Back to Journals » Infection and Drug Resistance » Volume 15
Molecular Surveillance of Antimicrobial Resistance of Mycobacterium leprae from Leprosy Patients in Zhejiang Province, China
Authors Shi Y , Kong W, Jiang H, Zhang W, Wang C, Wu L, Shen Y, Yao Q, Wang H
Received 30 March 2022
Accepted for publication 6 July 2022
Published 27 July 2022 Volume 2022:15 Pages 4029—4036
DOI https://doi.org/10.2147/IDR.S368682
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Ying Shi1– 3*, Wenming Kong1 *, Haiqin Jiang,2,3 Wenyue Zhang,2,3 Chen Wang,2– 4 Limei Wu,1 Yunliang Shen,1 Qiang Yao,1 Hongsheng Wang1– 3,5
1Department of Leprosy Control, Zhejiang Provincial Institute of Dermatology, Huzhou, People’s Republic of China; 2Hospital of Skin Diseases, Institute of Dermatology, Chinese Academy of Medical Sciences & Peking Union Medical College, Jiangsu Key Laboratory of Molecular Biology for Skin Diseases and STIs, Nanjing, People’s Republic of China; 3National Centre for STD and Leprosy Control, China CDC, Nanjing, People’s Republic of China; 4Department of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, People’s Republic of China; 5Centre for Global health, School of Public Health, Nanjing Medical University, Nanjing, People’s Republic of China
* These authors contributed equally to this work
Correspondence: Qiang Yao, Department of Leprosy Control, Zhejiang Provincial Institute of Dermatology, St 61, Zhejiang, People’s Republic of China, Email [email protected] Hongsheng Wang, Institute of Dermatology, Chinese Academy of Medical Sciences & Peking Union Medical College, No. 12 Jiangwangmiao Street, Nanjing, People’s Republic of China, Tel: +86 025 8547 8953, Email [email protected]
Purpose: Reports on antimicrobial resistance (AMR) of Mycobacterium leprae (M. leprae) in Zhejiang Province are limited. Thus, this study aimed to investigate the drug resistance of new leprosy cases within several years and analyse the emergence of AMR mutations from Zhejiang Province.
Methods: This study enrolled 34 leprosy cases in Zhejiang Province, China, from 2018 to 2021. Gene mutation of WHO-recommended DRDRs (folP1, rpoB and gyrA) and genes of compensatory AMR-associated DRDRs, including nth, rpoA, rpoC, gyrB and 23S rRNA, were detected by amplification. Clinical data analysis was performed to investigate the epidemiological association of leprosy.
Results: Of the 34 samples, 2 (5.9%) strains showed drug resistance, which were mutated to dapsone and ofloxacin, separately. Two single mutations in gyrB were detected in different strains (5.9%), whereas one of the rpoC mutation was also detected in one strain each (2.9%), which were proved to be polymorphs. No correlation of drug resistance proportion was identified in male vs female, nerve vs no nerve involvement, deformity vs no deformity and reaction vs non-reaction cases.
Conclusion: Results showed well control of leprosy patients in Zhejiang Province. Gene mutations of WHO-recommended DRDRs folP1 and gyrA confirmed the resistance to dapsone and ofloxacin. Compensatory AMR-associated mutations confirmed to be polymorphs still require further study to determine their phenotypic outcomes in M. leprae. The results demonstrated that drug-resistant strains are not epidemic in this area. Given the few cases of leprosy, analysing the AMR of M. leprae in Zhejiang Province more comprehensively is difficult. However, regular MDT treatment and population management in the early stage may contribute to the low prevalence of leprosy.
