. doi:10.1371/journal.pone.0137891
9. Letchumanan I, Md Arshad MK, Balakrishnan SR, et al. Gold-nanorod enhances dielectric voltammetry detection of c-reactive protein: a predictive strategy for cardiac failure. Biosens Bioelectron. 2019;130:40–47. doi:10.1016/j.bios.2019.01.042
10. Balakrishnan SR, Hashim U, Gopinath SCB, et al. Polysilicon nanogap lab-on-chip facilitates multiplex analyses with single analyte. Biosens Bioelectron. 2015;1–9. Available from: http://www.sciencedirect.com/science/article/pii/S0956566315305340. Accessed November 23, 2020.
11. Ghobaei Namhil Z, Kemp C, Verrelli E, et al. A label-free aptamer-based nanogap capacitive biosensor with greatly diminished electrode polarization effects. Phys Chem Chem Phys. 2019;21(2):681–691. doi:10.1039/C8CP05510F
12. Miwa S, Shirai T, Yamamoto N, et al. Current and emerging targets in immunotherapy for osteosarcoma. J Oncol. 2019;7035045.
13. Czarnecka AM, Synoradzki K, Firlej W, et al. Molecular biology of osteosarcoma. Cancers. 2020;12(8):1–27. doi:10.3390/cancers12082130
14. Sah NK, Khan Z, Khan GJ, et al. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006;244(2):164–171. doi:10.1016/j.canlet.2006.03.007
15. Ramanathan S, Gopinath SCB, Md Arshad MK, et al. Aluminosilicate nanocomposites from incinerated chinese holy joss fly ash: a potential nanocarrier for drug cargos. Sci Rep. 2020;10(1):1–14. doi:10.1038/s41598-020-60208-x
16. Zhong G, Liu A, Xu X, et al. Detection of femtomolar level osteosarcoma-related gene via a chronocoulometric DNA biosensor based on nanostructure gold electrode. Int J Nanomedicine. 2012;7:527
17. Ramanathan S, Gopinath SCB, Arshad MKM, et al. Aluminosilicate nanocomposite on genosensor: a prospective voltammetry platform for epidermal growth factor receptor mutant analysis in non-small cell lung cancer. Sci Rep. 2019;9(1):17013. doi:10.1038/s41598-019-53573-9
18. Sonawane MD, Nimse SB. Surface modification chemistries of materials used in diagnostic platforms with biomolecules. J Chem. 2016;2016:9241378.
19. Cao Y, Deng Q, Liu Z, et al. Enhanced thermal properties of poly(vinylidene fluoride) composites with ultrathin nanosheets of MXene. RSC Adv. 2017;7(33):20494–20501. doi:10.1039/C7RA00184C
20. Luo J, Zhang W, Yuan H, et al. Pillared structure design of MXene with ultralarge interlayer spacing for high-performance lithium-ion capacitors. ACS Nano. 2017;11(3):2459–2469. doi:10.1021/acsnano.6b07668
21. Yang Q, Jiao T, Li M, et al. In situ formation of NaTi2(PO4)3 cubes on Ti3C2 MXene for dual-mode sodium storage. J Mater Chem A. 2018;6(38):18525–18532. doi:10.1039/C8TA06995F
22. Iqbal MA, Tariq A, Zaheer A, et al. Ti3C2-MXene/bismuth ferrite nanohybrids for efficient degradation of organic dyes and colorless pollutants. ACS Omega. 2019;4(24):20530–20539. doi:10.1021/acsomega.9b02359
23. Li Z, Wang L, Sun D, et al. Synthesis and thermal stability of two-dimensional carbide MXene Ti3C2. Mater Sci Eng B Solid State Mater Adv Technol. 2015;191:33–40. doi:10.1016/j.mseb.2014.10.009
24. Satheeshkumar E, Makaryan T, Melikyan A, et al. One-step solution processing of Ag, Au and Pd@MXene hybrids for SERS. Sci Rep. 2016;6(1):32049. doi:10.1038/srep32049
25. Shah SA, Habib T, Gao H, et al. Template-free 3D titanium carbide (Ti3C2Tx) MXene particles crumpled by capillary forces. Chem Commun. 2017;53(2):400–403. doi:10.1039/C6CC07733A
26. Wang X, Shen X, Gao Y, et al. Atomic-scale recognition of surface structure and intercalation mechanism of Ti3C2X. J Am Chem Soc. 2015;137(7):2715–2721. doi:10.1021/ja512820k
27. Halim J, Lukatskaya MR, Cook KM, et al. Transparent conductive two-dimensional titanium carbide epitaxial thin films. Chem Mater. 2014;26(7):2374–2381. doi:10.1021/cm500641a
28. Nie Y, Huang J, Ma S, et al. MXene-hybridized silane films for metal anticorrosion and antibacterial applications. Appl Surf Sci. 2020;527:146915. doi:10.1016/j.apsusc.2020.146915
29. Ratajczak K, Krazinski BE, Kowalczyk AE, et al. Optical biosensing system for the detection of survivin mRNA in colorectal cancer cells using a graphene oxide carrier-bound oligonucleotide molecular beacon. Nanomaterials. 2018;8(7):510. doi:10.3390/nano8070510
30. Giannetti A, Barucci A, Cosi F, et al. Optical fiber nanotips coated with molecular beacons for DNA detection. Sensors. 2015;15(5):9666–9680. doi:10.3390/s150509666
31. Stobiecka M, Chalupa A. DNA strand replacement mechanism in molecular beacons encoded for the detection of cancer biomarkers. J Phys Chem B. 2016;120(21):4782–4790. doi:10.1021/acs.jpcb.6b03475
32. Ratajczak K, Krazinski BE, Kowalczyk AE, et al. Hairpin–hairpin molecular beacon interactions for detection of survivin mRNA in malignant SW480 cells. ACS Appl Mater Interfaces. 2018;10(20):17028–17039. doi:10.1021/acsami.8b02342
33. Stobiecka M, Dworakowska B, Jakiela S, et al. Sensing of survivin mRNA in malignant astrocytes using graphene oxide nanocarrier-supported oligonucleotide molecular beacons. Sens Actuators B Chem. 2016;235:136–145. doi:10.1016/j.snb.2016.04.176
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.