Back to Journals » Infection and Drug Resistance » Volume 11
Multidrug-resistant Pseudomonas aeruginosa from sputum of patients with cystic fibrosis demonstrates a high rate of susceptibility to ceftazidime–avibactam
Authors Atkin SD , Abid S, Foster M, Bose M, Keller A, Hollaway R, Sader HS, Greenberg DE, Finklea JD, Castanheira M, Jain R
Received 18 May 2018
Accepted for publication 24 July 2018
Published 17 September 2018 Volume 2018:11 Pages 1499—1510
DOI https://doi.org/10.2147/IDR.S173804
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eric Nulens
Stan D Atkin,1 Shadaan Abid,1 Michael Foster,1 Moumita Bose,1 Ashley Keller,1 Rita Hollaway,2 Helio S Sader,3 David E Greenberg,1,4 James D Finklea,1 Mariana Castanheira,3 Raksha Jain1,4
1Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX, USA; 2Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA; 3JMI Laboratories, North Liberty, IA, USA; 4Department of Microbiology, University of Texas Southwestern Medical Center, Dallas, TX, USA
Purpose: Ceftazidime–avibactam is a novel antimicrobial combining a third-generation cephalosporin with a non-β-lactam β-lactamase inhibitor that was recently approved to treat Gram-negative hospital- and ventilator-acquired pneumonia. The use of ceftazidime–avibactam to treat Pseudomonas aeruginosa respiratory infections in patients with cystic fibrosis (CF) has not been evaluated. In this study, we assessed the ceftazidime–avibactam susceptibility of multidrug-resistant (MDR) P. aeruginosa sputum isolates from adults with CF.
Methods: Sputum was collected from individuals with CF, aged ≥18 years, known to be colonized with MDR P. aeruginosa, and tested for susceptibility to 11 different antipseudomonal antimicrobial agents. Isolates were included in the analysis if they were resistant to both ceftazidime and at least one agent in ≥3 different antimicrobial categories routinely used to treat P. aeruginosa. Subject demographics and clinical characteristics were collected. Ceftazidime–avibactam-resistant isolates were screened for the presence of β-lactam-resistant mechanisms.
Results: Thirty-two P. aeruginosa isolates were analyzed, of which 23 isolates were sensitive to ceftazidime–avibactam (71.9%). Ten of the isolates were mucoid and 22 isolates were nonmucoid, both demonstrating >70% susceptibility to ceftazidime–avibactam. The most notable difference in the subjects with resistant strains was an older age and lower body mass index (BMI). Ceftazidime–avibactam-resistant strains showed elevated AmpC expression in >60% of the strains and loss of OprD detection in >70% of the strains.
Conclusion: Ceftazidime–avibactam demonstrated a significant in vitro activity against highly resistant P. aeruginosa sputum isolates from individuals with CF. Further evaluation of the cause of resistance and clinical impact of ceftazidime–avibactam in CF patients with MDR P. aeruginosa is warranted.
Keywords: ceftazidime, avibactam, cystic fibrosis, Pseudomonas aeruginosa, multidrug-resistant
Introduction
Cystic fibrosis (CF) is a multisystem autosomal recessive genetic disorder caused by mutations in the CF transmembrane conductance regulator (CFTR) ion channel. The disease manifests predominantly with gastrointestinal and respiratory complications, including chronic bacterial infections. Airway obstruction from chronic infection and inflammation is the leading cause of premature morbidity and mortality in patients with CF. More than 50% of CF individuals aged 18 years and older in the USA are infected with Pseudomonas aeruginosa, of whom approximately one-third is multidrug-resistant (MDR).1 MDR P. aeruginosa may be treated with an array of available antibiotics, but the effectiveness of these antibiotics in practice has been quite variable. Clinicians and researchers have, therefore, been seeking newer antibiotics to treat infections in CF. Ceftazidime–avibactam is a novel antimicrobial that combines a third-generation cephalosporin, ceftazidime, with a non-β-lactam β-lactamase inhibitor.2,3
