Back to Journals » Journal of Inflammation Research » Volume 16
Mutations Status of NOTCH Signaling Pathway Predict Prognosis of Immune Checkpoint Inhibitors in Colorectal Cancer
Authors Lin A, Yao J , Cheng Q , Liu Z, Luo P , Zhang J
Received 26 October 2022
Accepted for publication 11 April 2023
Published 17 April 2023 Volume 2023:16 Pages 1693—1709
DOI https://doi.org/10.2147/JIR.S394894
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Monika Sharma
Anqi Lin,1,* Jiarong Yao,1,* Quan Cheng,2,3 Zaoqu Liu,4 Peng Luo,1 Jian Zhang1
1Department of Oncology, Zhujiang Hospital, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China; 2Department of Neurosurgery, Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China; 3National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China; 4Department of Interventional Radiology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Peng Luo; Jian Zhang, Department of Oncology, Zhujiang Hospital, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China, Tel +86-18588447321 ; +86-13925091863, Email [email protected]; [email protected]
Purpose: In recent years, tumour immunotherapy has ushered in a new era of oncology treatment. However, the use of immune checkpoint inhibitors (ICIs) in the treatment of CRC remains limited. There is an urgent clinical need for precise biomarkers that can aid in the screening and treatment of CRC subtypes. Therefore, we focused on the NOTCH pathway mutation status and conducted a systematic analysis for its predictive value of ICI therapy efficacy.
Methods: We collected mutational and clinical data from cohorts of CRC patients treated with ICIs. The relationship between NOTCH pathway mutations (NOTCH-MT) and CRC immunotherapy prognosis was analysed using univariate and multivariate Cox regression models. CRC cohort data from The Cancer Genome Atlas (TCGA) database were combined to obtain a comprehensive overview of immunogenicity and tumour microenvironment (TME) differences among different NOTCH pathway mutation statuses.
Results: We observed greater infiltration of M1 macrophages, CD8+ T cells, neutrophils, and activated natural killer (NK) cells with NOTCH-MT status. Immunogenicity was also significantly higher in patients with NOTCH-MT, as were tumour mutational burden (TMB), neoantigen load (NAL), and the number of mutations in DNA damage repair (DDR) pathways.
Conclusion: NOTCH-MT status was strongly associated with the prognosis of CRC patients treated with ICIs and is expected to serve as a novel biomarker and therapeutic target for CRC.
Keywords: NOTCH, CRC, ICIs, biomarker, tumour microenvironment
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in men worldwide and the second leading cause of cancer death.1 Approximately 41% of CRC cases occur in the proximal colon, 22% in the distal colon, and 28% in the rectum.2,3 Notably, 50% of patients develop distant metastasis, referred to as metastatic CRC (mCRC), which has a high mortality rate. Usually, chemoradiotherapy can be used to treat locally advanced CRC before surgery. However, some studies have shown that fractional radiation of tumour cells also leads to chemoresistance.4,5 Therefore, new effective treatment strategies for advanced CRC patients are urgently needed. Immunotherapy, a treatment option that employs the body’s immune system to fight cancer, has shown promise in treating certain cancer types.6 While patients with some cancers, such as melanoma7 and lung cancer,8 benefit from immune checkpoint inhibitor (ICI)treatment, others do not. Recently, it was discovered that ICIs are effective in a subgroup of CRC patients with mismatch repair defect (dMMR) and high microsatellite instability (MSI-H) tumours (dMMR-MSI-H tumours) but are ineffective in another subgroup of patients with pMMR and low microsatellite instability (pMMR-MSI-L tumour).9 The results indicated that patients with dMMR-MSI-H tumours had a 40% objective response rate when treated with pembrolizumab, compared to 0% in patients with pMMR-MSI-L tumors and a 78% immune-related progression-free survival rate when treated with pembrolizumab.10 In addition, previous studies show that carcinoembryonic antigen (CEA) levels can predict prognosis in CRC patients.11 However, elevated CEA levels have also been observed in many nonmalignant conditions, such as alcoholism, cigarette smoking, pancreatitis, and liver disease.12 Therefore, it is critical to identify biomarkers with a high specificity and detection rates for predicting PD-1/PD-L1 ICI efficacy in patients with CRC.
Important predictive molecular markers for ICI treatment of CRC include d-MMR-MSI-H, tumour mutational burden (TMB), tumour microenvironment (TME), tumour-infiltrating lymphocytes (TILs), programmed death-ligand 1(PD-L1), DNA polymerase epsilon (POLE), and polymerase delta 1 (POLD1).13 However, these biomarkers have some limitations. For example, although TMB is a useful molecular marker for CRC, there is no reliable evidence to support the use of universal critical values to define TMB-H and TMB-L tumours.14 Furthermore, although whole-exome sequencing (WES) is the gold standard for TMB detection, the associated high cost and uniformity of detection standards represent significant limitations.15 Different methods for detecting MMR and microsatellites may produce inconsistent results.16 For example, the expression of PD-L1 is highly variable between tumours with varying microsatellite states.17 As a result, there is an urgently need for new biomarkers to predict the prognosis and efficacy of immunotherapy in patients with CRC.
