Back to Journals » Infection and Drug Resistance » Volume 7
Mortality and molecular epidemiology associated with extended-spectrum β-lactamase production in Escherichia coli from bloodstream infection
Authors Leistner R, Sakellariou C, Gürntke S, Kola A , Steinmetz I, Kohler C, Pfeifer Y, Eller C, Gastmeier P, Schwab F
Received 2 November 2013
Accepted for publication 10 December 2013
Published 13 March 2014 Volume 2014:7 Pages 57—62
DOI https://doi.org/10.2147/IDR.S56984
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Rasmus Leistner,1 Christian Sakellariou,1 Stephan Gürntke,1 Axel Kola,1 Ivo Steinmetz,2 Christian Kohler,2 Yvonne Pfeifer,3 Christoph Eller,3 Petra Gastmeier,1 Frank Schwab1
1Institute of Hygiene and Environmental Medicine, National Reference Center for the Surveillance of Nosocomial Infections, Charité Universitätsmedizin Berlin, Berlin, Germany; 2Friedrich Löffler Institute of Medical Microbiology, Universitätsmedizin Greifswald, Greifswald, Germany; 3Robert Koch Institute, FG13 Nosocomial Pathogens and Antibiotic Resistance, Wernigerode, Germany
Background: The rate of infections due to extended-spectrum β-lactamase (ESBL)-producing Escherichia coli is growing worldwide. These infections are suspected to be related to increased mortality. We aimed to estimate the difference in mortality due to bloodstream infections (BSIs) with ESBL-positive and ESBL-negative E. coli isolates and to determine the molecular epidemiology of our ESBL-positive isolates.
Materials and methods: We performed a cohort study on consecutive patients with E. coli BSI between 2008 and 2010 at the Charité University Hospital. Collected data were ESBL production, basic demographic parameters, and underlying diseases by the Charlson comorbidity index (CCI). The presence of ESBL genes was analyzed by polymerase chain reaction (PCR) and sequencing. Phylogenetic groups of ESBL-positive E. coli were determined by PCR. Risk factors for mortality were analyzed by multivariable regression analysis.
Results: We identified 115 patients with BSI due to E. coli with ESBL phenotype and 983 due to ESBL-negative E. coli. Fifty-eight percent (n=67) of the ESBL-positive BSIs were hospital-acquired. Among the 99 isolates that were available for PCR screening and sequencing, we found mainly 87 CTX-M producers, with CTX-M-15 (n=55) and CTX-M-1 (n=21) as the most common types. Parameters significantly associated with mortality were age, CCI, and length of stay before and after onset of BSI.
Conclusion: The most common ESBL genotypes in clinical isolates from E. coli BSIs were CTX-M-15 (58%) and CTX-M-1 (22%). ESBL production in clinical E. coli BSI isolates was not related to increased mortality. However, the common occurrence of hospital-acquired BSI due to ESBL-positive E. coli indicates future challenges for hospitals.
Keywords: BSI, mortality, ESBL-genotype, sepsis
Background
Infections with extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae are increasing worldwide.1 In particular, bloodstream infections (BSIs) due to ESBL-producing Enterobacteriaceae have been reported to be related to increased mortality in the past 10 years.2–4 However, recent studies showed no significant difference in mortality comparing ESBL-positive and ESBL-negative cases of BSI due to Enterobacteriaceae.5,6 There is still a lack of current studies with large numbers of patients and adequate control cohorts consisting of infections with susceptible Enterobacteriaceae. Therefore, we present the results of a large study with more than 100 ESBL-positive patients and respective control patients with ESBL-negative Escherichia coli BSI.
Materials and methods
Setting, study design, and definitions
The study was carried out at the Charité University Hospital in Berlin, a 3,213-bed tertiary care center. The study was performed in conformity with the ethical guidelines of the Declaration of Helsinki. It is based on secondary data, and ethical approval was not required.