Ceftazidime–avibactam has shown a significant in vitro activity against a number of Gram-negative bacteria including Proteus, Citrobacter, Escherichia coli, Klebsiella species, extended spectrum beta lactamase (ESBL)-producing organisms, and Burkholderia cepacia.4–7 In fact, B. cepacia, an organism well described to be resistant to antibiotics in patients with CF, showed 90% susceptibility to ceftazidime–avibactam.8 Ceftazidime–avibactam was first approved for use in USA in 2015 for complicated intra-abdominal and urinary tract infections caused by MDR Gram-negative organisms.8 More recently, ceftazidime–avibactam was approved and showed beneficial outcomes for use in patients with severe respiratory infections, including hospital- and ventilator-associated pneumonia, suggesting appropriate lung penetration and utility in respiratory disease.9–11 Ceftazidime–avibactam has not been thoroughly evaluated in patients with CF. However, a pharmacokinetic–pharmacodynamic study of ceftazidime–avibactam use in patients with CF exacerbations did show similar total body clearance and total volume of distribution as published in healthy adults along with notable concentrations in sputum.12
P. aeruginosa is a common pathogen in the lungs of those with CF and is associated with frequent pulmonary exacerbations and high morbidity and mortality.13 The lungs of patients with CF can harbor this organism for decades. With increasing levels of P. aeruginosa drug resistance, treatment of pulmonary exacerbations can be increasingly difficult over time. P. aeruginosa has several mechanisms of resistance that lead to eradication failure and chronic infections, including porin loss and overexpression of efflux pumps as well as production of inactivating enzymes, such as β-lactamases.14,15 Another key mechanism of resistance is the generation of alginate polysaccharide biofilms; these are complex structures, which provide resistance by barrier protection and diffusion limitations.15 Although difficult to eradicate, certain organisms leading to chronic infection in CF mandate antimicrobial therapy during acute pulmonary exacerbations in patients with CF.16
There are limited studies on the use of ceftazidime–avibactam against MDR P. aeruginosa in sputum specimens from CF patients. The purpose of this study is to evaluate the in vitro activity of ceftazidime–avibactam against MDR P. aeruginosa isolates from sputum samples of adult CF patients with highly drug-resistant chronic P. aeruginosa infection and to understand the mechanisms involved in β-lactamase resistance.
Methods
Study design and population
The University of Texas Southwestern adult CF clinic population was queried using the electronic medical record and local Cystic Fibrosis Foundation patient registry database to generate a list of eligible subjects for the study. The study was approved by the Institutional Review Board at the University of Texas Southwestern Medical Center (STU 052011-020). Inclusion criteria were subjects with a confirmed diagnosis of CF by sweat or genetic testing, the age of ≥18 years, and ≥2 sputum cultures positive for P. aeruginosa prior to January 1, 2015.17 Exclusion criteria included subjects who had undergone lung transplantation. With informed and written consent, sputum was collected from eligible subjects. Isolates were included in the analysis if they were resistant to ceftazidime and to at least one agent in ≥3 different antimicrobial categories routinely used to treat P. aeruginosa including fluoroquinolones, aminoglycosides, β-lactams, carbapenems, and polymyxins.
Demographic information acquisition
Age, sex, race, and CFTR genetic information were collected from the University of Texas Southwestern electronic medical record. Body mass index (BMI) was calculated based on height and weight taken at the time of sputum sample collection using standard formulae. Percent predicted forced expiratory volume in 1 second (ppFEV1) was calculated using the NHANES methodology from spirometry measurements taken at the time of sputum sample collection. Inpatient and outpatient oral and intravenous antibiotic exposures for each subject were collected for 2 years prior to sample collection.
