Back to Journals » OncoTargets and Therapy » Volume 7
Multidrug resistance protein 1 localization in lipid raft domains and prostasomes in prostate cancer cell lines
Authors Gomà A, Mir R, Martínez-Soler F, Tortosa A, Vidal A , Condom E, Pérez–Tomás R, Giménez-Bonafé P
Received 17 June 2014
Accepted for publication 23 August 2014
Published 5 December 2014 Volume 2014:7 Pages 2215—2225
DOI https://doi.org/10.2147/OTT.S69530
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Faris Farassati
Alba Gomà,1,* Roser Mir,1–3,* Fina Martínez-Soler,1,4 Avelina Tortosa,4 August Vidal,5,6 Enric Condom,5,6 Ricardo Pérez–Tomás,6 Pepita Giménez-Bonafé1
1Departament de Ciències Fisiològiques II, Faculty of Medicine, Campus of Health Sciences of Bellvitge, Universitat de Barcelona, IDIBELL, Barcelona, Spain; 2División de Investigación Básica, Instituto Nacional de Cancerología, México DF, Mexico; 3Instituto de Física, Universidad Nacional Autónoma de México (UNAM), México DF, Mexico; 4Department of Basic Nursing, School of Nursing of the Health Campus of Bellvitge, Universitat de Barcelona, 5Department of Pathology, Hospital Universitari de Bellvitge, 6Department of Pathology and Experimental Therapeutics, Universitat de Barcelona, IDIBELL, Barcelona, Spain
*These authors contributed equally to this work
Background: One of the problems in prostate cancer (CaP) treatment is the appearance of the multidrug resistance phenotype, in which ATP-binding cassette transporters such as multidrug resistance protein 1 (MRP1) play a role. Different localizations of the transporter have been reported, some of them related to the chemoresistant phenotype.
Aim: This study aimed to compare the localization of MRP1 in three prostate cell lines (normal, androgen-sensitive, and androgen-independent) in order to understand its possible role in CaP chemoresistance.
Methods: MRP1 and caveolae protein markers were detected using confocal microscopy, performing colocalization techniques. Lipid raft isolation made it possible to detect these proteins by Western blot analysis. Caveolae and prostasomes were identified by electron microscopy.
Results: We show that MRP1 is found in lipid raft fractions of tumor cells and that the number of caveolae increases with malignancy acquisition. MRP1 is found not only in the plasma membrane associated with lipid rafts but also in cytoplasmic accumulations colocalizing with the prostasome markers Caveolin-1 and CD59, suggesting that in CaP cells, MRP1 is localized in prostasomes.
Conclusion: We hypothesize that the presence of MRP1 in prostasomes could serve as a reservoir of MRP1; thus, taking advantage of the release of their content, MRP1 could be translocated to the plasma membrane contributing to the chemoresistant phenotype. The presence of MRP1 in prostasomes could serve as a predictor of malignancy in CaP.
Keywords: prostate cancer, MRP1, Caveolin-1, lipid rafts, caveolae, prostasomes
Introduction
In the developed world, prostate cancer (CaP) is the most common non-skin cancer in men (899,000 new cases each year) and the fifth most common cancer overall. Nearly three-quarters of registered cases occur in developed countries (64,000 cases), and it is estimated to be the second leading male cancer in Spain in 2015.1
Localized CaP is the most commonly diagnosed stage. However, metastatic CaP still remains a major oncological problem in its follow-up. Despite initial response, most tumors relapse within 2 years to an incurable androgen-independent state.2
Chemotherapy is the main treatment to deal with hormonoresistant metastatic CaP. Unfortunately, CaP is also resistant to a broad range of antineoplastic agents, a phenomenon known as multidrug resistance (MDR) phenotype,3,4 which plays an important role in the progressive resistant phenotype observed in CaP.
