Back to Journals » Infection and Drug Resistance » Volume 15
Mutation of TonB-Dependent Receptor Encoding Gene MCR_0492 Potentially Associates with Macrolides Resistance in Moraxella catarrhalis Isolates
Authors Zhang Z, Yang Z, Xiang X, Liao P, Niu C
Received 7 March 2022
Accepted for publication 23 April 2022
Published 4 May 2022 Volume 2022:15 Pages 2419—2426
DOI https://doi.org/10.2147/IDR.S364397
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Zhen Zhang,1 Zhulan Yang,2 Xiaohong Xiang,3 Pu Liao,1 Changchun Niu1
1Department of Clinical Laboratory, Chongqing General Hospital, University of Chinese Academy of Sciences, Chongqing, People’s Republic of China; 2Department of Clinical Laboratory, Southwest Hospital, Army Medical University, Chongqing, People’s Republic of China; 3School of Pharmacy, Chongqing Medical and Pharmaceutical College, Chongqing, People’s Republic of China
Correspondence: Changchun Niu; Pu Liao, Email [email protected]; [email protected]
Introduction: Moraxella catarrhalis, which is an opportunistic pathogen and is one of the three major pathogens of community-acquired pneumonia, causes a variety of infections in clinic. In recent years, the isolation rate of Moraxella catarrhalis has gradually increased. In China, due to the clinical empirical use of antibiotics, the resistance rate of Moraxella catarrhalis isolated from children to β-lactam antibiotics has reached 99%. The non-susceptible rate of Moraxella catarrhalis to macrolide antibiotics has also increased significantly.
Methods: Two isolates of Moraxella catarrhalis (R17123922_R and R18013231_R) were isolated from in-patients and were confirmed to be resistant to macrolide antibiotics using the standard disk diffusion and broth microdilution method recommended by CLSI. Whole-genome sequencing (WGS) analysis was performed in these two resistant strains.
Results: A total of 696 SNVs (single nucleotide variations), and 79 indels (Insertion and Deletion) were found in R17123922_R and R18013231_R. These SNVs and indels were distributed evenly in the genome, and no centralized distribution occurred. Moreover, two isolates did not harbor any previously reported mutations in the 23S rRNA and ribosomal proteins.
Conclusion: A novel indel in the MCR_0492 gene encoding TonB-dependent receptor protein was identified, and we speculated that TonB-dependent protein receptor may play an important role in macrolide resistance of Moraxella catarrhalis.
Keywords: Moraxella catarrhalis, macrolides, mechanism of drug resistance, whole-genome sequencing
Introduction
Moraxella catarrhalis is a gram-negative diplococcus1 and is the cause of otitis media,2,3 sinusitis4 and community-acquired pneumonia in children,5 and acute exacerbation of chronic bronchitis in adults.6 Moraxella catarrhalis is also one of the pathogens leading to keratitis.7 Limited published data have reported Moraxella catarrhalis causing bacteremia8 and suppurative arthritis.9
It was reported that Moraxella catarrhalis was very sensitive to macrolide antibiotics,10 and drug-resistant isolates were rare.11 However, in China, the situation has changed, uneven economic development is the reason for the diversity of Moraxella catarrhalis detection ability among the clinical laboratories in different regions, which led to the unreasonable use of antibiotics in clinic. As a result, the resistance of Moraxella catarrhalis to antibiotics increased significantly: tetracycline resistance rate reached 65.7%,12 and the proportion of producing β-lactamases reached 99%.13 It has been reported that 60% Moraxella catarrhalis isolated from children were non-susceptible to macrolide antibiotics.14 Our unpublished data showed that the resistance Moraxella catarrhalis isolated from adults to macrolide antibiotics has also increased in recent years. Therefore, the drug resistance of Moraxella catarrhalis is still worth paying attention to in China.
Two Moraxella catarrhalis isolates with high-level resistance to macrolide antibiotics were isolated from clinical specimens, and accurate identification and antimicrobial susceptibility testing were carried out. Whole-genome sequencing analysis was performed in these resistant isolates to explore the resistance mechanism, which will lay a foundation for the rational application of macrolide antibiotics in clinic.
Materials and Methods
Isolates Source
The isolate R17123922_R was from the bronchoalveolar lavage fluid of a 68-year-old woman in the respiratory department of a hospital; The isolate R18013231_R was from the lower respiratory secretion of a 1-year-old girl in the pediatrics department of a hospital; The isolate S18010071_S was from the lower respiratory secretion of a 55-year-old woman in the respiratory department of a hospital.