The NOTCH-signalling pathway was first discovered in Drosophila melanogaster.18 It is a highly conserved signalling system that is regulated by Notch receptors (Notch1-4) and ligands (Jagged1, Jagged2, DLL1, DLL3, DLL4) through cell-to-cell interactions or through the activation of other pathways (NF-κB, WNT, TGFβ, and STAT3).19 The NOTCH pathway is involved in the differentiation and development of various tissues and cells. First, NOTCH signalling can affect the activation of CD8+T cells20 and the polarization of macrophages,21 thereby regulating the TME. Second, inhibiting Notch1 can increase the immunogenicity of DDR-regulated tumours.22 Additionally, Li et al found that highly-mutated NOTCH signalling indicated higher immunotherapy efficacy in non-small cell lung cancer (NSCLC).23 Notch1 signalling is essential for maintaining intestinal homeostasis, however, aberrant activation of the associated receptor disrupts the dynamic balance of the Notch1-mediated regulatory pathway, ultimately promoting CRC proliferation.24,25 Epithelial NOTCH signalling also rewires the TME of CRC cells to drive poor-prognosis subtypes and metastasis.26 However, the effect of NOTCH pathway mutation status on the clinical prognosis of patients with CRC receiving immunotherapy is unknown and warrants further investigation.
In this study, we used a CRC immunotherapy cohort (Samstein-CRC-cohort), TCGA-CRC, the TCGA-CRC cohort, and a local CRC cohort to evaluate the relationship between the mutation status of the NOTCH pathway and clinical prognosis in CRC patients receiving ICI therapy. We also elucidated the clinical and TME characteristics of CRC patients with NOTCH pathway mutations.
Methods
CRC Data Collection
The CRC cohort dataset published by Samstein et al comprised patients treated with anti-PD-1/PD-L1 therapy or anti-CTLA4 therapy or a combination of the two datasets from the public database (hereafter referred to as Samstein-CRC).27 The Samstein-CRC cohort data on mutations and immunotherapy prognosis from 109 patients with advanced CRC treated with ICIs.
TCGA-COAD and TCGA-READ clinical, mutation, and expression data were retrieved from the TCGA database.28 The TCGA-COAD and TCGA-READ cohorts were combined to form the TCGA-CRC cohort.We obtained mutation data on 103 CRC samples from Zhujiang Hospital, Southern Medical University, using targeted sequencing (HapOncoTM680 Panel). Supplementary Table 1 contains information about the panel on targeted sequencing. All participants provided written informed consent, and this study was approved by the Zhujiang Hospital Research Ethics Committee of Southern Medical University.
Supplementary Tables 2–4 contain the baseline characteristics of the three CRC cohorts.
Evaluation of NOTCH Pathway Mutation State
The NOTCH signalling gene set (KEGG Notch signalling pathway) was downloaded from the Molecular Signatures Database (MSI GDB),29 Supplementary Table 5 contains information about the NOTCH signalling gene set. We excluded synonymous mutation data from somatic mutation data for the three CRC cohorts, thereby retaining only nonsynonymous mutation data. We counted the number of NOTCH pathway gene mutations in each CRC patient. Then, based on whether each CRC patient had zero NOTCH pathway gene mutations, we divided the patients into mutant-type (MT) and wild-type (WT) groups.
Analysis of the Tumour Immune Microenvironment
TME immunogenicity analysis comprised immune-related gene expression, immune cell, immune-related score, and pathway enrichment analyses. Furthermore, the TMB, neoantigen loads (NAL), and number of mutations in several the DNA damage repair (DDR) pathways were used in the immunogenicity analysis. TMB and NAL were derived from a published study in the TCGA-CRC.30 The TMB score of the Samstein-CRC cohort was directly obtained from the public datasets, and the TMB scores of Local-CRC cohort and TCGA-CRC cohort were quantified by dividing the number of somatic mutations by 38 Mb. Moreover, DDR pathways were obtained from a previously published study.31 In TCGA-CRC cohort, we selected the number of nonsynonymous mutations in DDR pathways in each patient. Immune-related genes and their functional classifications were obtained from articles published by Rooney et al32 and Thorsson et al.30 Additionally, we calculated the immune cell abundance of each CRC patient using the CIBERSORT, EPIC, and IPS immune cell algorithms33,34 in the TCGA-CRC cohort. Path enrichment analysis comprised two algorithms: gene set enrichment analysis (GSEA) and single sample GSEA (ssGSEA).35,36 The signalling gene set was downloaded from the MSI GDB.Based on transcriptome data from the TCGA-CRC cohort, ssGSEA and GSEA were used to enrich and analyse the functional gene sets.
Statistical Analysis
The Mann–Whitney U-test to compare continuous variables between the two groups . Fisher’s exact test was used to compare categorical variables. Furthermore, univariate and multivariate Cox proportional hazards regression models, as well as Kaplan-Meier analysis, were used to determine prognosis. All statistics and visualizations in this study were performed and created using the R programming language (Version. 4.0). In this study, a two-tailed P value less than 0.05 was deemed statistically significant.