Keywords: M. leprae, antimicrobial resistance, gene mutation
Introduction
Leprosy, which is caused by Mycobacterium leprae (M. leprae), remains a public health problem associated with disability, deformity, stigma and discrimination in untreated individuals. Based on the official World Health Organization (WHO) records, a total of 127,396 new leprosy patients were reported globally in 2020,1 which indicates an endemicity of leprosy, particularly in underdeveloped countries. The current strategy for leprosy control is multidrug therapy (MDT), which is recommended by WHO in 1982.2 The combination therapy with dapsone, rifampicin and clofazimine has decreased the prevalence of leprosy globally, which is recommended for the treatment of multibacillary (MB) leprosy, whereas therapy with dapsone and rifampicin should be used for paucibacillary (PB) leprosy. In addition, ofloxacin is a key drug in the treatment of single-lesion leprosy cases and drug-resistant cases.3 Studies in lepromatous leprosy patients have demonstrated bactericidal activity killing 99.99% of viable bacteria but not acceptable as a single drug due to side effects in human.4,5 However, dead M. leprae are cleared from tissue very slowly, and the bacterial carcasses may therefore remain visible in biopsies for years after they have been killed. The antigens of these dead organisms may continue to elicit immunological and inflammatory responses.6,7
Since 1964, the resistance of M. leprae strains to dapsone was reported,8 and drug resistance cases increased gradually. Apart from single-drug resistance, an increasing number of resistances to multiple drugs has been reported in recent years. Based on previous studies, drug resistance detection of drug resistance-determining regions (DRDRs) containing folP1, rpoB and gyrA associated with dapsone, rifampicin and ofloxacin resistance,9–11 respectively, has been well established and incorporated in the WHO guidelines for AMR surveillance. For example, position 53, 55 of folP1 gene are known to confer drug resistance to dapsone, while position 89, 91 of gyrA gene conferring to ofloxacin and position 441, 451, 456, 458 of rpoB gene conferring to rifampicin.12 In addition to mutations in WHO-recommended DRDRs, compensatory AMR-associated mutations, including nth (DNA repair), rpoA (rifampicin), rpoC (rifampicin), gyrA, gyrB (ofloxacin) and 23S rRNA (clarithromycin), have been reported in M. tuberculosis and M. leprae.13–15 However, the roles of these mutations in M. leprae are yet to be confirmed.
In China, leprosy is still at a relatively low endemic level, primarily in Yunnan, Sichuan, Hunan and Guizhou. A total of 406 new cases were detected in China in 2020, with a case detection rate of 0.032 per 100,000 people. The registered cases were 704 by the end of 2020, accounting for a prevalence rate of 0.054 per 100,000 population.1 Zhejiang is one of the leprosy-endemic provinces in China with a population of 57.37 million, about 1% of China.16 A total of 16,000 cases have been registered until the end of 2019. Since the implementation of MDT recommended by WHO in the 1980s, Zhejiang achieved leprosy elimination by Chinese level (less than 1 per 100,000) in 1995. Nevertheless, as an important economic province, Zhejiang has a large number of floating populations, which has become the main source of newly detected cases. A total of 167 new leprosy cases were identified in Zhejiang Province from 2011 to 2019, including 128 cases of floating population in other provinces (accounting for 76.65%). In recent years, a few new cases still can be detected in Zhejiang every year. However, almost no studies on the drug resistance of Zhejiang leprosy cases have been reported.
In the current study, we collected 34 newly diagnosed leprosy cases from Zhejiang Province and analysed the AMR of M. leprae by direct PCR sequencing of the WHO-recommended DRDRs containing folP1, rpoB and gyrA. We also investigated the sequence of regions and genes of compensatory AMR-associated DRDRs, including nth, rpoA, rpoC, gyrB and 23S rRNA, to evaluate the necessity to complement the current surveillance system for AMR mutations. Finally, combined with clinical characteristics of leprosy cases, the prevalence of leprosy resistance in Zhejiang Province was obtained.
Materials and Methods
Sample Collection
During the study period, 34 newly diagnosed cases of leprosy in Zhejiang Province were isolated from skin biopsy (75% alcohol-soaked tissues), including eight local cases and 26 floating population from other provinces from 2018 to 2021. Of the 34 cases, 85.3% were MB, whereas the remaining cases were PB. All patients were examined by experienced dermatologists to ensure the quality and uniformity of these procedures. Skin biopsies were collected based on standard recommendations. Diagnosis and prescription of PB and MB-MDT regimen were performed in accordance with the WHO guidelines.
PCR and Sequencing of DRDRs
The presence of M. leprae genomic DNA was confirmed by PCR detection of the 16S rRNA gene. PCR and nested PCR were performed to amplify the WHO-recommended DRDRs (folP1, rpoB and gyrA) and genes of compensatory AMR-associated DRDRs, including nth, rpoA, rpoC, gyrB and 23S rRNA, by using primers and conditions listed in Supplementary Tables 1 and 2. The quality of amplified products was assessed using 1–2% agarose gel. PCR products were purified and sequenced at a local commercial sequencing company (Tsingke Biological Technology Co., Ltd., Nanjing, China). Alignments to NCBI nucleotide databases were performed using a basic local alignment sequence tool (NCBI-BLAST) to detect mutations.