Identification of Isolates
The isolates (S18010071_S, R17123922_R, R18013231_R) were removed from the freezer at −80 °C and placed in the biosafety cabinet (BIOBASE BSC-1500IIB2-X), inoculated with 90mm Columbia blood plate (Chongqing Pangtong Medical Instrument Co., LTD) and cultured at 35 °C for 24 hours in 5% CO2 environment. All of the isolates were previously identified by VITEK® 2 Compact (BioMérieux Corporate, France). In order to ensure the accuracy of identification, the isolates were identified again by matrix-assisted laser desorption time-of-flight mass spectrometry (Bioyong Technologies Inc./Clin-ToF-II). And then, some of the remaining colonies were collected and used for extracting bacterial DNA by the rapid bacterial genomic DNA isolation kit (Shanghai Sangon Biotech Co., Ltd.), according to the instructions of the manufacturer (http://www.sangon.com/productDetail?productInfo.code=B518225). Genomic DNA were amplified by 16sRNA primers (16SF27 5’-AGAGTTTGATCCTGGCTCAG-3’;16S-R1522 5’-AAGGAGGTGATCCAGCCGCA-3’), and the amplicons were sent to Shanghai Sangon Biotech Co., Ltd for sequencing.
Antimicrobial Susceptibility Testing
Disk Diffusion Method
The antibiotic susceptibility testing was performed with disk diffusion according to the CLSI M45 (Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd Edition) document. Briefly, the isolates were cultured in 5% CO2 for 24 hours at 35 °C in Columbia blood plate and the turbidity was adjusted to 0.5McFarland. Following the standard method of Clinical & Laboratory Standards Institute (CLSI M100, 30TH Edition), the bacterial suspension was evenly coated on MH Agar. After the suspension was fully absorbed, erythromycin, azithromycin, and clarithromycin (ThermoFisher, Oxoid) were applied respectively. After incubating in 5% CO2 incubator at 35°C for 20–24 hours, the diameter of the zones of complete inhibition was measured, including the diameter of the disk by a vernier caliper. The susceptibility was evaluated according to the CLSI M45 breakpoints for Moraxella catarrhalis. Staphylococcus aureus ATCC25923 was used as the quality control strain.
Broth Microdilution Method
According to the standard method for dissolution of antibiotic in M100 30TH Edition published by the Clinical and Laboratory Standards Institute, different concentrations of erythromycin, azithromycin, and clarithromycin were prepared in sterile 96-well plate using Cation-adjusted Mueller-Hinton broth (CAMHB). Three Moraxella catarrhalis isolates were cultured in 35 °C and 5% CO2 for 24 hours in Columbia blood plate, and were adjusted to corresponding concentrations and inoculated into 96-well plates containing different concentrations of antibiotics (final inoculation concentration 1–4*105CFU/mL), incubation for 20 – 24 hours under ambient air conditions. According to the breakpoints of Moraxella catarrhalis in the third edition of CLSI M45, the results of antimicrobial susceptibility testing were obtained, and Staphylococcus aureus ATCC29213 was used as the quality control strain.
Genome Sequencing
Whole-genome sequencing was performed by Shanghai Sangon Biotech Co., Ltd. The HiSeq PE150 model (Pair-end sequencing) was used to analyze the three isolates. Quality assessment of raw data was performed using BBTools, Phred quality score (Q score) was used to assess the accuracy of all the sequencing. The sequencing data were analyzed and processed by R (“circlize” package)15 and python (“biopython” package), Moraxella catarrhalis BBH18 (GeneBank accession number: NC_014147) was used as the reference sequence.
Results
Three Moraxella catarrhalis Isolates Were Confirmed Using MALDI-TOF MS and 16s rRNA Sequencing
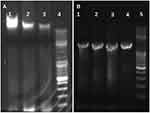
The isolates were identified by Matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF MS), and the results showed that all isolates were Moraxella catarrhalis (Figure 1). The genomes of three isolates (S18010071_S, R17123922_R and R18013231_R) were further extracted (Figure 2A), and 16s rRNA was amplified by universal primers (Figure 2B). The amplified products were sent to Shanghai Sangon Biotech Co., Ltd. for sequencing and NCBI Blast (http://ncbi.nlm.nih.gov/). Results showed that the three isolates were also identified as Moraxella catarrhalis, which was consistent with MALDI-TOF MS.