Antibiotic susceptibility testing
Isolation of P. aeruginosa from sputum samples was performed in the University of Texas Southwestern microbiology laboratory. Sputum samples were inoculated onto MacConkey agar, sheep blood agar, chocolate agar, B. cepacia selective media, mannitol salt agar, and inhibitory mold agar. P. aeruginosa was identified as oxidase-positive, nonlactose-fermenting colonies on MacConkey agar and reported as mucoid vs nonmucoid. The isolates were identified definitively as P. aeruginosa by MicroScan Neg Urine Combo Panel Type 61 (Beckman Coulter, Inc., Brea, CA, USA). P. aeruginosa isolates were subsequently sent to JMI Laboratories (North Liberty, IA, USA) for susceptibility testing to ceftazidime–avibactam along with other standard antipseudomonal antibiotics including ceftazidime, cefepime, aztreonam, meropenem, piperacillin–tazobactam, amikacin, gentamicin, colistin, levofloxacin, and ciprofloxacin. JMI Laboratories was blinded to any patient data. All isolates were tested for susceptibility using the reference broth microdilution method as described by the Clinical and Laboratory Standards Institute (CLSI).18,19 Ceftazidime was combined with avibactam at a fixed concentration of 4 mg/L. Ceftazidime–avibactam breakpoints approved by the US-Food and Drug Administration (FDA) (≤8/4 mg/L for susceptible and ≥16/4 mg/L for resistant) when testing P. aeruginosa were applied. Susceptibility interpretations for comparator agents were those found in CLSI document M100-S2619 and/or US-FDA package insert.20 Quality control was performed using E. coli ATCC 25922 and 35218, Klebsiella pneumoniae ATCC 700603 and BAA-1705, and P. aeruginosa ATCC 27853. MIC50 and MIC90 calculations were made as previously described.21 Drug-resistant categories were defined as follows: multidrug-resistant (MDR) strains were defined as isolates resistant to at least one agent in ≥3 different antimicrobial categories;22 extensive drug-resistant (XDR) strains were defined as those resistant to at least one agent in all but two or fewer antimicrobial categories; and pan drug-resistant (PDR) strains were defined as those resistant to all classes except colistin.22,23
Biofilm assay
Biofilm breakdown assays were performed in 96-well minimum biofilm eradication concentration (MBEC) plates (Innovotech, Inc., Edmonton, AB, Canada) as described previously.24 Briefly, P. aeruginosa strains were inoculated at 5×105 CFU/mL in Mueller Hinton II broth followed by exposure to ceftazidime–avibactam at concentrations ranging from 1 to 256 µg/mL for 24 hours. Biofilm was quantified using a crystal violet assay.24 The OD of the eluted stain was then measured at 570 nm to quantify the biofilm.
Detection of acquired β-lactamases
Isolates displaying resistance to ceftazidime–avibactam with MIC values (≥16 µg/mL) were screened by PCR for the presence of blaIMP, blaVIM, blaKPC, blaNDM, blaOXA-48, blaGES (blaGES-2, blaGES-4, blaGES-5, blaGES-6, and blaGES-8), blaNMC-A, blaSME, blaIMI, blaFRI-1, blaBKC-1, blaGIM-1/-2, blaSIM-1, blaSPM-1, blaKHM-1, blaAIM-1, blaBIC-1, and blaDIM-1 genes.
Expression analysis of the chromosomally encoded AmpC and efflux pumps
The expression of ampC, mexA (MexAB-OprM), mexC (MexCD-OprJ), mexE (MexEF-OprN), and mexX (MexXY-OprM) was determined by quantitative real-time PCR (qRT-PCR) using DNA-free RNA preparations. Total RNA was extracted from mid-log-phase bacterial cultures (cell density at OD600 0.3–0.5) using RNA Protect Reagent and RNeasy Mini Kit (Qiagen NV, Venlo, the Netherlands) in the QIAcube workstation (Qiagen NV), and residual DNA was eliminated with RNase-free DNase (Promega Corporation, Fitchburg, WI, USA). Quantification and quality of mRNA were done using the RNA 6000 Pico kit on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) according to manufacturer’s instructions. Only preparations with RNA integrity number (RIN) >8 that showed no visual degradation were used for experiments.25 Relative quantification of target genes was performed in triplicate by normalization to an endogenous reference gene (rpsL) on the StepOnePlus instrument (Thermo Fisher Scientific, Waltham, MA, USA) using Power SYBR® Green RNA-to-CT™ kit (Thermo Fisher Scientific) and custom-designed primers showing efficiency >96.0% previously validated.26,27 Transcription levels were considered significantly different if at least 5- or 10-fold differences were noted compared with P. aeruginosa PAO1 for efflux pumps and AmpC expression, respectively.26,28,29
Porin detection
Outer membrane proteins were purified by using the FastPrep®-24 instrument (MP Biomedicals, Solon, OH, USA), according to the manufacturer’s instructions. Normalized concentrations of purified outer membrane proteins were electrophoretically separated and transferred onto PVDF membranes. Western blots were probed with an affinity-purified polyclonal antibody raised in rabbits using the synthetic OprD peptide N″-SDKTGTGNLPVMNDGKPPD-C″ (Thermo Fisher Scientific) and revealed with the WesternBreeze Chromogenic Kit (Thermo Fisher Scientific).30 P. aeruginosa PAO1 and two OprD downregulated laboratory constructs were used as positive and negative controls for comparative analysis.