Results
NOTCH-MT is Related to Improvement in OS After ICI Treatment
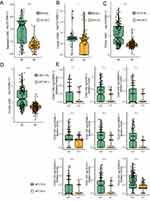
To investigate the relationship between NOTCH-MT and ICI efficacy in CRC patients, we downloaded the CRC cohort comprising patients treated with ICIs (Samstein-CRC). A detailed breakdown of the analytical process is shown in Figure 1A. Next, we divided all patients into two groups according to the nonsynonymous mutation status of the NOTCH pathway, namely the NOTCH-MT group and the NOTCH-WT group. Univariate Cox regression analysis revealed that while clinical characteristics such as age (old vs young) and sample type (metastatic vs primary) were unrelated to the survival rate of ICI patients, the mutation status of the NOTCH pathway was closely related to clinical prognosis (Supplementary Figure 1A and B). Additionally, multivariate Cox regression analysis revealed that NOTCH pathway mutation status is an independent protective factor for CRC patients undergoing immunotherapy (Supplementary Figure 1A and C).To determine the gene mutant phenotype had an opposing effect on prognosis, we performed univariate Cox proportional hazards regression analysis to assess the effects of the mutation status of six genes and NOTCH pathway on prognosis in CRC patients receiving ICI immunotherapy (Supplementary Figure 1D).The results suggest that the mutant phenotype of each relevant gene does not indicate worse prognosis after immunotherapy. Therefore, gene mutation status and NOTCH pathway mutation status do not affect the prognosis of CRC patients receiving ICI therapy. The overall survival (OS) of NOTCH-MT in CRCpatients was significantly longer than that of NOTCH-WT patients (P < 0.001, HR = 0.42, 95% CI 0.23–0.77; Figure 1B). Next, we explored the prognostic value of NOTCH pathway mutations in CRCpatients who did not undergo therapy with ICIs. Notably, in the TCGA-CRC cohort, there was no significant difference in the OS between NOTCH-MT and NOTCH-WT CRC patients (Figure 1C). Finally, we evaluated the NOTCH pathway mutation status of patients in the PanCancer cohort treated with ICIs using six genes included in the Samstein-CRC cohort and performed an immunotherapy prognostic analysis. We found a significant difference in the OS of NOTCH-MT PanCancer patients (P = 0.001, HR = 0.79, 95% CI: 0.69–0.91;Supplementary Figure 2A).
Landscape of Gene Mutation in Different NOTCH Signalling Pathway States
To investigate differences in the frequency of somatic mutations between the NOTCH-MT and NOTCH-WT groups, we analysed the top 20 somatic mutations in an ICI treatment cohort. First, we identified that higher mutation rates among the top 20 mutant genes, KMT2D (48.1% vs 10.5%; P<0.05), ARID1A (50% vs 7%; P<0.05), PTPRS (40.4% vs 5.2%; P<0.05), RNF43 (38.5% vs 1.8%; P<0.05), TCF7L2 (30.8% vs 8.8%; P<0.05), KMT2C (34.6% vs 3.5%; P<0.05), FAT1 (28.8% vs 5.3%; P<0.05), SMARCA4 (26.9% vs 7.0%; P<0.05), ARID1B (30.8% vs 1.8%; P<0.05), FBXW7 (25.0% vs 7.0%; P<0.05), PTCH1 (26.9% vs 5.3%; P<0.05), ZFHX3 (32.7% vs 0%; P<0.05), BRCA2 (23.1% vs 7.0%; P<0.05), CREBBP (30.8% vs 0%; P<0.05) and NF1 (25.0% vs 5.3%; P<0.05). Furthermore, APC (63.5% vs 82.5%; P<0.05) and TP53 (40.4% vs 61.4%; P<0.05) had lower mutation rates in the NOTCH-MT group. The NOTCH-MT group had a higher TMB value than the NOTCH-WT group (P<0.05). There were no significant differences in other clinical characteristics, such as age and sample type between the NOTCH-MT and NOTCH-WT groups (Figure 2A). We also integrated the NOTCH pathway gene set in the 3 cohorts (Supplementary Figure 2B) and found CREBBP, EP300, NOTCH1, NOTCH2, and NOTCH3 to be shared genes.In addition, we further investigated NOTCH pathway gene mutations in the TCGA-CRC cohort, and found that the mutation frequencies of CREBBP(26%), EP300(17%), NOTCH1(11%), NOTCH2(15%), and NOTCH3(18%) ranked very high in the TCGA-CRC cohort (Supplementary Figure 2C). We also identified NOTCH1-4 as critical genes in the NOTCH pathway after analysing the NOTCH signalling using the pathway visualization function in the KEGG database (Supplementary Figure 3).
Next, we examined the mutual exclusion co occurrence of the top 20 mutant genes in a cohort of ICI patients. In Figure 2B, brown represents co occurrence while yellow represents mutually exclusive relationships. We found high correlations between several genes ARID1A and KMT2D(P<0.01), PTPRS and KMT2D(P<0.01), ARID1B and ARID1A(P<0.01) and ZFHX3 and PTPRS(P<0.01) showed high correlation. In addition, (RNF43 vs TP53; P<0.01) had the most exclusive relationships (Figure 2B). The immunogenicity of the NOTCH-MT group was higher than that of the NOTCH-WT group.