 |
Figure 1 MALDI-TOF MS results of three isolates; green curve was S18010071_S, the blue curve was R17123922_R, and the red curve was R18013231_R. |
Two Isolates of Moraxella catarrhalis Were Highly Resistant to Macrolides
The standard disk diffusion method recommended by CLSI was followed to analyze the susceptibility of S18010071_S, R17123922_R, and R18013231_R to macrolides. The result showed that the zone diameters of S18010071_S to erythromycin, azithromycin, and clarithromycin were more than 30mm. The zone diameters of R17123922_R and R18013231_R to erythromycin, azithromycin, and clarithromycin were 6mm, suggesting that the two isolates were resistant to macrolides (Table 1). Then, CLSI to recommend another experimental method (Broth microdilution method) was used to analyze the susceptibility of R17123922_R and R18013231_R to macrolide antibiotics, and the results showed that the minimum inhibitory concentrations (MIC) of R17123922R and R18013231R to erythromycin and azithromycin were higher than 512μg/mL, which confirmed that isolates R17123922_R and R18013231_R were highly resistant to macrolide antibiotics (Table 2).
 |
Table 1 The Results of Antimicrobial Susceptibility Testing by Disk Diffusion Method |
 |
Table 2 The Results of Antimicrobial Susceptibility Testing by Broth Microdilution Method |
SNVs (Single Nucleotide Variations) and Indels (Insertion and Deletion) Analysis Related to Macrolide Resistance
Whole-genome sequencing results showed that the average sequencing depth was 1379.4 (Figure 3) and the average GC content was 42.77%. The sequencing data were aligned to the BBH18 (GeneBank accession number: CP002005.1) reference genome, and 8949 SNVs, and 757 indels were found in the isolates. Venn Diagram analysis showed that there were 696 SNVs in R17123922_R and R18013231_R (Figure 4A), including 407 genes (excluding synonymous mutations), and 79 indels (including 40 genes) in R17123922_R and R18013231_R (Figure 4B). Our results also showed that SNVs and indels were evenly distributed in the whole genome (Figure 5).
 |
Figure 3 Sequencing depth distribution of Moraxella catarrhalis. |
 |
Figure 5 Distribution of SNV and indel in genomes of two macrolide-resistant isolates. |
Identifying the Genes and Proteins That May Contribute to Macrolide Resistance in Moraxella catarrhalis
There were 407 genes involved in the 696 SNVs identified above, excluding synonymous mutations, involved 163 genes (Table S1). There were 41 genes involved in the 79 indels (Table S2). Among them, there was no change in the 23SrRNA gene which has been reported to be associated with macrolide resistance, there was no SNV which was related to ribosomal protein-coding genes. Similarly, there was also no indel was associated with reported macrolide resistance. Interestingly, our analysis showed that the indel in MCR_0492, a gene that encodes a TonB-dependent receptor protein (TBDR), occurred in both macrolide-resistant isolates (Table 3). TonB-dependent receptor proteins are located at the cell wall, and TonB-dependent receptors mediate substrate-specific transmembrane transport using the proton power transmitted by the TonB-ExbB-ExbD complex (TonB system) located in the inner membrane.16 It has been reported that antimicrobials can be transported through TonB-dependent receptors.17 Frameshift mutation of TonB-ExbB-ExbD complex in Acinetobacter baumannii can significantly reduce the susceptibility of monocyclic β-lactam antibiotics.18 Therefore, we speculated that the indel in the MCR_0492 gene led to the amino acid change, and then result in a conformational change or loss of the function of TonB-dependent receptor proteins, which affects the entry of macrolide antibiotics into bacterial cells from extracellular, resulting in the resistance of two isolates of Moraxella catarrhalis to macrolide antibiotics.