Statistical analysis
Differences in ceftazidime–avibactam-sensitive vs -resistant strains were compared using a chi-squared test and Fisher’s exact test for categorical variables and unpaired t-test for continuous variables. A P-value of <0.05 was considered statistically significant. Statistical analysis was conducted with GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Subjects
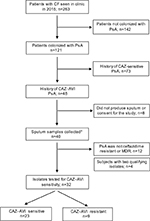
From January 1, 2015, to December 31, 2015, a total of 263 patients with CF who had not undergone lung transplant were seen in the University of Texas Southwestern adult CF clinic, of whom 142 patients were excluded due to the lack of P. aeruginosa in ≥2 previous cultures and a further 73 patients were excluded due to the lack of ≥2 ceftazidime-resistant P. aeruginosa strains (Figure 1). Forty-eight subjects were eligible for the study. Forty of these patients were stable at the time of their visit, produced sputum, and consented to participate in the study. A total of 32 isolates were MDR, and ceftazidime-resistant P. aeruginosa isolates were tested, of which 23 (71.9%) isolates were ceftazidime–avibactam sensitive while nine (28.1%) isolates were ceftazidime–avibactam resistant.
Patients’ characteristics
We compared demographic data of subjects who had P. aeruginosa isolates found to be ceftazidime–avibactam sensitive vs those who had resistant strains (Table 1). Given that four subjects had two P. aeruginosa isolates each, some of which had differing susceptibility profiles to ceftazidime–avibactam, a subject’s demographics was included twice if they had two isolates. Individuals with ceftazidime–avibactam-sensitive P. aeruginosa were significantly younger than those with resistant strains (27.8±8.3 vs 36.9±8.1 years, respectively, P=0.017). There was no significant difference in sex or racial characteristics in ceftazidime–avibactam-sensitive vs -resistant samples. Approximately 55.6% of ceftazidime–avibactam-resistant isolates came from patients with a BMI of <19, which correlates clinically with more severe disease burden in patients with CF.31 There was no significant genotypic difference or mean ppFEV1 difference in the two groups (39.2±15.0 vs 35.6±22.1; P=0.208). Overall, these data demonstrate a higher association between individual’s colonization with β-lactamase inhibitor-resistant P. aeruginosa strains who are older and less well nourished. Antibiotic history demonstrated no notable differences in the average number of antibiotic exposure days between subjects with ceftazidime–avibactam-resistant vs -sensitive strains in all tested antibiotics other than meropenem (S=16.4±5.5 days; R=53.1±16.7 days, P=0.018) (Figure 2). Average antibiotic days for other antibiotics are as follows: tobramycin (S=29.6±6.8 days; R=51.3±19.8 days, P=0.224), ceftazidime (S=14.7±4.8 days; R=15.9±7.8 days, P=0.886), piperacillin–tazobactam (S=21.2±8.2 days; R=25.0±14.4 days, P=0.805), levofloxacin (S=38.0±12.2 days; R=51.0±16.6 days, P=0.528), colistin (S=12.4±6.67 days; R=12.7±4.9 days, P=0.976), ciprofloxacin (S=74.2±18.2 days; R=44.9±13.1 days, P=0.268), aztreonam (S=2.12±1.18 days; R=3.5±2.39 days, P=0.567), and cefepime (S=5.24±3.97 days; R=3.40±2.31 days, P=0.741).
Ceftazidime–avibactam susceptibility testing results
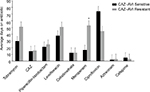
P. aeruginosa isolates from sputum samples were tested for susceptibility to ceftazidime–avibactam and to a panel of 10 commonly used antipseudomonal antibiotics (Table 2). US-FDA clinical breakpoints were applied for ceftazidime–avibactam, ceftazidime, and cefepime while CLSI breakpoints were applied for the remaining antimicrobials. In our cohort of sputum isolates, 71.9% were susceptible to ceftazidime–avibactam, 3.1% were susceptible to aztreonam, 34.3% were susceptible to meropenem, and 3.1% were susceptible to piperacillin/tazobactam. Susceptibility to fluoroquinolones such as levofloxacin and ciprofloxacin was 21.9 and 25.0%, respectively, and susceptibility to aminoglycosides such as amikacin and gentamicin was 25.0 and 12.5%, respectively. Analysis of MIC data across all isolates demonstrated that ceftazidime–avibactam had the highest rate of susceptibility of all antimicrobial classes tested outside of colistin, which demonstrated 93.7% susceptibility to the ceftazidime-resistant P. aeruginosa respiratory isolates. Full antimicrobial susceptibility results are shown in Table S1. The MIC50 for ceftazidime–avibactam was 4 µg/mL, which was also the lowest of all antimicrobials tested other than colistin, which was found to have an MIC50 of 1 µg/mL. We evaluated the percentage of ceftazidime–avibactam-sensitive and -resistant isolates that were MDR, XDR, and PDR and found that isolates resistant to ceftazidime–avibactam had a significantly higher rate of XDR and PDR phenotypes than the ceftazidime–avibactam-sensitive strains (P=0.0054) (Figure 3A). This suggests that ceftazidime–avibactam-resistant isolates show an extensive antimicrobial resistance to multiple drug classes.