One of the mechanisms leading to the MDR phenotype is the activation of efflux proteins belonging to the ATP-binding cassette (ABC) transporter superfamily;5 P-glycoprotein (Pgp)6 and the multidrug resistant protein 1 (MRP1) are members of this family with a putative role in CaP chemoresistance.7 Although ABC transporters are mainly localized in the plasma membrane,8 some of them have also been described in other cellular localizations such as lysosomes,9 cytoplasmic vesicles,10,11 and secretory vesicles putatively derived from the Golgi apparatus.5,12,13 A more recent localization of ABC transporters is in specific regions of the plasma membrane rich in cholesterol and sphingolipids, called lipid rafts. Several functions have been attributed to lipid rafts such as signal transduction,14 vesicle trafficking,15,16 and cell adhesion and motility.17 There is strong evidence that the lipid composition of biological membranes is closely related to the presence of ABC transporters.6,18–20 In the case of MRP1, there are some controversies about the presence of this transporter in lipid rafts.21–23
A subset of lipid rafts is found in cell surface invaginations known as caveolae. Caveolae are formed from lipid rafts by polymerization of caveolins, integral membrane proteins that tightly bind cholesterol.24 Elevated expression of caveolin-1 (Cav-1) is accompanied by the acquisition of MDR in various chemotherapy-resistant prostate tumor cells25 and has been found in late stages of CaP progression.26 Cav-1 is secreted by CaP cells, and the results of recent studies showed that secreted Cav-1 can stimulate cell survival and angiogenic activities, defining a role for Cav-1 in the CaP microenvironment.27–30
Additionally, Cav-1 has also been related to prostasomes, secretory granules stored and released by the glandular epithelial prostate cells. Cav-1 has been observed in prostasomes of the prostate cancer cell 3 line (PC3) human CaP cell line together with CD59,31 a specific marker of these vesicles. Prostasomes have also been related to cell transformation, proliferation, and angiogenesis.32–35
In addition, there are studies that suggest that prostasomes may be implicated in CaP, and they are proposed as a prognostic indicator of tumor progression.36
Our studies have been focused on the analysis of a new localization for MRP1. We show that MRP1 is localized both in cytoplasmic accumulations and in plasma membranes of cell lines colocalizing with Cav-1. We also found MRP1 together with Cav-1 in lipid rafts, and this localization is more obvious in the cancer cell lines. The presence of caveolae is more abundant in the cancer cell lines compared with the normal cell line. Finally, we have demonstrated that MRP1 colocalizes with Cav-1 and CD59 in the accumulations found in the cytoplasm, concluding that MRP1 is found in prostasomes. The function of MRP1 in prostasomes needs to be studied in more detail, but one of the hypotheses we propose is that prostasomes could serve as a reservoir/storage of MRP1 and, taking advantage of the fusion of prostasomes to the plasma membrane, MRP1 could be transported to the plasma membrane, participating in the development of CaP.
Methods
Cell culture
The normal human prostate epithelial cell line (PNT2) was provided by Dr Maitland (YCR Cancer Research Unit, University of York). The two tumor cell lines used were the androgen-dependent cell line lymph node carcinoma of the prostate (LNCaP) and the androgen-independent cell line PC3, both provided by The Prostate Center (Vancouver, Canada). All three cell lines were maintained at 37°C in a 5% CO2 atmosphere and cultured in Roswell Park Memorial Institute (RPMI) 1640 supplemented with inactivated 10% fetal bovine serum, 1% l-glutamine, and 1% penicillin–streptomycin (Biological Industries, Israel Beit Haemek LTD). All cell cultures were mycoplasma free. Cancer cell lines LNCaP and PC3 derive from metastasis to lymph nodes and bone, respectively (American Type Culture Collection [ATCC], Manassas, VA, USA). They present intrinsic MDR phenotype, expressing MRP1, and acquired MDR when exposed to increasing doses of chemotherapeutics.