We conducted a cohort study of all consecutive patients with BSI caused by E. coli between January 1, 2008 and December 31, 2010. Patients were retrospectively identified in the Charité microbiology database as patients with blood cultures positive for E. coli. For all patients enrolled in this study, the following demographic characteristics were collected from their electronic files: age, sex, in-hospital death, day of BSI onset, and underlying comorbidities by the Charlson comorbidity index (CCI) on the basis of the patients’ International Classification of Diseases (ICD)-10-coded diagnoses.7 If a patient had more than one episode of E. coli BSI within the analyzed period, the first episode was analyzed. Each patient was included in the analysis only once. Onset of BSI was defined as the date of the first blood culture positive for E. coli. BSI was considered hospital-acquired when the onset occurred at least 48 hours after admission.
Microbiological methods, resistance-gene screening, and bacterial typing
The VITEK 2 automated system was used for the identification of E. coli species and antimicrobial susceptibility testing. The results were interpreted according to the Clinical and Laboratory Standards Institute standards.8 Confirmation of ESBL production was performed by a minimum inhibitory concentration dilution test on a multiwell microtiter plate. Three third-generation cephalosporins (cefoxitin, ceftazidime, cefpodoxime) were tested alone and in combination with ESBL inhibitor clavulanic acid (standard operating procedure according to the German Accreditation Council, registration DGA-ML-6243.03). All E. coli isolates with ESBL phenotype were screened for the presence of different ESBL genes (blaTEM-type, blaSHV-type, blaCTX-M-1/2/9 group) by polymerase chain reaction (PCR) and subsequent sequencing.9 If none of these ESBL genes could be identified, additional PCR tests for the presence of plasmid-mediated AmpC β-lactamases10 and further ESBL genes (blaCTX-M-8-type, blaCTX-M-26-type, blaVEB-type, blaPER-type, blaGES-type, blaOXA-1/2/10 group) were performed. Furthermore, basic bacterial typing of all ESBL-positive E. coli isolates was performed by a PCR-based method for determination of the four major E. coli phylogenetic groups.11
Statistical methods
Parameters in the descriptive analysis of patients with ESBL-positive and ESBL-negative E. coli BSIs were tested using the Wilcoxon rank-sum test for continuous variables and Fisher’s exact test for categorical variables. A multivariable analysis was performed to estimate the effects of multiple factors associated with mortality using a stepwise forward regression. Included variables were ESBL production, age, sex, CCI, hospital acquisition, and length of stay before and after BSI onset. Variables with P-values<0.05 were included, and variables with P≥0.05 were excluded. Odds ratios and their 95% confidence intervals were calculated. All tests of significance were two-tailed, with a P-value<0.05 considered to be significant. Data were analyzed using PASW Statistics 18 (IBM, Armonk, NY, USA).
Results
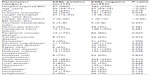
We identified 1,098 consecutive patients with E. coli BSI. One hundred and fifteen patients (10.5%) with ESBL-positive due to E. coli, and 983 patients with BSI due to ESBL-negative E. coli (89.5%). The baseline characteristics of patients with ESBL-positive and ESBL-negative E. coli BSI are shown in Table 1. Overall and regardless of ESBL production, 449 (40%) E. coli BSIs were hospital-acquired and associated with an in-house mortality of 24% (n=107), compared to 16% (n=104, P=0.001) among community-acquired cases. Fifty-eight percent (n=67) of the ESBL-positive BSIs and 38% (n=382) of ESBL-negative E. coli BSIs (P<0.001) were hospital-acquired.