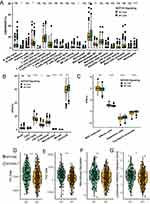
As illustrated in Figure 3A–C, we analysed TMB in an ICI treatment cohort, TCGA-CRC cohort, and local treatment cohort based on NOTCH pathway mutation status. TMB was significantly greater in the NOTCH-MT group than in the NOTCH-WT group (all p<0.05). The accumulation of cancer genome mutations may generate “new antigens” specific to tumours. As a result, we analysed the NAL in the TCGA-CRC cohort and found that the NOTCH-MT group had a higher NAL (p<0.05, Figure 3D). TMB and NAL levels may have increased in NOTCH-MT patients as a result of their improved response to ICIs. Numerous studies have established a link between DDR pathway mutations and the curative effect of ICIs. Thus, we compared the number of DDR pathway mutations in NOTCH-MT and NOTCH-WT tumours. The number of DDR pathway mutations in the TCGA-CRC cohort increased significantly in the NOTCH-MT group (including BER, HR, MMR, SSB, DSB, NER, NHEJ, FA, and DDR, all p<0.05, Figure 3E).
Difference in the Immune Microenvironment Between the NOTCH-MT and NOTCH-WT Groups in the TCGA-CRC Cohort
There are obvious therapeutic differences between patients receiving immunotherapy are largely due heterogeneity in the immune microenvironment. To measure differences in the immune microenvironment between the NOTCH-MT and NOTCH-WT groups, we compared immune cell characteristics, immune-related scores, and immune-related genes. We calculated the immune cell infiltration state of patients in the TCGA-CRC cohort using the CIBERSORT, EPIC, and IPS algorithms and compared differences in immune cell infiltration patterns between NOTCH-MT and NOTCH-WT patients. The results indicated that memory B cells, CD8+ T cells, activated NK cells, M1 macrophages, and neutrophils were more abundant in the NOTCH-MT group than in the NOTCH-WT group using the CIBERSORT algorithm (p<0.05, Figure 4A).
In addition, CD4+ T cells, CD8+ T cells, macrophages, and NK cells were more abundant in the NOTCH-MT group than in the NOTCH-WT group using the EPIC algorithm (p<0.05, Figure 4B). MHC molecules, effector cells, and checkpoint molecules were more abundant in the NOTCH-MT group than in the NOTCH-WT group using the IPS algorithm (p<0.05, Figure 4C). This indicated that the NOTCH-MT group had a significantly higher proportion of immunocompetent cells than the NOTCH-WT group. We calculated immune-related scores (Th1 cells, Th2 cells, macrophage regulation, and lymphocyte infiltration characteristic scores) and discovered that the NOTCH-MT group had significantly higher immune-related scores than the NOTCH-WT group (p<0.05, Figure 4D–G).
Immune-related genes regulate the immune status of tumours, and their expression affects the results of ICI treatment. In the TCGA-CRC cohort, we compared the relative expression of nine immune checkpoint-related genes between the NOTCH-WT and NOTCH-MT groups. The results indicated that CD274, HAVCR2, LAG3, IDO1, CTLA4, TIGIT, PDCD1, and PDCD1LG2 expression levels were significantly higher in the NOTCH-MT group than in the NOTCH-WT group (all P<0.05, Figure 5A). The differences in the expression patterns of immune-related genes between NOTCH-MT and NOTCH-WT are depicted in Figure 5B. The results indicate that NOTCH-MT patients had significantly increased expression of antigen presentation-related genes, cytolysis-related genes, stimulating immune-related genes, activated immune cell-related genes (CD4+ regulatory T-cells, CD8+ T-cells, NK cells), and inhibition-related genes.
Gene Enrichment Analysis Between the NOTCH-MT and NOTCH-WT Groups
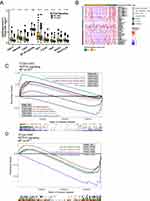
GSEA and ssGSEA were used to identify functional gene sets enriched in the NOTCH-MT and NOTCH-WT groups. JAK-STAT, Toll-like receptor, B-cell receptor, T-cell receptor, and Fcγ receptor were all significantly upregulated in the NOTCH-MT group (p<0.05, ES>0). Wnt, FGFR4 ligand binding, and associated action pathways were significantly downregulated in the NOTCH-MT group (all p<0.05, ES<0, Figure 5C). Some immune exhaustion-related pathways, such as fatty acid catabolism and glucose metabolism regulation, were more abundant in the NOTCH-WT group (all p<0.05, ES<0, Figure 5D). Certain pathways involved in the killing function of immune cells were found to be upregulated in the NOTCH-MT group, including leukocyte migration, antigen treatment cross-presentation, and natural killer cell activation (all p<0.05, ES>0, Figure 6A). Certain cytokine-related pathways, such as interleukin, tumour necrosis factor, interferon, colony-stimulating factor, and chemokine-related pathways, are also significantly enriched in NOTCH-MT (all p<0.05, ES>0, Figure 6B). Similarly, the results of ssGSEA showed that the ssGSEA scores of several cytokines and chemokines were significantly higher in the NOTCH-MT group. In contrast, the ssGSEA scores of FGFR ligand binding and fatty acid metabolism pathways in the NOTCH-MT group were significantly lower than those in the NOTCH-WT group (all p<0.05, Figure 7).