Relationship to mucoid phenotype
P. aeruginosa in CF sputum has the unique ability to secrete alginate and acquire a mucoid phenotype, which can demonstrate high drug resistance in some cases.15 Ceftazidime–avibactam-sensitive vs -resistant isolates in our study were stratified by mucoid and nonmucoid phenotypes (Figure 3B). There was no significant difference in the number of mucoid and nonmucoid isolates in these groups (seven of the 23 ceftazidime–avibactam-sensitive isolates were mucoid [30.4%], while three of the nine ceftazidime–avibactam-resistant isolates were mucoid [33.3%], P=0.874), suggesting that mucoid phenotype may not play a role in resistance mechanisms to ceftazidime–avibactam.
Ceftazidime–avibactam can eliminate P. aeruginosa biofilm
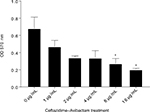
We studied whether ceftazidime–avibactam would have activity in the biofilm setting. We tested four sensitive strains with an MIC of 1 or 2 µg/mL and four resistant strains with an MIC of 16 or 64 µg/mL. At a concentration of 1 µg/mL, there was a 69% decrease in biofilm biomass and this effect was dose dependent with a >50% reduction at 16 µg/mL (Figure 4). The resistant strains did not show antibiofilm activity even at doses of ceftazidime–avibactam as high as 256 µg/mL. These results overall demonstrate potential benefit of ceftazidime–avibactam in eradicating MDR P. aeruginosa even in a biofilm setting.
Mechanisms of resistance to β-lactams among ceftazidime–avibactam-resistant strains
The nine ceftazidime–avibactam-resistant isolates were evaluated further for the presence of acquired carbapenemases, AmpC gene expression, Mex-family efflux pumps, and detection of OprD (Table 3). Acquired β-lactamase, including the most common metallo-β-lactamases, was not detected among ceftazidime–avibactam-resistant isolates. OprD protein loss was noted in 77.78% of strains (Figure S1), elevated AmpC gene expression was noted in 66.67% of strains, elevated MexC expression was noted in 22.22% of strains, moderate expression of MexX and MexA was noted in 22.2% of strains, and no expression of MexE was noted.
Discussion
This study demonstrates for the first time that MDR P. aeruginosa respiratory isolates from adults with CF have a high rate of in vitro susceptibility to ceftazidime–avibactam (71.9%). Ceftazidime–avibactam demonstrated an MIC50 of 4 and an MIC90 of 64 (MIC range 0.5–128 µg/mL) against these highly antibiotic-resistant strains. There was no notable difference in susceptibility patterns to ceftazidime–avibactam when assessing mucoid vs nonmucoid strains. Ceftazidime–avibactam use also inhibited biofilm formation in a dose-dependent manner in strains noted to be ceftazidime–avibactam sensitive. Analysis of MIC data across the isolates collected demonstrated that ceftazidime–avibactam had the highest rate of susceptibility of all antimicrobial classes tested outside of colistin. The ceftazidime–avibactam-resistant P. aeruginosa strains also showed a high expression of AmpC and the protein detection of OprD and interestingly no expression of tested β-lactamases.
Ceftazidime is a β-lactam cephalosporin with a robust history in treating P. aeruginosa and other Gram-negative lung infections, including in individuals with CF. Avibactam is a member of a novel class of non-β lactam-β lactamase inhibitors, the diazabicyclooctanes (DBOs).32 Compared with the inhibitors currently available for clinical use, DBOs are more potent and have a broader spectrum and different mechanisms of action. While co-administration of a β-lactamase inhibitor can help restore antibacterial activity to cephalosporins, previously approved β-lactamase inhibitors such as tazobactam and clavulanic acid do not inhibit certain important β-lactamases.33–35 Avibactam effectively inactivates class A (including K. pneumoniae carbapenemase [KPC]), class C (AmpC), and some class D (OXA) lactamases, with a 50% inhibitory concentration (IC50) value and low turnover numbers, but is not active against New Delhi metallo-b-lactamase 1 (NDM-1).36,37 Thus, avibactam extends the antibacterial activity of ceftazidime against many ceftazidime-resistant organisms that produce the above-cited enzymes, including P. aeruginosa.