Isolation of lipid rafts
Lipid rafts were isolated from 5×107 cells of each cell line. Pelleted cells were disrupted with 1 mL of raft II buffer (50 mM Tris HCl pH =8.0, 10 mM MgCl2, 0.15 M NaCl, 1% Triton X-100, 5% Glycerol, 50 mM PMSF, 25× cocktail of protease inhibitors [Complete™ EDTA-free; Biological Industries, Israel Beit Haemek LTD] [to a final 1× concentration] and 0.03% of β-mercaptoethanol). The disrupted pellet was centrifuged at 500× g for 5 minutes at 4°C. The supernatant was transferred into a 15 mL falcon tube and rotated for 1 hour at 4°C. Afterward, 1 mL of raft I buffer (50 mM Tris HCl pH =8, 10 mM MgCl2, 0.15 M NaCl) containing 80% sucrose was added to achieve a final concentration of 40% sucrose, followed by 7.5 mL of the same buffer containing 38% sucrose and 2 mL of buffer with 15% sucrose, thereby achieving a discontinuous sucrose gradient.
Finally, the gradient was ultracentrifuged at 100,000× g for 18 hours at 4°C in a SW41 rotor (Ti70 Beckman rotor). Fractions of 1 mL (12 fractions) were collected from the top of the gradient to the bottom (F1–F12 respectively). Immunoblot analysis was performed to confirm the fractions that contained lipid raft microdomains.
The isolation of lipid rafts was performed at least three times from three independent biological replicates to ensure reproducibility.
Western blot analysis
Fractions obtained from the sucrose gradient were analyzed by SDS/PAGE on 7.5% or 12% (w/v) gel, transferred onto a nitrocellulose membrane, blocked in 5% nonfat dry milk and 0.05% Tween-20 in phosphate buffered saline (PBS). The primary antibodies used were Cav-1 (BD Transduction Laboratories), MRP1 (provided by Dr G Sheffer and Dr R Skepper of Free University Medical Center, Amsterdam, the Netherlands), and Flotillin-1 (BD Transduction Laboratories). Secondary antibodies were HRP-conjugated goat antimouse and goat antirat immunoglobulin, and the blots were developed using the ECL Western blotting kit (Amersham Biosciences) and visualized using LAS-3000 Fujifilm.
Confocal microscopy
Cells were grown in 24-well plates on sterilized coverslips. Fixation and permeabilization was with methanol (−20°C for 3 minutes), followed by two washes with PBS and blocked with 0.2% bovine serum albumin. The cells were incubated for 1 hour at room temperature (RT) with Cav-1 (1/500), MRP1 (1/200), or CD59 (1/1,000) (Exbio antibodies). After three washes with PBS, cells were incubated with: Alexa Fluor® 488 goat antirat IgG (molecular probes; Invitrogen) (1/400), Cys™3 conjugated Affini Pure, fragment donkey antimouse IgG (Jackson ImmunoResearch, Laboratories, Inc, West Grove, PA, USA) (1/500), for 45 minutes at RT. The nuclei were stained with Topro III (Calbiochem) for 30 minutes at RT. Controls were done incubating the fixed cells with only the secondary antibodies. Finally, the cells were mounted on Mowiol (Calbiochem-Novabiochem) and visualized in a Leica TCS-SL confocal microscope.
MRP1 quantification
To quantify the presence of MRP1 in both the membrane and the cytoplasm, the images from the confocal microscopy were examined and assessed by two investigators independently. By prior agreement, the intensity of MRP1 antibody was scored depending on the staining of cells in strongly positive (+++), moderate (++), or weak (+) staining. To obtain a numerical value with which to do the statistical analysis, a constant =1 has been added to the number of + and has been multiplied by the percentage of stained cells. For instance: a sample that has stained 30% of the cells with moderate intensity (++) would be: (2 + 1) × 30 = 90. Statistical analyses were performed using the Mann–Whitney U test.