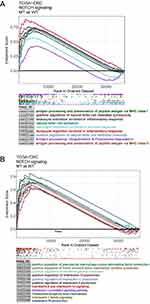 |
Figure 6 (A) In the TCGA-CRC cohort, differences in immune cells (A) and cytokines (B) were observed between NOTCH-WT and NOTCH-MT CRC patients (identified by GSEA). |
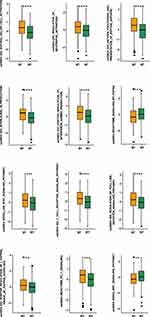 |
Figure 7 Comparison of NOTCH-MT and NOTCH-WT tumours in the TCGA-CRC cohort using ssGSEA (**p<0.01; ***p<0.001; and ****p<0.0001). |
Discussion
This study sought to determine the relationship between NOTCH-MT and prognosis in patients withCRC treated with ICIs. We discovered that NOTCH-MT is associated with a favour prognosis in patients receiving ICIs, indicating that NOTCH pathway mutations can be serve as an independent predictive factor for CRC immunotherapy. We also sought to determine reasons why NOTCH-MT is associated with improved clinical benefits, which were particularly notable in the immune microenvironment (Figure 8). The NOTCH-MT group demonstrated increased immunogenicity, a greater number of immune-activated cells, and higher expression of immune point-related genes, all of which are typically associated with a better prognosis following immunotherapy. These findings suggest that NOTCH pathway status may be used as a biomarker to predict prognosis of CRC patients treated with ICIs.
 |
Figure 8 Potential mechanism underlying the prognostic value of NOTCH-MT. |
The TME may be one mechanism by which NOTCH -MT patients experience improved prognosis with immunotherapy. It comprises tumour cells, fibroblasts, endothelial cells, immune infiltrating cells, and extracellular matrix components. Immune infiltrating cells are the main components of the TME and are shown to be related to the efficacy of immunotherapy.37 According to the fundings on the immune cell infiltration state of patients in the TCGA-CRC cohort, we inferred that patients with NOTCH-MT had a higher proportion of memory B cells, CD8+ T cells, activated NK cells, and M1 macrophages. Macrophages exhibit two distinct phenotypes. M1 macrophages typically express proinflammatory cytokines and contribute to the antitumor immune response, whereas, M2 macrophages express anti-inflammatory cytokines and chemokines, inhibit CD8+ T-cell activation, promote Treg recruitment, and contribute to tumour immune escape.38 NOTCH signalling promotes macrophage polarization to the M1 phenotype, thereby enhancing antitumour immunity.21 Macrophages can mediate T-cell activation through the production of IL-12 and the expression of costimulatory molecules such as CD86.39 Furthermore, T-cell infiltration, particularly of CD8+ T-cells into the TME, has been associated with better prognosis in various malignant tumours types, including breast cancer, lung cancer, melanoma, colon cancer, and colorectal cancer.40,41
Similarly, the NOTCH pathway can promote CD8+ T-cell activation by expressing granzyme B and IFN-γ.20 NK cells are innate cytotoxic lymphocytes that play a role in cancer surveillance and elimination.42 IL-12 is primarily produced by antigen-presenting cells (APCs) with the assistance of Toll-like receptors and stimulates NK cells to exert various physiological effects on peripheral blood lymphocytes.42,43 The majority of the effects induced by IL-12 are mediated by IFN-γ secretion.44,45 IFN-γ has been shown to reduce Treg infiltration, thus, enhancing the antitumour immune effect.45,46 NK cells have recently been shown to enhance the efficacy of PD-1/PD-L1 immunotherapy.46 Taken together, we hypothesize that the NOTCH signalling pathway also regulates NK cell activation via IFN-γ, thereby enhancing the effect of immunotherapy.
In addition to the TME, increased tumour immunogenicity may contribute to an improved immunotherapy prognosis. Immunogenicity is the body’s capacity to promote immunity,47 which has been linked to immunotherapy efficacy48 and can be assessed through TMB, NAL, and mutations in the DDR pathways.49,50 TMB appears to be a predictive biomarker of tumour response to ICIs in various cancer types.51,52 In the KEYNOTE-158 study, a high TMB (≥10 mut/Mb) was associated with a better response to anti-PD-1 treatment.53 TMB is the primary regulator of tumour-specific antigen expression (new antigen).54 The new antigen can enhance antitumour immunity, thereby enhancing the efficacy of immunotherapy.55 The DDR pathway is critical for genome integrity.49 Increased DDR mutation accumulation results in increased TMB and NAL levels, which can induce an antitumour response mediated by T cells.56 In recent studies, NOTCH signalling has been linked to the antitumour immune process mediated by T cells.57 Thus, the higher mutation rates in the TMB, NAL and DDR pathways observed in the NOTCH-MT group demonstrates that CRC patients with NOTCH-MT respond better to ICI treatment.Some tumour signalling pathways can interact with the NOTCH transduction pathway, affecting the immunotherapy prognosis. Studies have shown that Wnt/β-catenin signaling, the upstream Notch pathway, may inhibit the therapeutic effect of ICIs.58,59 By interacting with the Notch pathway ligand Jagged1, β-catenin activates the Notch pathway, thereby contributing to the development of colon cancer.60,61 We discovered that the Wnt/β-catenin pathway, FGFR4 ligand binding, and associated pathways were significantly down-regulated in the NOTCH-MT group via GSEA.62,63 According to previous reports, activation of the FGFR4 signalling pathway is intimately linked with the development and progression of cancer.64,65 By activating the Ras-Raf-MAPK and PI3K-AKT pathways, FGFR4 signalling can promote tumour growth. Interestingly, it has been demonstrated that FGFR4 inhibition indirectly inhibits PD-L1 expression on the surface of tumour cells by affecting the TME.66 Through the JAK-STAT pathway, IL-2 can regulate the development and maturation of NK cells, thereby affecting the antitumour effect.67–69 Some pathways associated with immune exhaustion, such as lipid and glucose metabolism, were also significantly downregulated in the NOTCH-MT group. Numerous studies have demonstrated that lipid and glucose metabolism can promote tumour growth.70 This study investigated the relationship between CRC immunotherapy prognosis and NOTCH pathway mutation status to elucidate the possible mechanism underlyingNOTCH pathway mutation as an independent prognostic marker for CRC immunotherapy. However, this study has some limitations. First, there was a lack of functional assays related to pathways in GSEA analysis. Due to the absence of hot spot mutations among various gene mutations, it is currently difficult to perform experimental verification of in pathway mutation research. Currently, functional enrichment analysis can assess correlations in the NOTCH pathway as closely as possible through associations identified in previously published pathways, ICIs, and the immune microenvironment. We must also concede that the evidence supporting such arguments are relatively weak and thus, we can only propose hypotheses that may explain the observed phenomena. Second, given the small size of the CRC cohort receiving ICIs, we examined the relationship between NOTCH pathway expression and immunotherapy prognosis in the Samstein-CRC cohort. The TCGA-CRC cohort and a local cohort of 108 CRC patients from Zhujiang Hospital, Southern Medical University, were used for verification.