 |
Table 3 Common Site of Indel Mutation in Two Macrolide-Resistant Moraxella catarrhalis |
Discussion
There is no doubt that Moraxella catarrhalis is an important pathogen in clinic. As no vaccine for Moraxella catarrhalis was used clinically, Moraxella catarrhalis as a pathogen is threatening human health, especially in children. According to the data of China Center for Antibacterial Surveillance (http://y.chinadtc.cn), macrolides accounted for 24.86% of the total use of antibiotics in outpatient prescriptions in Chinese hospitals in 2018, ranking first. Similarly, macrolides ranked first in the use of antibiotics in children, which will inevitably lead to a sharp increase in the resistance rate of macrolide antibiotics. Macrolide antibiotics inhibit the extension of the peptide chain by binding to the large subunit of ribosomal 50s by consisting a 5S rRNA, 23S rRNA, and ribosomal protein.19,20 A Japanese study found that Moraxella catarrhalis with high erythromycin resistance contained three or more 23S rRNA alleles with A2058T mutation.21 Some erythromycin-resistant isolates found mutations in rplD gene and rplV gene encoding ribosomal proteins L4 (V27A/R161C/ Q61R) and L22 (K68T/ Insertion98SRADRIS).22,23 A Chinese study have shown that 23SrRNA gene A2982T, A2796T and A2330T mutations are associated with high resistance to macrolides of Moraxella catarrhalis (MICs:24–256μg/mL), and A2983T mutations are associated with low drug resistance (MICs:0.19–16μg/mL).14 However, there was no resistance mutation above was observed in two highly resistant isolates of Moraxella catarrhalis in our study. Genome sequencing analysis showed that the two isolates of macrolide resistance were associated with the indel mutation of MCR_0492 gene encoding TonB-dependent receptors, which are pore-like channel proteins across the inner membrane. It is well known that the envelope of Gram-negative bacteria is composed of inner and outer membranes; therefore, macrolide antibiotics must be transported through two membranes to function in bacterial cells. We speculated that the loss of TonB-dependent receptor protein function affects the entry of macrolide antibiotics into the cells of Moraxella catarrhalis, which may be a novel mechanism of resistance of Moraxella catarrhalis to macrolide antibiotics.
Conclusion
In this study, two isolates of Moraxella catarrhalis with high resistance to macrolide antibiotics were isolated from clinical specimens, and the whole genome was sequenced. Using Moraxella catarrhalis BBH18 as the reference genome, the whole gene sequencing was compared and analyzed. Two isolates did not harbor any previously reported mutations in the 23S rRNA and ribosomal proteins, we speculate that the two macrolide-resistant isolates are related to the inactivation of MCR_0492 gene function and the exact relationship needs to be confirmed by further experiments, such as gene knockout.
Ethics Approval
This study was approved by the Ethics Committee of Chongqing General Hospital, University of Chinese Academy of Sciences. All data were anonymized to maintain participant’s privacy. In light of retrospective nature of the study, the Ethics Committee did not require written informed consent provided by participants. We certify that the study was performed in accordance with the 1964 Declaration of Helsinki and later amendments.
Acknowledgments
This work was funded by innovation projects of Chongqing General Hospital (Grant No. 2016MSXM28), the general program of Chongqing Science and Technology Commission (Grant No. cstc2018jcyjAX0667).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ariza-Prota MA, Pando-Sandoval A, Garcia-Clemente M, Fole-Vazquez D, Casan P. Community-acquired Moraxella catarrhalis bacteremic pneumonia: two case reports and review of the literature. Case Rep Pulmonol. 2016;2016:5134969. doi:10.1155/2016/5134969
2. Nagai K, Kimura O, Domon H, Maekawa T, Yonezawa D, Terao Y. Antimicrobial susceptibility of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis clinical isolates from children with acute otitis media in Japan from 2014 to 2017. J Infect Chemother. 2019;25(3):229–232. doi:10.1016/j.jiac.2018.08.018
3. Sillanpaa S, Oikarinen S, Sipila M, et al. Moraxella catarrhalis might be more common than expected in acute otitis media in young Finnish children. J Clin Microbiol. 2016;54(9):2373–2379. doi:10.1128/JCM.01146-16
4. Farajzadeh Sheikh A, Ahmadi K, Nikakhlagh S. Detection of Streptococcus pneumoniae and Moraxella catarrhalis in patients with paranasal chronic sinusitis by polymerase chain reaction method. J Chin Med Assoc. 2016;79(8):440–444. doi:10.1016/j.jcma.2016.03.002
5. Sy MG, Robinson JL. Community-acquired Moraxella catarrhalis pneumonia in previously healthy children. Pediatr Pulmonol. 2010;45(7):674–678. doi:10.1002/ppul.21243
6. Ferrer Marcelles A, Martinez Ojeda E, Falco Ferrer V, de la Torre Tejedor E, Gonzalez Fuente T. [Moraxella (Branhamella) catarrhalis: its isolation in the respiratory secretions of adult patients]. An Med Interna. 1997;14(11):554–558. Swedish.