Electron microscopy
Cell monolayers were fixed with 2.5% glutaraldehyde in 0.1 M PBS for 2 hours at 4°C. Cells were gently scraped, collected, and pelleted by centrifugation (5 minutes at 500× g). After three rinses in 0.1 M PBS, pellets were postfixed in 1% OsO4 −0.8% FeCNk for 90 minutes at RT. Finally, the three cell lines were embedded in Spurr resins (Sigma-Aldrich Co, St Louis, MO, USA), and ultrathin sections were analyzed with a Jeol1010 electron microscope. Grids were systematically screened, and the number of both caveolae and prostasomes was determined in random fields of sections from images acquired at the same magnification (25,000×). The images were examined and assessed by three investigators independently. For each cell line, over ten cells were examined with an estimated total linear surface of 500 μm. Finally, for each cell line an average number of caveolae/μm2 and prostasomes/μm2 perimeter was calculated. Statistical analyses were performed using the Mann–Whitney U test.
Results
Localization of MRP1 in prostate cancer cell lines
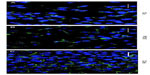
MRP1 was localized through immunocytochemistry techniques in two cellular compartments, with different distributions in normal versus cancer cell lines. In the normal PNT2 cell line, MRP1 was mainly localized in plasma membrane, more in some areas than others (Figures 1A, 2A, and 3A). On the other hand, in the cancer cell lines, MRP1 was observed not only in plasma membrane but also in cytoplasm regions (Figures 1B, C, 2D, G, 3D and J). These results were confirmed when the staining of MRP1 was quantified both in the membrane and in the cytoplasm of the three cell lines: the PNT2 cell line had more staining of MRP1 in the membrane (190.2±46.7) than in the cytoplasm (0.25±0.5). In addition, compared with malignant prostate cell lines, membrane MRP1 staining was higher in PNT2 than in LNCaP and PC3 (118.75±14.6 [P=0.029] and 98.7±12.7 [P=0.020], respectively), whereas in the cytoplasm, MRP1 expression was higher in LNCaP and PC3 cells (33±12.3 [P=0.019] and 22±6.9 [P=0.017], respectively).
Localization of MRP1 in lipid raft domains
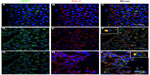
We analyzed the presence of MRP1 in lipid raft regions of the three cell lines. To that purpose, confocal microscopy was applied using Cav-1 as a typical lipid raft marker.
Cav-1 expression was found in plasma membrane of the three cell lines, and also in cytoplasm accumulations in the case of LNCaP and PC3 (Figure 2B, E, and H; Figure 4A, D, G, J, and N). When a merging of MRP1 and Cav-1 images was done, the two proteins colocalized both in the plasma membrane, especially in PNT2 (Figure 2C), and cytoplasm accumulations in LNCaP (Figure 2F) and PC3 (Figure 2I). Similar results were obtained when performing colocalization assays with Flotillin-1, another lipid raft marker (data not shown).
To verify the presence of MRP1 in lipid raft domains together with Cav-1, lipid rafts were isolated followed by MRP1 and Cav-1 detection in the different fractions (F) obtained (Figure 5). In all cell lines, Cav-1 localized in a different grade in the fractions that corresponded to lipid raft (F1–3), where Flotillin-1 (Flot-1), a protein also found in lipid rafts, was present. In PNT2, most of the MRP1 was observed in the soluble fractions (F5–12) and also in small amounts in the last fraction (slightly in F2, and mainly in F3) (Figure 5A). In LNCaP, MRP1 started to be detected in early insoluble fractions corresponding to lipid rafts (mainly in F3), and, similarly to PNT2, most MRP1 was found in more soluble fractions (F3 and on) (Figure 5B). By contrast, MRP1 entirely localized in the lipid raft fractions of PC3 (F1–2) and, to a lesser extent, in the soluble fractions (Figure 5C).
Presence of lipid raft-related structures: caveolae and prostasomes
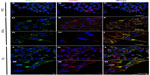
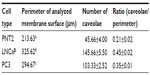
The presence of caveolae was detected in PNT2 (Figure 6A and B), LNCaP (Figure 6D and E), and PC3 (Figure 6G and H). The number was counted, and the ratio between the number and the perimeter of the cells was obtained. CaP cell lines contained twice as many caveolae as the normal prostate cell line (Table 1).