Conclusions
In this study, we found that the OS of CRC patients with NOTCH-MT was significantly longer than that of patients with NOTCH-WT patients in the Samstein-CRC cohort. Additionally, NOTCH-MT enriched activated immune cells had increased immunogenicity and enhanced immune-related characteristics. Therefore, NOTCH-MT status may be used as a biomarker to stratify CRC patients for immunotherapy.
Abbreviations
ICIs, Immune checkpoint inhibitors; CRC, colorectal cancers; TCGA, The Cancer Genome Atlas; MT, mutant-type; WT, wild-type; TME, tumor microenvironment; TMB, tumor mutational burden; NAL, neoantigen load; DDR, DNA damage repair; dMMR, mismatch repair defect; MSI-H, high microsatellite instability; CEA, carcinoembryonic antigen; TILs, tumor-infiltrating lymphocytes; PD-L1, programmed death-ligand 1; POLE, DNA polymerase epsilon; POLD1, polymerase delta 1; WES, whole-exome sequencing; NSCLC, non-small cell lung cancer; GSEA, gene set enrichment analysis; ssGSEA, single sample gene set enrichment analysis; APC, antigen-presenting cells; OS, overall survival.
Data Sharing Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Approval and Informed Consent
The patients/participants provided their written informed consent to participate in this study and the research presented here has been performed in accordance with the Declaration of Helsinki and has been approved by the ethics committee of the Zhujiang Hospital of Southern Medical University (NO. 2022-YW-041). All procedures performed in this study were undertaken as a part of routine clinical practice.
Acknowledgments
Special thanks to Tianqi Gu, Yushan Huang, and Shuang Hou for their contribution to data analysis. Special thanks to the funding providers of the Natural Science Foundation of Guangdong Province, the Science and Technology Planning Project of Guangdong Province and the National Natural Science Foundation of China.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. Anqi Lin and Jiarong Yao share first authorship.
Funding
This work was supported by the Natural Science Foundation of Guangdong Province (Grant No. 2018A030313846 and 2021A1515012593), the Science and Technology Planning Project of Guangdong Province (Grant No. 2019A030317020) and the National Natural Science Foundation of China (Grant No. 81802257, 81871859, 81772457, 82172750 and 82172811).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Cheng L, Eng C, Nieman LZ, Kapadia AS, Du XL. Trends in colorectal cancer incidence by anatomic site and disease stage in the United States from 1976 to 2005. Am J Clin Oncol. 2011;34(6):573–580. doi:10.1097/COC.0b013e3181fe41ed
3. Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients. 2019;11(1):164. doi:10.3390/nu11010164
4. Afshar S, Sedighi Pashaki A, Najafi R, et al. Cross-resistance of acquired radioresistant colorectal cancer cell line to gefitinib and regorafenib. Iran J Med Sci. 2020;45(1):50–58. doi:10.30476/ijms.2019.44972
5. Tanzadehpanah H, Bahmani A, Hosseinpour Moghadam N, et al. Synthesis, anticancer activity, and β-lactoglobulin binding interactions of multitargeted kinase inhibitor sorafenib tosylate (SORt) using spectroscopic and molecular modelling approaches. Luminescence. 2021;36(1):117–128. doi:10.1002/bio.3929
6. Eggermont AMM, Blank CU, Mandala M, et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N Engl J Med. 2018;378(19):1789–1801. doi:10.1056/NEJMoa1802357
7. Schachter J, Ribas A, Long GV, et al. Pembrolizumab versus ipilimumab for advanced melanoma: final overall survival results of a multicentre, randomised, open-label Phase 3 study (KEYNOTE-006). Lancet. 2017;390(10105):1853–1862. doi:10.1016/S0140-6736(17)31601-X
8. Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378(22):2078–2092. doi:10.1056/NEJMoa1801005
9. Ganesh K, Stadler ZK, Cercek A, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–375. doi:10.1038/s41575-019-0126-x
10. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi:10.1056/NEJMoa1500596