The study design for this work has some inherent limitations. We stringently limited our analysis to P. aeruginosa isolates from individuals with CF with a known history of resistance to ceftazidime and MDR P. aeruginosa. As a result, our sample size was small. Our study samples were also collected from a single-center study, so practice patterns on antibiotic use in the CF population may impact susceptibility and gene expression results seen. However, our data are consistent with the evaluation of ceftazidime–avibactam against another fastidious respiratory pathogen in CF, Burkholderia species, in which >90% of the isolates were sensitive to avibactam.8 Finally, we acknowledge that results of susceptibility testing in vitro do not always correlate with treatment efficacy in individuals with CF38 and, thus, use of this data primarily serves as a foundation to support the evaluation of ceftazidime–avibactam for use in treating P. aeruginosa respiratory infections in CF.39,40
Despite limitations, this is the first evaluation of susceptibility testing of MDR P. aeruginosa strains from the sputum of patients with CF with an evaluation of potential resistance mechanisms. In our study, ceftazidime–avibactam-resistant P. aeruginosa isolates predominantly displayed the loss of OprD and elevated AmpC enzyme, suggesting the role of porins and β-lactamases in the resistance mechanisms involved. In support of these findings, others have similarly shown that P. aeruginosa resistance to ceftazidime–avibactam is through modification of the Ω loop region of AmpC, though these isolates were not from patients with CF.41 Others report resistance mechanisms that include AmpC and efflux pump overexpression but not OprD involvement; these data were from P. aeruginosa isolates taken from blood stream infections again in patients without CF.42 Finally, in samples taken from blood, urine, tracheal aspirates, and wounds from a variety of patients, investigators showed ceftazidime–avibactam resistance mechanism that predominantly involved the overexpression of efflux pumps.43 Taken together, these data suggest that ceftazidime–avibactam resistance in P. aeruginosa isolates is likely multifactorial and may vary depending on the source of the specimen along with the underlying disease process of the patient. Our data provides initial mechanistic insight into the characteristics of ceftazidime–avibactam resistance in P. aeruginosa strains isolated from the sputum of patients with CF.
Conclusion
Ceftazidime–avibactam is a novel antimicrobial drug targeting difficult to treat Gram-negative organisms. This study demonstrates excellent susceptibility profiles for ceftazidime–avibactam against highly drug-resistant P. aeruginosa strains collected from the sputum of individuals with CF. Strains resistant to ceftazidime–avibactam demonstrated high rates of OprD loss and elevated AmpC expression. Future studies are needed to determine the efficacy of ceftazidime–avibactam in vivo in individuals with CF being actively treated for pulmonary exacerbations who harbor highly drug-resistant P. aeruginosa.
Acknowledgment
We would like to thank the patients with CF who participated in this study.
Author contributions
SA, SDA, MF, and AK contributed to subject identification, sample collection, and writing and editing the article. RH contributed to sputum processing and editing the article. DEG, MB, and JDF contributed to writing and editing the article. HS and MC contributed to the susceptibility studies and gene expression data along with editing the article. RJ contributed to oversight of the project along with sample collection, subject identification, manuscript writing, and editing. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Cystic Fibrosis Foundation Patient Registry. 2016 Annual Data Report. 2016. | ||
Aktaş Z, Kayacan C, Oncul O. In vitro activity of avibactam (NXL104) in combination with β-lactams against Gram-negative bacteria, including OXA-48 β-lactamase-producing Klebsiella pneumoniae. Int J Antimicrob Agents. 2012;39(1):86–89. | ||
Zhanel GG, Lawson CD, Adam H, et al. Ceftazidime-avibactam: a novel cephalosporin/β-lactamase inhibitor combination. Drugs. 2013;73(2):159–177. | ||