Due to the fact that MRP1 and Cav-1 also colocalized in cytoplasm accumulations (Figure 2), and that lipid rafts are related to prostasomes,31 the presence of prostasomes was also analyzed by electron microscopy. The three cell lines showed prostasomes, structures formed of vesicles approximately 0.5 μm containing an electron-lucent substance and small vesicles and dense bodies in the inside (Figure 6C, F, and I). The shape of the storage vesicles was mostly circular. Two kinds of prostasomes were observed: “dense” prostasomes often filled with small dark particles or inclusions (Figure 6C and F), and “clear” prostasomes filled with less electro dense content. Prostasomes had thin flexible walls, which rendered them irregular in outline in those cellular areas where the vesicles were crowded (Figure 6I). When the number of prostasomes was counted, the androgen-independent and thus more aggressive PC3 cell line displayed a higher number of prostasomes compared with the other two cell lines (the ratio of prostasomes/μm2 was 2.77, 2.75, and 3.76 in the PNT2, LNCaP, and PC3 cell lines, respectively), the differences between the PNT2 and the PC3 cell lines being statistically significant (P<0.01).
Localization of MRP1 in prostasomes
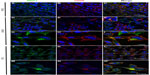
To verify the presence of Cav-1 in prostasomes of tumor cell lines, colocalization studies of Cav-1 and CD59 (a positive marker of prostasomes) were performed in PNT2, LNCaP, and PC3 (Figure 4). As already observed, Cav-1 was localized in the plasma membrane of PNT-2 (Figure 2), where some portions of the membrane presented stronger staining (Figure 4A). CD59 was also present in the plasma membrane, and some cells had some specific dots in the cytoplasm (Figure 4B). But the merge showed that in general, there was no colocalization (Figure 4C). In LNCaP and PC3, Cav-1 was found along the plasma membrane, stronger in specific regions than in others (Figure 4D, G and J, N, respectively) and, in contrast to PNT2, it was also found in cytoplasmic regions (Figure 4A). CD59 was found along the plasma membrane, also stronger in some regions and in cytoplasmic accumulations (Figure 4E, H, L, and O). Cav-1 and CD59 colocalized in both cancer cell lines, in regions of the membrane where the individual staining for each of the proteins was strongest (Figure 4I and P). The cytoplasmic accumulations where Cav-1 and CD59 colocalized were, in most cases, close to the plasma membrane, although in a few cases these accumulations were not close to it (Figure 4F and upper left M).
The next step was to analyze if the cytoplasmic accumulations where Cav-1 and MRP1 colocalized were prostasomes, and for that purpose, the prostasome marker CD59 was used.
In PNT2, MRP1 and CD59 were localized only in the plasma membrane, with stronger staining in specific membrane regions (Figure 3A and B). CD59 was expressed only in some cells as nonhomogeneous cytoplasmic accumulations, and in these localizations, Cav-1 did not colocalize with MRP1 (Figure 3C). Thus, in PNT2, MRP1 was not found in prostasomes. In LNCaP, MRP1 and CD59 were found in the plasma membrane (Figure 3D, E, G, and H) but not in a continuous way; curiously, the regions positive for MRP1 within a cell were negative for CD59, and all the way around. There were also some cytoplasmic accumulations where both proteins colocalized (Figure 3F and I). This colocalization was very specific and observed only in a few cells. Finally, in PC3, MRP1 and CD59 were found along the plasma membrane in continuous staining in all cells (Figure 3J, K, M, and N). In a quarter of the cells observed, both proteins were also found in cytoplasmic accumulations where both colocalized (Figure 3L and O). Thus, in LNCaP and PC3, MRP1 colocalized in CD59 positive cytoplasmic regions, also positive for Cav-1, markers typically used to describe prostasomes.