The blood culture specimens of 99 (86%) BSI patients were available for molecular analysis. ESBL genes were identified in 95 E. coli isolates. One isolate was positive for AmpC β-lactamase CMY-2, and three isolates were only positive for β-lactamase type TEM-1 (n=2) and TEM-181 (n=1), respectively. All four of these patients were removed from the analysis, leaving 95 analyzed patients. Among the remaining 95 ESBL-producing isolates, CTX-M enzymes (n=87, 92%) were predominant, with CTX-M-15 (n=55, 58%), CTX-M-1 (n=21, 22%), and CTX-M-14 (n=4, 4%) the most common ESBL-types (Figure 1A). Furthermore, 43 (45%) of all ESBL-producing E. coli additionally harbored TEM-type β-lactamases. Among the 54 hospital-acquired ESBL-positive E. coli isolates that were available for molecular analysis, the most common genotypes were CTX-M-15 (n=33, 61%), CTX-M-1 (n=10, 19%) and CTX-M-14 (n=3, 6%).
Bacterial typing (Figure 1B) revealed that the 95 ESBL-producing E. coli isolates belonged to phylogenetic groups B2 (n=31, 33%), D (n=28, 29%), A (n=26, 27%), and B1 (n=10, 10%). The distribution of the phylogenetic groups among the 54 hospital-acquired E. coli isolates was nearly identical. However, considering only the isolates with CTX-M-15 (n=55) and CTX-M-1 (n=21), phylogenetic group B2 was mainly associated with CTX-M-15 (n=24, 44%) in contrast to isolates of phylogenetic group A producing mainly CTX-M-1 (n=10, 48%) (Figure 1C and D).
The results of the regression analysis are shown in Table 2. Age, Charlson comorbidity index, and length of stay before and after onset of BSI were significantly associated with hospital mortality. ESBL production was not a significant risk factor in the multivariable analysis.
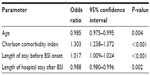  | Table 2 Results of the multivariable regression analysis of risk factors for mortality in cases of Escherichia coli bloodstream infection (BSI) |
Discussion
In the present single-center cohort study of patients with E. coli BSI, we analyzed 115 patients with ESBL-positive and 983 patients with ESBL-negative E. coli. BSI due to ESBL-positive E. coli was significantly more often hospital-acquired than ESBL-negative E. coli BSI. However, in this study, ESBL production was not associated with increased mortality risk. In our study, ESBL E. coli BSI was related to an in-house mortality of 26%. This result is comparable to several recent studies that reported ESBL BSI mortality (30-day or in-hospital) between 20% and 25%.6,12–14 Nevertheless, few published studies have assessed the mortality of ESBL or third-generation cephalosporin-resistant E. coli BSI in comparison to patients with BSI due to susceptible E. coli.3–6 Our data are comparable with the results of a study by Nasa et al that analyzed cases of Enterobacteriaceae BSI on intensive care units.5 Another study used a multistate approach to analyze the burden of ESBL-positive BSI.6 Even though they found increased costs and length of stay, the mortality of their ESBL-positive cases was not significantly increased.
In contrast, de Kraker et al analyzed data from 13 European tertiary care centers.4 They found high mortality rates of BSI cases due to third-generation cephalosporin-resistant E. coli that were significantly increased compared to their susceptible controls (36% versus 17%). However, many of their data derived from countries with high prevalence of carbapenem nonsusceptible Enterobacteriaceae. Since the study included no information on additional carbapenem resistance of the isolates, the possibility that carbapenemase production influenced the calculated mortality rate cannot be excluded.15 A further study conducted in 2010 found a significantly increased mortality of patients with ESBL E. coli BSI compared to patients with non-ESBL E. coli BSI (30% versus 6%).3 Here, the majority of ESBL-positive patients received