Results
Clinical Details of Collected Cases
The detailed study profile is shown in Figure 1. Clinical data confirmed that 34 patients were newly diagnosed from 2018 to 2021, and their characteristics are summarised in Table 1 and Supplementary Table 3. Twenty-nine of the patients (85.3%) were MB, whereas four cases (11.8%) were PB, and one case (2.9%) was not defined. The initial bacilloscopy index of the MB cases ranged between 0 and 5.5, with an average of 2.96. Gender analysis showed that males (67.6%) were more affected than females (32.4%) with age ranging from 17 to 83. Of the 34 cases, eight (23.5%) were born in Zhejiang Province, and 26 (76.5%) were floating population (from other provinces in China). The average age of all the patients was 46.9 years old, whereas floating population cases (33.8 years old) were generally younger than native cases (60 years old) based on the data. All patients had different time delays in diagnosis, ranging from 1 month to 13 years. However, most delay time of all cases was within 24 months (85.3%). Based on the MDT scheme, patients diagnosed from 2018 to 2020 completed the official treatment regimen, whereas cases in 2021 were still under treatment.
 |
Table 1 Clinical Characteristics of Patients Involved in the Study |
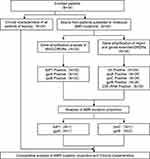 |
Figure 1 Schematic procedure and structure of the study. |
Molecular AMR Analysis
In this study, all the DRDR-associated genes were based on the annotations of M. leprae TN, relative accession numbers of M. leprae TN and codon numbers are listed in Supplementary Table 4. The results of amplification and DNA sequencing of WHO-recommended DRDRs containing folP1, rpoB and gyrA are presented in Table 2. All the 34 leprosy samples (100%) yielded sequence results for at least one gene fragment; informative sequences were obtained in 32 cases (94.1%) for folP1, 33 (97.1%) for rpoB and 34 (100%) for gyrA. Of the 34 samples, two (5.9%) strains showed drug resistance, which had a mutation in folP1 gene (linked with dapsone resistance) with CCC-to-CTC transversion (Proline to Leucine) at amino acid position 55 and a mutation in gyrA gene (linked with ofloxacin resistance) with GCA-to-GTA transversion (Alanine to Valine) at amino acid position 91. No correlation of drug resistance proportion was identified in male vs female, nerve vs no nerve involvement, deformity vs no deformity and reaction vs non-reaction cases. Single-drug resistance to dapsone was detected in LL cases (Tables 1 and 2).
 |
Table 2 Results of PCR and DNA Sequencing of WHO-Recommended AMR Genes |
Among 34 (100%) cases, at least one gene fragment of sequence results was obtained, and informative sequences were found in 29 cases (85.3%) for nth, 29 (85.3%) for rpoA, 25 (73.5%) for rpoC, 24 (70.6%) for gyrB and 33 (97.1%) for 23S rRNA. No mutations of associated DRDRs were identified in nth, rpoA and 23S rRNA. The two single mutations in gyrB were detected in different strains (5.9%), whereas one rpoC mutation was detected in each strain (2.9%, Table 3).
 |
Table 3 Results of PCR and DNA Sequencing of Compensatory AMR-Associated DRDRs |
Discussion
In 1982, WHO recommended that all registered patients with leprosy should receive combination therapy with three antibiotics, including rifampicin, clofazimine and dapsone, which remedied many of the problems associated with monotherapy.17 Moreover, further studies have demonstrated that ofloxacin-containing MDT is both safe and effective for treatment of leprosy.18,19 WHO declared that MDT treatment can effectively control leprosy incidence and contribute to the elimination of leprosy burden in several countries, including China. The emergence of drug resistance in leprosy is a major concern for the implementation of disease intervention programmes. As limited studies described the prevalence of gene mutations associated with AMR in Zhejiang Province, in this study, we analysed the prevalence of drug-resistant leprosy cases by sequencing WHO-recommended DRDRs containing sites in three genes and potential compensatory AMR-associated DRDR genes, using samples of 34 patients with leprosy from 2018 to 2021.