Discussion
The present study is focused on the cellular localization of MRP1 in CaP cancer cells with particular attention to lipid rafts. Lipid rafts lead to plasma membrane invaginations called caveolae, and cytoplasmic vesicles called prostasomes. MRP1 was found in the three prostate cell lines, both in the plasma membrane and also in the cytoplasm. The fact that MRP1 had stronger staining in specific regions of the plasma membrane and that previous studies have reported the presence of other ABC transporters in lipid raft microdomains, led us to investigate the presence of MRP1 in these membrane regions. Lipid rafts have been implicated in many cellular functions, including protein and lipid transport and signal transduction. MRP1 and glucosylceramide have been described to be overexpressed and enriched in lipid rafts during the acquisition of the MDR phenotype in colon cancer cells.22 We found colocalization of Cav-1 and MRP1 in the plasma membrane, suggesting that MRP1 could also be localized in lipid rafts in CaP cell lines. Lipid raft isolation showed the presence of MRP1 since MRP1 was found in Cav-1 (and Flot-1) enriched fractions. In addition, MRP1 colocalized with Cav-1 not only in the plasma membrane but also in cytoplasmic accumulations. This phenomenon was observed only in CaP cell lines and not in PNT2, normal prostate cell line. Furthermore, the number of caveolae was counted and it was found to be double in LNCaP and PC3 compared with PNT-2, as Quest et al. observed in 2004.26 MRP1 is known to have a major role in CaP.37 Although previous studies described the presence of MRP1 in lipid rafts domains38 an extended work needed to be done in CaP.
Studies by Llorente et al31 showed that Cav-1 colocalized in cytoplasmic accumulations with the prostasome marker CD59, in PC3, and concluded that Cav-1 was localized in prostasomes. Prostasomes are vesicles/granules secreted by prostate epithelial cells to the prostatic fluid that release Cav-1, contributing to cell survival and metastasis.25,28,31,39,40 Our results showed that MRP1 colocalized with Cav-1 mostly in cytoplasmic accumulations where CD59 was also present. Confocal microscopy results of LNCaP and PC3 with Cav-1 and CD59 showed that the two proteins colocalized. Parallel experiments with MRP1 and CD59 also showed a colocalization between the two proteins, suggesting that MRP1 was in prostasomes. Our results showed a higher colocalization of MRP1 and Cav-1 in the androgen-independent PC3 cell line compared with the androgen-sensitive LNCaP cell line. Thus, PC3 malignancy seems to be in accordance with higher colocalization/expression of the two proteins.
When performing the colocalizations of MRP1 and CD59, in LNCaP there was a sort of “mirror effect”: most of the cells that expressed MRP1 did not express CD59, and those who expressed CD59, did not expressed MRP1, in LNCaP, a small percentage of cells presented colocalization of both proteins. This could be explained by the fact that perhaps there might be two different populations of cells within LNCaP: those cells less aggressive that do not express MPR1 in prostasomes, and those cells that have MRP1 in prostasomes colocalizing with MRP1, these being more aggressive cells. We could conclude that both CD59 and Cav-1 could act as prostasome markers when the cells are acquiring malignancy.
The fusion of prostasomes to the plasma membrane could enrich the plasma membrane with MRP1 while releasing Cav-1, contributing to the chemoresistant phenotype.29,41 The presence of MRP1 in prostasomes could serve as a reservoir of the protein. Cancer cells that have more prostasomes would have less MRP1 in the plasma membrane, and all the way around.
Acknowledgments
The authors thank Tom Yohannan for language assistance. We also thank Benjamin Torrejón Escribano from the Scientific and Technological Centers (CCiTUB) of the Health Sciences Campus of Bellvitge; Valenti for his help in the immunohistochemistry protocols, and Dr Núria Cortadelles and Dr Almudena from the Serveis Cientifico-Tècnics of the Clinic Hospital. This study was supported by grants from the Instituto Carlos III (PI 11/01377; ISCIII-RETIC RD12/0036/0029) and from the Catalunya Government (2009SGR-00395). The publication of this paper has been supported by the Vicerrectorat de Política Científica of the University of Barcelona.
Disclosure
The authors report no conflicts of interest in this work.