inadequate initial antimicrobial chemotherapy. Moreover, in a meta-analysis on BSI cases due to ESBL-producing Enterobacteriaceae between 1996 and 2003, significantly increased mortality (pooled crude mortality 34% versus 20%) and a significant delay in administration of appropriate antimicrobial therapy was associated with ESBL production.2 A meta-analysis by Rottier et al furthermore found that increased mortality in cases of BSI due to ESBL-positive Enterobacteriaceae is strongly influenced by the administration of empirically adequate antimicrobial therapy.16 In the present study, data on antimicrobial therapy were not available. Hence we cannot evaluate the definite influence of appropriate antimicrobial therapy in the present cohort. However, internal antibiotic policy was communicated to us as the following: for patients with severe sepsis or septic shock, empirical treatment was initiated with carbapenems and less severe infections with third-generation cephalosporins or piperacillin/tazobactam. Furthermore, the mortality rate in our cohort is comparable with many other recent studies, and documents the current outcome associated with ESBL E. coli BSI.6,12–14
The most common ESBL genotypes identified in the present study (CTX-M-15, 58%; CTX-M-1, 22%) were also found in other studies from Europe. However, differences in distribution of these three CTX-M-types and other ESBLs in European countries have been reported, eg, the dominant ESBL type in Spain is CTX-M-14.12,17 A possible explanation could be delivered by the ongoing discussion on ESBL distribution through the food chain. While recent studies from Germany have shown that diet could play an important role in the distribution of CTX-M-1,18,19 a study from Spain showed in contrast that CTX-M-14 is the most common ESBL enzyme in E. coli from Spanish retail meat.20
The finding that four E. coli isolates with ESBL phenotype in our study harbored no ESBL type is most likely due to overexpression of TEM β-lactamases in three isolates and one false-positive ESBL confirmation test for one CMY-producing isolate. Typing of our ESBL E. coli isolates revealed a high proportion of CTX-M-15-positive isolates belonging to phylogenetic group B2. This may partly be due to the spread of strains belonging to the internationally disseminated clonal lineage E. coli O25b:H4-ST131.21 In contrast, the majority of CTX-M-1-positive isolates (48%) belonged to phylogenetic group A, which is generally associated with less virulence than group B2 or D isolates. CTX-M-1-producing E. coli are more often described in animals, food, or community-acquired isolates than CTX-M-15, indicating an association of distinct CTX-M-types with different settings due to various ways of transmission.3,18,21–25
In conclusion, although there was no difference in mortality of BSIs with ESBL-positive or ESBL-negative E. coli in our study, the significantly higher number of hospital-acquired BSIs due to E. coli with ESBLs indicates the importance of hygiene precautions. Measures like high compliance in hand hygiene, preemptive isolation of high-risk patients, and prudent use of antibiotic agents are needed to prevent further dissemination and selection of these drug-resistant bacteria.
Acknowledgments
We would like to thank Georg Pilarsky, Sebastian Kruse, and Oleg Jarychkivski for database programming, Ryan Plocher for language consultation, Gabriele Rose and Petra Dem for the preparation of the isolates at the Charité laboratory, and Janine Zweigner for counseling while writing this manuscript. We also thank Sybille Müller-Bertling and Christine Günther for excellent technical assistance. This work was partly funded by grants from the Federal Ministry of Education and Health, Germany (01KI1013H to RL and PG and 01KI1013E to CE).
Disclosure
The authors report no conflicts of interest in this work.