All the cases were newly diagnosed leprosy patients, and they finished MDT treatment based on the WHO guidelines. A mutation at codon 55 (Pro-Leu) of folP1 (Figure S1A), known to cause resistance to dapsone among 34 samples, was identified.9,20 The frequency of drug resistance to dapsone (2.9%) was lower than that in Hunan (10.7%) but higher than that in Sichuan (0%), Guizhou (1.7%) and Yunnan (1.0%).21 The gyrA sequence also showed a single mutation at codon 91 with amino acid changes from Alanine to Valine (Figure S1B), which demonstrated ofloxacin resistance.22 Whatever, the frequency (2.9%) was lower than that in Yunnan (3.8%) for ofloxacin. In the present study, no mutation associated with rifampicin (rpoB gene) was confirmed. Both mutations found in the folP1 and gyrA gene are single mutations. No significant correlation in AMR mutations in M. leprae was found in male vs female, nerve vs no nerve involvement, deformity vs no deformity, and reaction vs non-reaction. Although dapsone monotherapy has been used in the pre-MDT era across China,23 the irregular use of this drug resulted in high dapsone resistance. Based on previous reports, these resistant strains were continuously transmitted in the community.24
Compensatory mutations in rpoA and rpoC, encoding the alpha and beta-prime subunits of RNA polymerase, can occur in rifampicin-resistant M. tuberculosis,13 whereas only nth and rpoC mutations were found in M. leprae.14 We identified only one mutation in rpoC gene at codon 879 (Val-Met) (Figure S1C), which seems to be a polymorph other than compensatory mutation since no rifampicin-resistant strains was identified here. A hypothesis was proposed that compensatory mutations may be associated with mutations in rifampicin resistance-determining regions (RRDRs)25,26 in rifampicin-resistant M. tuberculosis. Rifampicin-susceptible isolates or rifampicin-resistant strains without rpoB mutations can hardly occur compensatory mutations.13 However, further study is required to determine the relationship between phenotype and genotype. As mentioned by previously reported literature, two single mutations of gyrB gene were also identified at codon 368 (Pro-Leu) and 215 (Lys-Asn) (Figure S1D and E). The substitutions in gyrB are based on the gene numbering system of M. tuberculosis gyrB.22 The Quinolone-Resistance Determining Region (QRDR) of gyrB stretches from amino acid 426 to 464, from 461 to 499 and from 464 to 504 in E. coli, M. tuberculosis and M. leprae, respectively, while an extension of the M. tuberculosis QRDR to amino acid 501 has been proposed previously.27 In our findings of gyrB, position 215 and 368 are not included in the QRDR, which might indicate polymorphs only, and drug resistance to ofloxacin cannot develop. Endonuclease III (Nth) is central to the base excision repair pathway in bacteria.28 Mycobacterial genomes usually contain a single nth gene and two fpg/nei genes, but M. leprae has lost fpg/nei orthologues and retained the nth gene. It has been reported that all nth mutants were also drug-resistant so Nth loss likely favors emergence of drug resistance, and nth mutations might serve as a surrogate marker for potential drug resistance and treatment failure.29,30 However, no nth mutation was identified among 34 leprosy cases. 23S rRNA mutation conferring clarithromycin resistance was also not found in Zhejiang leprosy patients. Furthermore, the role of these mutations still requires further study to determine their phenotypic outcomes in M. leprae.
In general, mutations of folP1 gene and gyrA gene were showed single-drug resistance to dapsone and ofloxacin, respectively, among all the strains with mutations. Although the substitution of amino acid in rpoC was a non-synonymous mutation, while no compensatory was occurred rifampicin resistance was not involved. Two mutations in gyrB were also confirmed to be independent of drug resistance since outside the DRDRs. No cases of multidrug resistance were found. Since the establishment of national policies and socioeconomic factors, the transmission of AMR and non-AMR cases of leprosy has been reduced in China. In addition, rifampicin drug resistance is low in Chinese population. Irregular MDT may be the main factor causing drug resistance.31 Notably, standardised treatment can achieve good therapeutic effects based on our study. Fluoroquinolones are promising and widely used antibiotics, which are introduced to routine clinical practice to treat infectious diseases in China. Given the easy availability and inappropriate use of these drugs, AMR to fluoroquinolones has emerged in China.32 Clarithromycin is another antibiotic used to treat various bacterial infections33,34 and it is used as an alternative or combinational drug that can result in clarithromycin resistance of leprosy.35
The movement of people within nations and around the globe has always been a factor in shaping the epidemiological portrait of leprosy. Migration has unique challenges to leprosy control in the 21st century. Among the 34 cases enrolled in this study, 26 (76.5%) were immigrants from other provinces. Given the movement of people affected by leprosy to provinces that have relatively few cases of the disease, services for leprosy treatment have become scarce.36 General practitioners and dermatologists may be unlikely to diagnose the disease in its early stages. When they associate person’s symptoms with leprosy, they may have little knowledge of where to begin with treatment, thereby leading to misdiagnosis and delayed diagnosis.