References
Cris Foundation for Cancer Research. Cancer incidence in Spain. Available from: http://www.criscancer.org/en/why-cris.php?zona=1&seccion=2&apartado=5. Accessed November 24, 2014. | |
Damber JE, Aus G. Prostate cancer. Lancet. 2008;371(9625):1710–1721. | |
van Brussel JP, Mickisch GH. Multidrug resistance in prostate cancer. Onkologie. 2003;26(2):175–181. | |
van Brussel JP, van Steenbrugge GJ, Romijn JC, Schroder FH, Mickisch GH. Chemosensitivity of prostate cancer cell lines and expression of multidrug resistance-related proteins. Eur J Cancer. 1999;35(4):664–671. | |
Zhang JT. Use of arrays to investigate the contribution of ATP-binding cassette transporters to drug resistance in cancer chemotherapy and prediction of chemosensitivity. Cell Res. 2007;17(4):311–323. | |
Modok S, Heyward C, Callaghan R. P-glycoprotein retains function when reconstituted into a sphingolipid- and cholesterol-rich environment. J Lipid Res. 2004;45(10):1910–1918. | |
Sanchez C, Mercado A, Contreras HR, et al. Chemotherapy sensitivity recovery of prostate cancer cells by functional inhibition and knock down of multidrug resistance proteins. Prostate. 2011;71(16):1810–1817. | |
Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. | |
Rajagopal A, Simon SM. Subcellular localization and activity of multidrug resistance proteins. Mol Biol Cell. 2003;14(8):3389–3399. | |
Bakker M, Renes J, Groenhuijzen A, et al. Mechanisms for high methoxymorpholino doxorubicin cytotoxicity in doxorubicin-resistant tumor cell lines. Int J Cancer. 1997;73(3):362–366. | |
Slapak CA, Mizunuma N, Kufe DW. Expression of the multidrug resistance associated protein and P-glycoprotein in doxorubicin-selected human myeloid leukemia cells. Blood. 1994;84(9):3113–3121. | |
Kaufmann AM, Toro-Ramos AJ, Krise JP. Assessment of golgi apparatus versus plasma membrane-localized multi-drug resistance-associated protein 1. Mol Pharm. 2008;5(5):787–794. | |
Van Luyn MJ, Muller M, Renes J, et al. Transport of glutathione conjugates into secretory vesicles is mediated by the multidrug-resistance protein 1. Int J Cancer. 1998;76(1):55–62. | |
Holowka D, Gosse JA, Hammond AT, et al. Lipid segregation and IgE receptor signaling: a decade of progress. Biochim Biophys Acta. 2005;1746(3):252–259. | |
Parton RG. Caveolae–from ultrastructure to molecular mechanisms. Nat Rev Mol Cell Biol. 2003;4(2):162–167. | |
Salaun B, de Saint-Vis B, Pacheco N, et al. CD208/dendritic cell-lysosomal associated membrane protein is a marker of normal and transformed type II pneumocytes. Am J Pathol. 2004;164(3):861–871. | |
Manes S, Viola A. Lipid rafts in lymphocyte activation and migration. Mol Membr Biol. 2006;23(1):59–69. | |
Meyer dos Santos S, Weber CC, Franke C, Muller WE, Eckert GP. Cholesterol: coupling between membrane microenvironment and ABC transporter activity. Biochem Biophys Res Commun. 2007; 354(1):216–221. | |
Storch CH, Ehehalt R, Haefeli WE, Weiss J. Localization of the human breast cancer resistance protein (BCRP/ABCG2) in lipid rafts/caveolae and modulation of its activity by cholesterol in vitro. J Pharmacol Exp Ther. 2007;323(1):257–264. | |
Klappe K, Dijkhuis AJ, Hummel I, et al. Extensive sphingolipid depletion does not affect lipid raft integrity or lipid raft localization and efflux function of the ABC transporter MRP1. Biochem J. 2010;430(3):519–529. | |
Cerf E, Gasper R, Belani JD, et al. Multidrug resistance protein 1 is not associated to detergent-resistant membranes. Biochem Biophys Res Commun. 2007;355(4):1025–1030. | |