11. Sisik A, Kaya M, Bas G, Basak F, Alimoglu O. CEA and CA 19-9 are still valuable markers for the prognosis of colorectal and gastric cancer patients. Asian Pac J Cancer Prev. 2013;14(7):4289–4294. doi:10.7314/APJCP.2013.14.7.4289
12. van der Schouw YT, Verbeek AL, Wobbes T, Segers MF, Thomas CM. Comparison of four serum tumour markers in the diagnosis of colorectal carcinoma. Br J Cancer. 1992;66(1):148–154. doi:10.1038/bjc.1992.233
13. Du F, Liu Y. Predictive molecular markers for the treatment with immune checkpoint inhibitors in colorectal cancer. J Clin Lab Anal. 2022;36(1):e24141. doi:10.1002/jcla.24141
14. Schrock AB, Ouyang C, Sandhu J, et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol. 2019;30(7):1096–1103. doi:10.1093/annonc/mdz134
15. Zeng Z, Yang B, Liao Z. Biomarkers in immunotherapy-based precision treatments of digestive system tumors. Front Oncol. 2021;11:650481. doi:10.3389/fonc.2021.650481
16. Cohen R, Hain E, Buhard O, et al. Association of primary resistance to immune checkpoint inhibitors in metastatic colorectal cancer with misdiagnosis of microsatellite instability or mismatch repair deficiency status. JAMA Oncol. 2019;5(4):551–555. doi:10.1001/jamaoncol.2018.4942
17. Llosa NJ, Luber B, Tam AJ, et al. Intratumoral adaptive immunosuppression and type 17 immunity in mismatch repair proficient colorectal tumors. Clin Cancer Res. 2019;25(17):5250–5259. doi:10.1158/1078-0432.CCR-19-0114
18. Chance O. La valeur des statistiques dans l'etude du cancer du cot de l'utérus [Value of statistics in the study of cancer of the uterine cervix]. C R Soc Fr Gyncol. 1951;21(7):305–311. French.
19. Ayaz F, Osborne BA. Non-canonical notch signaling in cancer and immunity. Front Oncol. 2014;4:345. doi:10.3389/fonc.2014.00345
20. Cho OH, Shin HM, Miele L, et al. Notch regulates cytolytic effector function in CD8+ T cells. J Immunol. 2009;182:3380–3389. doi:10.4049/jimmunol.0802598
21. Wang YC, He F, Feng F, et al. Notch signaling determines the M1 versus M2 polarization of macrophages in antitumor immune responses. Cancer Res. 2010;70(12):4840–4849. doi:10.1158/0008-5472.CAN-10-0269
22. Licciulli S, Avila JL, Hanlon L, et al. Notch1 is required for Kras-induced lung adenocarcinoma and controls tumor cell survival via p53. Cancer Res. 2013;73(19):5974–5984. doi:10.1158/0008-5472.CAN-13-1384
23. Li X, Wang Y, Li X, Feng G, Hu S, Bai Y. The impact of NOTCH pathway alteration on tumor microenvironment and clinical survival of immune checkpoint inhibitors in NSCLC. Front Immunol. 2021;12:638763. doi:10.3389/fimmu.2021.638763
24. Tyagi A, Sharma AK, Damodaran C. A review on notch signaling and colorectal cancer. Cells. 2020;9(6):1549. doi:10.3390/cells9061549
25. Vinson KE, George DC, Fender AW, Bertrand FE, Sigounas G. The notch pathway in colorectal cancer. Int J Cancer. 2016;138(8):1835–1842. doi:10.1002/ijc.29800
26. Jackstadt R, van Hooff SR, Leach JD, et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell. 2019;36(3):319–336.e317. doi:10.1016/j.ccell.2019.08.003
27. Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi:10.1038/s41588-018-0312-8
28. Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. 2015;19(1A):A68–77. doi:10.5114/wo.2014.47136
29. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi:10.1093/bioinformatics/btr260
30. Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e814. doi:10.1016/j.immuni.2018.03.023
31. Luo P, Lin A, Li K, Wei T, Zhang J. DDR pathway alteration, tumor mutation burden, and cisplatin sensitivity in small cell lung cancer: difference detected by whole exome and targeted gene sequencing. J Thorac Oncol. 2019;14(12):e276–e279. doi:10.1016/j.jtho.2019.08.2509
32. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi:10.1016/j.cell.2014.12.033
33. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259.
34. Racle J, Gfeller D. EPIC: a tool to estimate the proportions of different cell types from bulk gene expression data. Methods Mol Biol. 2020;2120:233–248.
35. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
36. Hnzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7.
37. Balkwill FR, Capasso M, Hagemann T. The tumor microenvironment at a glance. J Cell Sci. 2012;125(Pt 23):5591–5596. doi:10.1242/jcs.116392
38. Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233(9):6425–6440. doi:10.1002/jcp.26429
39. Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11(11):723–737. doi:10.1038/nri3073
40. Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298–306. doi:10.1038/nrc3245
41. Reiser J, Banerjee A. Effector, memory, and dysfunctional CD8(+) T cell fates in the antitumor immune response. J Immunol Res. 2016;2016:8941260. doi:10.1155/2016/8941260
42. Hodgins JJ, Khan ST, Park MM, Auer RC, Ardolino M. Killers 2.0: NK cell therapies at the forefront of cancer control. J Clin Invest. 2019;129(9):3499–3510. doi:10.1172/JCI129338
43. Trinchieri G, Rengaraju M, D’Andrea A, et al. Producer cells of interleukin 12. Parasitol Today. 1993;9:97. doi:10.1016/0169-4758(93)90215-2
44. Ma X, Chow JM, Gri G, et al. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. J Exp Med. 1996;183(1):147–157. doi:10.1084/jem.183.1.147
45. Overacre-Delgoffe AE, Chikina M, Dadey RE, et al. Interferon-gamma drives treg fragility to promote anti-tumor immunity. Cell. 2017;169(6):1130–1141.e1111. doi:10.1016/j.cell.2017.05.005
46. Hsu J, Hodgins JJ, Marathe M, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest. 2018;128(10):4654–4668. doi:10.1172/JCI99317
47. Blankenstein T, Coulie PG, Gilboa E, Jaffee EM. The determinants of tumour immunogenicity. Nat Rev Cancer. 2012;12(4):307–313. doi:10.1038/nrc3246
48. Wang S, He Z, Wang X, Li H, Liu XS. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. Elife. 2019;8. doi:10.7554/eLife.49020
49. Wang Z, Zhao J, Wang G, et al. Comutations in DNA damage response pathways serve as potential biomarkers for immune checkpoint blockade. Cancer Res. 2018;78(22):6486–6496. doi:10.1158/0008-5472.CAN-18-1814
50. Tran E, Ahmadzadeh M, Lu YC, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387–1390. doi:10.1126/science.aad1253
51. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi:10.1126/science.aaa1348
52. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi:10.1056/NEJMoa1406498
53. Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, Phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21(10):1353–1365. doi:10.1016/S1470-2045(20)30445-9
54. Desrichard A, Snyder A, Chan TA. Cancer neoantigens and applications for immunotherapy. Clin Cancer Res. 2016;22:807–812. doi:10.1158/1078-0432.CCR-14-3175
55. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi:10.1126/science.aaa4971
56. Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Nat Med. 2019;25:389–402. doi:10.1038/s41591-019-0382-x
57. Kelliher MA, Roderick JE. NOTCH signaling in T-cell-mediated anti-tumor immunity and T-cell-based immunotherapies. Front Immunol. 2018;9:1718. doi:10.3389/fimmu.2018.01718
58. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic ¦Â-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi:10.1038/nature14404
59. Kaler P, Augenlicht L, Klampfer L. Activating mutations in ¦Â-catenin in colon cancer cells alter their interaction with macrophages; the role of snail. PLoS One. 2012;7:e45462. doi:10.1371/journal.pone.0045462
60. Rodilla V, Villanueva A, Obrador-Hevia A, et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci USA. 2009;106:6315–6320. doi:10.1073/pnas.0813221106
61. Moradi M, Najafi R, Amini R, et al. Remarkable apoptotic pathway of Hemiscorpius lepturus scorpion venom on CT26 cell line. Cell Biol Toxicol. 2019;35(4):373–385. doi:10.1007/s10565-018-09455-3
62. Chen J, Du F, Dang Y, et al. Fibroblast growth factor 19-mediated up-regulation of SYR-related high-mobility group box 18 promotes hepatocellular carcinoma metastasis by transactivating fibroblast growth factor receptor 4 and Fms-related tyrosine kinase 4. Hepatology. 2020;71:1712–1731. doi:10.1002/hep.30951
63. Manoochehri H, Jalali A, Tanzadehpanah H, Taherkhani A, Saidijam M. Identification of key gene targets for sensitizing colorectal cancer to chemoradiation: an integrative network analysis on multiple transcriptomics data. J Gastrointest Cancer. 2022;53(3):649–668. doi:10.1007/s12029-021-00690-2
64. Boonstra J, Rijken P, Humbel B, Cremers F, Verkleij A, van Bergen En Henegouwen P. The epidermal growth factor. Cell Biol Int. 1995;19:413–430. doi:10.1006/cbir.1995.1086
65. Bahmani A, Tanzadehpanah H, Hosseinpour Moghadam N, Saidijam M. Introducing a pyrazolopyrimidine as a multi-tyrosine kinase inhibitor, using multi-QSAR and docking methods. Mol Divers. 2021;25(2):949–965. doi:10.1007/s11030-020-10080-8
66. Katoh M. FGFR inhibitors: effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review). Int J Mol Med. 2016;38:3–15. doi:10.3892/ijmm.2016.2620
67. Pulliam SR, Uzhachenko RV, Adunyah SE, Shanker A. Common gamma chain cytokines in combinatorial immune strategies against cancer. Immunol Lett. 2016;169:61–72. doi:10.1016/j.imlet.2015.11.007
68. Wrangle JM, Patterson A, Johnson CB, et al. IL-2 and beyond in cancer immunotherapy. J Interferon Cytokine Res. 2018;38(2):45–68. doi:10.1089/jir.2017.0101
69. Abel AM, Yang C, Thakar MS, Malarkannan S. Natural killer cells: development, maturation, and clinical utilization. Front Immunol. 2018;9:1869. doi:10.3389/fimmu.2018.01869
70. Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. 2016;73(2):377–392. doi:10.1007/s00018-015-2070-4
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.