King M, Heil E, Kuriakose S, et al. Multicenter study of outcomes with ceftazidime/avibactam in patients with carbapenem-resistant Enterobacteriaceae (CRE) infections. Antimicrob Agents Chemother. 2017;61(7):1–4. | ||
Castón JJ, Lacort-Peralta I, Martín-Dávila P, et al. Clinical efficacy of ceftazidime/avibactam versus other active agents for the treatment of bacteremia due to carbapenemase-producing Enterobacteriaceae in hematologic patients. Int J Infect Dis. 2017;59:118–123. | ||
Qin X, Tran BG, Kim MJ, et al. A randomised, double-blind, phase 3 study comparing the efficacy and safety of ceftazidime/avibactam plus metronidazole versus meropenem for complicated intra-abdominal infections in hospitalised adults in Asia. Int J Antimicrob Agents. 2017;49(5):579–588. | ||
Chalhoub H, Tunney M, Elborn JS, et al. Avibactam confers susceptibility to a large proportion of ceftazidime-resistant Pseudomonas aeruginosa isolates recovered from cystic fibrosis patients. J Antimicrob Chemother. 2015;70(5):1596–1598. | ||
Papp-Wallace KM, Becka SA, Zeiser ET, et al. Overcoming an Extremely Drug Resistant (XDR) Pathogen: Avibactam Restores Susceptibility to Ceftazidime for Burkholderia cepacia Complex Isolates from Cystic Fibrosis Patients. ACS Infect Dis. 2017;3(7):502–511. | ||
Xipell M, Bodro M, Marco F, Losno RA, Cardozo C, Soriano A. Clinical experience with ceftazidime/avibactam in patients with severe infections, including meningitis and lung abscesses, caused by extensively drug-resistant Pseudomonas aeruginosa. Int J Antimicrob Agents. 2017;49(2):266–268. | ||
Sader HS, Castanheira M, Flamm RK, Mendes RE, Farrell DJ, Jones RN. Ceftazidime/avibactam tested against Gram-negative bacteria from intensive care unit (ICU) and non-ICU patients, including those with ventilator-associated pneumonia. Int J Antimicrob Agents. 2015;46(1):53–59. | ||
Torres A, Zhong N, Pachl J, et al. Ceftazidime-avibactam versus meropenem in nosocomial pneumonia, including ventilator-associated pneumonia (REPROVE): a randomised, double-blind, phase 3 non-inferiority trial. Lancet Infect Dis. 2018;18(3):285-295. | ||
Bensman TJ, Wang J, Jayne J, et al. Pharmacokinetic-Pharmacodynamic Target Attainment Analyses To Determine Optimal Dosing of Ceftazidime-Avibactam for the Treatment of Acute Pulmonary Exacerbations in Patients with Cystic Fibrosis. Antimicrob Agents Chemother. 2017;61(10):1–12. | ||
Sriramulu D. Evolution and impact of bacterial drug resistance in the context of cystic fibrosis disease and nosocomial settings. Microbiol Insights. 2013;6:29–36. | ||
Kung VL, Ozer EA, Hauser AR. The accessory genome of Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2010;74(4):621–641. | ||
Lambert PA. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med. 2002;95(Suppl 41):22–26. | ||
Hyatt AC, Chipps BE, Kumor KM, Mellits ED, Lietman PS, Rosenstein BJ. A double-blind controlled trial of anti-Pseudomonas chemotherapy of acute respiratory exacerbations in patients with cystic fibrosis. J Pediatr. 1981;99(2):307–311. | ||
Lee TW, Brownlee KG, Conway SP, Denton M, Littlewood JM. Evaluation of a new definition for chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. J Cyst Fibros 2003;2(1):29–34. | ||
Wayne P. Clinical and Laboratory Standards Institute M07-A10. Methods for Dilution Antimicrobial Susceptibility. 10th ed. 2015. | ||
Wayne P. Clinical and Laboratory Standards Institute M100-S26. Performance Standards for Antimicrobial Susceptibility Testing: 26th Informational Supplement. 2016. | ||
AVYCAZ (ceftazidime-avibactam) package insert: United States Food and Drug Administration Full Prescribing Information. 2015. | ||
Drummond LJ, Mccoubrey J, Smith DG, Starr JM, Poxton IR. Changes in sensitivity patterns to selected antibiotics in Clostridium difficile in geriatric in-patients over an 18-month period. J Med Microbiol. 2003;52(Pt 3):259–263. | ||
Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. | ||
Sievert DM, Ricks P, Edwards JR, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol. 2013;34(1):1–14. | ||