Klappe K, Hinrichs JW, Kroesen BJ, Sietsma H, Kok JW. MRP1 and glucosylceramide are coordinately over expressed and enriched in rafts during multidrug resistance acquisition in colon cancer cells. Int J Cancer. 2004;110(4):511–522. | |
Radeva G, Perabo J, Sharom FJ. P-Glycoprotein is localized in intermediate-density membrane microdomains distinct from classical lipid rafts and caveolar domains. FEBS J. 2005;272(19):4924–4937. | |
Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1(1):31–39. | |
Thompson TC, Timme TL, Li L, Goltsov A. Caveolin-1, a metastasis-related gene that promotes cell survival in prostate cancer. Apoptosis. 1999;4(4):233–237. | |
Quest AF, Leyton L, Parraga M. Caveolins, caveolae, and lipid rafts in cellular transport, signaling, and disease. Biochem Cell Biol. 2004;82(1):129–144. | |
Thompson TC, Tahir SA, Li L, et al. The role of caveolin-1 in prostate cancer: clinical implications. Prostate Cancer Prostatic Dis. 2010; 13(1):6–11. | |
Corn PG, Thompson TC. Identification of a novel prostate cancer biomarker, caveolin-1: implications and potential clinical benefit. Cancer Manag Res. 2010;2:111–122. | |
Gumulec J, Sochor J, Hlavna M, et al. Caveolin-1 as a potential high-risk prostate cancer biomarker. Oncol Rep. 2012;27(3):831–841. | |
Freeman MR, Yang W, Di Vizio D. Caveolin-1 and prostate cancer progression. Adv Exp Med Biol. 2012;729:95–110. | |
Llorente A, de Marco MC, Alonso MA. Caveolin-1 and MAL are located on prostasomes secreted by the prostate cancer PC-3 cell line. J Cell Sci. 2004;117(Pt 22):5343–5351. | |
Arienti G, Carlini E, Nicolucci A, Cosmi EV, Santi F, Palmerini CA. The motility of human spermatozoa as influenced by prostasomes at various pH levels. Biol Cell. 1999;91(1):51–54. | |
Kelly RW, Holland P, Skibinski G, et al. Extracellular organelles (prostasomes) are immunosuppressive components of human semen. Clin Exp Immunol. 1991;86(3):550–556. | |
Palmerini CA, Saccardi C, Carlini E, Fabiani R, Arienti G. Fusion of prostasomes to human spermatozoa stimulates the acrosome reaction. Fertil Steril. 2003;80(5):1181–1184. | |
Ronquist G. Prostasomes are mediators of intercellular communication: from basic research to clinical implications. J Intern Med. 2012;271(4):400–413. | |
Ronquist G, Nilsson BO. The Janus-faced nature of prostasomes: their pluripotency favours the normal reproductive process and malignant prostate growth. Prostate Cancer Prostatic Dis. 2004;7(1):21–31. | |
Zalcberg J, Hu XF, Slater A, et al. MRP1 not MDR1 gene expression is the predominant mechanism of acquired multidrug resistance in two prostate carcinoma cell lines. Prostate Cancer Prostatic Dis. 2000;3(2):66–75. | |
Marbeuf-Gueye C, Stierle V, Sudwan P, Salerno M, Garnier-Suillerot A. Perturbation of membrane microdomains in GLC4 multidrug-resistant lung cancer cells–modification of ABCC1 (MRP1) localization and functionality. FEBS J. 2007;274(6):1470–1480. | |
Bartz R, Zhou J, Hsieh JT, Ying Y, Li W, Liu P. Caveolin-1 secreting LNCaP cells induce tumor growth of caveolin-1 negative LNCaP cells in vivo. Int J Cancer. 2008;122(3):520–525. | |
Tahir SA, Yang G, Ebara S, et al. Secreted caveolin-1 stimulates cell survival/clonal growth and contributes to metastasis in androgen-insensitive prostate cancer. Cancer Res. 2001;61(10):3882–3885. | |
Lavie Y, Fiucci G, Liscovitch M. Upregulation of caveolin in multidrug resistant cancer cells: functional implications. Adv Drug Deliv Rev. 2001;49(3):317–323. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.