7. Kenny SE, Puig M, Salinas R, Johnson DA, Kheirkhah A. Moraxella Keratitis: a case series. Eye Contact Lens. 2021;47(12):674–676. doi:10.1097/ICL.0000000000000839
8. Funaki T, Inoue E, Miyairi I. Clinical characteristics of the patients with bacteremia due to Moraxella catarrhalis in children: a case-control study. BMC Infect Dis. 2016;16:73. doi:10.1186/s12879-016-1408-3
9. Leonardou A, Giali S, Daoussis D, Siambi V, Gogos H, Liossis SN. Moraxella catarrhalis-induced septic arthritis of a prosthetic knee joint in a patient with rheumatoid arthritis treated with anakinra: comment on the article by Schiff et al. Arthritis Rheum. 2005;52(4):
10. Hare KM, Seib KL, Chang AB, Harris TM, Spargo JC, Smith-Vaughan HC. Antimicrobial susceptibility and impact of macrolide antibiotics on Moraxella catarrhalis in the upper and lower airways of children with chronic endobronchial suppuration. J Med Microbiol. 2019;68(8):1140–1147. doi:10.1099/jmm.0.001033
11. Bandet T, Whitehead S, Blondel-Hill E, Wagner K, Cheeptham N. Susceptibility of clinical Moraxella catarrhalis isolates in British Columbia to six empirically prescribed antibiotic agents. Can J Infect Dis Med Microbiol. 2014;25(3):155–158. doi:10.1155/2014/370964
12. Sun X, Zhang B, Xu G, et al. In vitro activity of the novel tetracyclines, tigecycline, eravacycline, and omadacycline, against Moraxella catarrhalis. Ann Lab Med. 2021;41(3):293–301. doi:10.3343/alm.2021.41.3.293
13. Shi W, Wen D, Chen C, et al. beta-Lactamase production and antibiotic susceptibility pattern of Moraxella catarrhalis isolates collected from two county hospitals in China. BMC Microbiol. 2018;18(1):77. doi:10.1186/s12866-018-1217-5
14. Liu Y, Zhao C, Zhang F, Chen H, Chen M, Wang H. High prevalence and molecular analysis of macrolide-nonsusceptible Moraxella catarrhalis isolated from nasopharynx of healthy children in China. Microb Drug Resist. 2012;18(4):417–426. doi:10.1089/mdr.2011.0175
15. Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811–2812. doi:10.1093/bioinformatics/btu393
16. Fujita M, Mori K, Hara H, Hishiyama S, Kamimura N, Masai E. A TonB-dependent receptor constitutes the outer membrane transport system for a lignin-derived aromatic compound. Commun Biol. 2019;2:432. doi:10.1038/s42003-019-0676-z
17. Luscher A, Moynie L, Auguste PS, et al. TonB-dependent receptor repertoire of Pseudomonas aeruginosa for uptake of siderophore-drug conjugates. Antimicrob Agents Chemother. 2018;62(6). doi:10.1128/AAC.00097-18
18. Moynie L, Luscher A, Rolo D, et al. Structure and function of the PiuA and PirA siderophore-drug receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob Agents Chemother. 2017;61(4). doi:10.1128/AAC.02531-16
19. Champney WS, Tober CL, Burdine R. A comparison of the inhibition of translation and 50S ribosomal subunit formation in Staphylococcus aureus cells by nine different macrolide antibiotics. Curr Microbiol. 1998;37(6):412–417. doi:10.1007/s002849900402
20. Champney WS, Burdine R. Macrolide antibiotics inhibit 50S ribosomal subunit assembly in Bacillus subtilis and Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39(9):2141–2144. doi:10.1128/AAC.39.9.2141
21. Saito R, Kasai A, Ogihara S, Yamada K, Tao K. Rapid assay of A2058T-mutated 23S rRNA allelic profiles associated with high-level macrolide resistance in Moraxella catarrhalis. J Med Microbiol. 2015;64(9):990–992. doi:10.1099/jmm.0.000130
22. Kasai A, Ogihara S, Yamada K, Tanimichi Y, Nishiyama H, Saito R. Prevalence and molecular analysis of macrolide-resistant Moraxella catarrhalis clinical isolates in Japan, following emergence of the highly macrolide-resistant strain NSH1 in 2011. J Med Microbiol. 2015;64(7):708–713. doi:10.1099/jmm.0.000076
23. Kasai A, Ohta A, Maeda Y, Yamada K, Tao K, Saito R. Novel mechanism responsible for high-level macrolide resistance in Moraxella catarrhalis. Infect Drug Resist. 2018;11:2137–2140. doi:10.2147/IDR.S181714
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.