Howard JJ, Sturge CR, Moustafa DA, Daly SM, et al. Inhibition of Pseudomonas aeruginosa by Peptide-Conjugated Phosphorodiamidate Morpholino Oligomers. Antimicrob Agents Chemother. 2017;61(4):1–12. | ||
Jahn CE, Charkowski AO, Willis DK. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J Microbiol Methods. 2008;75(2):318–324. | ||
Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009-11 in 14 European and Mediterranean countries. J Antimicrob Chemother. 2014;69(7):1804–1814. | ||
Lomovskaya O, Warren MS, Lee A, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45(1):105–116. | ||
Xavier DE, Picão RC, Girardello R, Fehlberg LCC, Gales AC, et al. Efflux pumps expression and its association with porin down-regulation and β-lactamase production among Pseudomonas aeruginosa causing bloodstream infections in Brazil. BMC Microbiol. 2010;10(1):217–217. | ||
Hocquet D, Roussel-Delvallez M, Cavallo JD, Plésiat P, Mexab-Oprm- PP. MexAB-OprM- and MexXY-overproducing mutants are very prevalent among clinical strains of Pseudomonas aeruginosa with reduced susceptibility to ticarcillin. Antimicrob Agents Chemother. 2007;51(4):1582–1583. | ||
Carlone GM, Thomas ML, Rumschlag HS, Sottnek FO. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol. 1986;24(3):330–332. | ||
Courtney JM, Bradley J, Mccaughan J, et al. Predictors of mortality in adults with cystic fibrosis. Pediatr Pulmonol. 2007;42(6):525–532. | ||
Diazabicyclooctanes CK. DBOs): a potent new class of non-beta-lactam beta-lactamase inhibitors. Curr Opin Microbiol. 2011;14(5):550–555. | ||
Blazquez J, Baquero MR, Canton R, Alos I, Baquero F. Characterization of a new TEM-type beta-lactamase resistant to clavulanate, sulbactam, and tazobactam in a clinical isolate of Escherichia coli. Antimicrob Agents Chemother. 1993;37(10):2059–2063. | ||
Higgins PG, Wisplinghoff H, Stefanik D, Seifert H. In vitro activities of the beta-lactamase inhibitors clavulanic acid, sulbactam, and tazobactam alone or in combination with beta-lactams against epidemiologically characterized multidrug-resistant Acinetobacter baumannii strains. Antimicrob Agents Chemother. 2004;48(5):1586–1592. | ||
Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev. 2010;23(1):160–201. | ||
Ehmann DE, Jahic H, Ross PL, et al. Kinetics of avibactam inhibition against Class A, C, and D β-lactamases. J Biol Chem. 2013;288(39):27960–27971. | ||
Abboud MI, Damblon C, Brem J, et al. Interaction of Avibactam with Class B Metallo-β-Lactamases. Antimicrob Agents Chemother. 2016;60(10):5655–5662. | ||
Smith AL, Fiel SB, Mayer-Hamblett N, Ramsey B, Burns JL. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration: lack of association in cystic fibrosis. Chest. 2003;123(5):1495–1502. | ||
Lucasti C, Popescu I, Ramesh MK, Lipka J, Sable C. Comparative study of the efficacy and safety of ceftazidime/avibactam plus metronidazole versus meropenem in the treatment of complicated intra-abdominal infections in hospitalized adults: results of a randomized, double-blind, Phase II trial. J Antimicrob Chemother. 2013;68(5):1183–1192. | ||
Vazquez JA, González Patzán LD, Stricklin D, et al. Efficacy and safety of ceftazidime-avibactam versus imipenem-cilastatin in the treatment of complicated urinary tract infections, including acute pyelonephritis, in hospitalized adults: results of a prospective, investigator-blinded, randomized study. Curr Med Res Opin. 2012;28(12):1921–1931. | ||
Lahiri SD, Walkup GK, Whiteaker JD, et al. Selection and molecular characterization of ceftazidime/avibactam-resistant mutants in Pseudomonas aeruginosa strains containing derepressed AmpC. J Antimicrob Chemother. 2015;70(6):1650–1658. | ||
Torrens G, Cabot G, Ocampo-Sosa AA, et al. Activity of Ceftazidime-Avibactam against Clinical and Isogenic Laboratory Pseudomonas aeruginosa Isolates Expressing Combinations of Most Relevant β-Lactam Resistance Mechanisms. Antimicrob Agents Chemother. 2016;60(10):6407–6410. | ||
Winkler ML, Papp-Wallace KM, Hujer AM, et al. Unexpected challenges in treating multidrug-resistant Gram-negative bacteria: resistance to ceftazidime-avibactam in archived isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2015;59(2):1020–1029. |
Supplementary materials
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.