References
Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008;13(47):19044. | |
Schwaber MJ, Carmeli Y. Mortality and delay in effective therapy associated with extended-spectrum beta-lactamase production in Enterobacteriaceae bacteraemia: a systematic review and meta-analysis. J Antimicrob Chemother. 2007;60(5):913–920. | |
Tumbarello M, Spanu T, Di Bidino R, et al. Costs of bloodstream infections caused by Escherichia coli and influence of extended-spectrum-beta-lactamase production and inadequate initial antibiotic therapy. Antimicrob Agents Chemother. 2010;54(10):4085–4091. | |
de Kraker ME, Wolkewitz M, Davey PG, et al. Burden of antimicrobial resistance in European hospitals: excess mortality and length of hospital stay associated with bloodstream infections due to Escherichia coli resistant to third-generation cephalosporins. J Antimicrob Chemother. 2011;66(2):398–407. | |
Nasa P, Juneja D, Singh O, Dang R, Singh A. An observational study on bloodstream extended-spectrum beta-lactamase infection in critical care unit: incidence, risk factors and its impact on outcome. Eur J Intern Med. 2012;23(2):192–195. | |
Stewardson A, Fankhauser C, De Angelis G, et al. Burden of bloodstream infection caused by extended-spectrum β-lactamase-producing enterobacteriaceae determined using multistate modeling at a Swiss University Hospital and a nationwide predictive model. Infect Control Hosp Epidemiol. 2013;34(2):133–143. | |
Thygesen SK, Christiansen CF, Christensen S, Lash TL, Sorensen HT. The predictive value of ICD-10 diagnostic coding used to assess Charlson comorbidity index conditions in the population-based Danish National Registry of Patients. BMC Med Res Methodol. 2011;11:83. | |
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Seventeenth Informational Supplement. Wayne (PA): CLSI; 2007. | |
Gröbner S, Linke D, Schütz W, et al. Emergence of carbapenem-non-susceptible extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tübingen, Germany. J Med Microbiol. 2009;58(Pt 7):912–922. | |
Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. | |
Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66(10):4555–4558. | |
Rodríguez-Baño J, Mingorance J, Fernández-Romero N, Serrano L, López-Cerero L, Pascual A. Outcome of bacteraemia due to extended-spectrum β-lactamase-producing Escherichia coli: impact of microbiological determinants. J Infect. 2013;67(1):27–34. | |
Frakking FN, Rottier WC, Dorigo-Zetsma JW, et al. Appropriateness of empirical treatment and outcome in bacteremia caused by extended-spectrum-β-lactamase-producing bacteria. Antimicrob Agents Chemother. 2013;57(7):3092–3099. | |
Peralta G, Lamelo M, Alvarez-García P, et al. Impact of empirical treatment in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. bacteremia. A multicentric cohort study. BMC Infect Dis. 2012;12:245. | |
Cantón R, Akóva M, Carmeli Y, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18(5):413–431. | |
Rottier WC, Ammerlaan HS, Bonten MJ. Effects of confounders and intermediates on the association of bacteraemia caused by extended-spectrum β-lactamase-producing Enterobacteriaceae and patient outcome: a meta-analysis. J Antimicrob Chemother. 2012;67(6):1311–1320. | |
Rodríguez-Baño J, Picón E, Gijón P, et al. Risk factors and prognosis of nosocomial bloodstream infections caused by extended-spectrum-beta-lactamase-producing Escherichia coli. J Clin Microbiol. 2010;48(5):1726–1731. | |
Kola A, Kohler C, Pfeifer Y, et al. High prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae in organic and conventional retail chicken meat, Germany. J Antimicrob Chemother. 2012;67(11):2631–2634. | |
Leistner R, Meyer E, Gastmeier P, Pfeifer Y, Eller C, Dem P, et al. Risk factors associated with the community-acquired colonization of extended-spectrum beta-lactamase (ESBL) positive Escherichia coli. An exploratory case-control study. PLoS One. 2013;8(9):e74323. | |
Ojer-Usoz E, González D, Vitas AI, et al. Prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Sci. 2013;93(2):316–321. | |
Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother. 2008;61(2):273–281. | |
Voets GM, Platteel TN, Fluit AC, et al. Population distribution of beta-lactamase conferring resistance to third-generation cephalosporins in human clinical Enterobacteriaceae in The Netherlands. PLoS One. 2012;7(12):e52102. | |
Reuland EA, Overdevest IT, Al Naiemi N, et al. High prevalence of ESBL-producing Enterobacteriaceae carriage in Dutch community patients with gastrointestinal complaints. Clin Microbiol Infect. 2013;19(6):542–549. | |
Izdebski R, Baraniak A, Fiett J, et al. Clonal structure, extended-spectrum β-lactamases, and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob Agents Chemother. 2013;57(1):309–316. | |
Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect. 2012;18(7):646–655. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.