Conclusion
This study is the first systematic analysis of AMR surveillance of leprosy in Zhejiang Province in recent years. Two single mutations related to dapsone and ofloxacin resistance separately were identified and no multidrug resistance was found among 34 leprosy patients. The results presented in our study demonstrate that drug-resistant strains are not epidemic strains in this area. Given the few cases of leprosy, analysing the AMR of M. leprae in Zhejiang Province more comprehensively is difficult. However, regular MDT treatment and population management in the early stage may contribute to the low prevalence of leprosy.
Ethics Approval and Informed Consent
This study was approved by the institutional ethical committee of the Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, China (2014-KY-003) and this study was conducted in accordance with the Declaration of Helsinki. All participants provided written informed consent.
Acknowledgments
We thank all the staff for their support in sample and data collection.
Funding
This work was supported by Zhejiang Provincial Natural Science Foundation of China under Grant (LGF19H260001) and National Natural Science Foundation of China (grant 81371751, 81972950).
Disclosure
All authors declare that they have no conflicts of interest.
References
1. World Health Organization = Organisation mondiale de la Santé. Global leprosy (Hansen disease) update, 2020: impact of COVID-19 on global leprosy control –Situation de la lèpre (maladie de Hansen) dans Le Monde, 2020: impact de la COVID-19 sur les activités mondiales de lutte contre la lèpre. Wkly Epidemiol Rec Relevé Épidémiologique Hebd. 2021;96(36):421–444.
2. World Health Organization. Chemotherapy of leprosy for control programmes. World Health Organ Tech Rep Ser. 1982;675(1–33):1–33.
3. Ramu G. Single-dose rifampicin, oflaxicin and minocycline (ROM) therapy for single leprosy lesions. Lepr Rev. 1998;69(1):78–82.
4. Fish DN. Fluoroquinolone adverse effects and drug interactions. Pharmacotherapy. 2001;21(10 Pt 2):253S–272S. doi:10.1592/phco.21.16.253S.33993
5. Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis off Publ Infect Dis Soc Am. 2000;31(Suppl 2):S24–28. doi:10.1086/314056
6. Daniel E, Ebenezer GJ, Job CK. Pathology of iris in leprosy. Br J Ophthalmol. 1997;81(6):490–492. doi:10.1136/bjo.81.6.490
7. Sivaprasad N, Snehalatha S, Lobo D, Aschhoff M, Job CK. Viability of Mycobacterium leprae in lepromatous patients after five years of dapsone monotherapy supplemented with two years of multidrug therapy. Indian J Lepr. 1995;67(4):427–433.
8. Pearson JM, Rees RJ, Waters MF. Sulphone resistance in leprosy. A review of one hundred proven clinical cases. Lancet Lond Engl. 1975;2(7924):69–72. doi:10.1016/S0140-6736(75)90508-5
9. Cambau E, Carthagena L, Chauffour A, Ji B, Jarlier V. Dihydropteroate synthase mutations in the folP1 gene predict dapsone resistance in relapsed cases of leprosy. Clin Infect Dis. 2006;42(2):238–241. doi:10.1086/498506
10. Honore N, Cole ST. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob Agents Chemother. 1993;37(3):414–418. doi:10.1128/AAC.37.3.414
11. Emmanuelle C, Bonnafous P, Evelyne P, Sougakoff W, Baohong J, Vincent J. Molecular detection of rifampin and ofloxacin resistance for patients who experience relapse of multibacillary leprosy. Clin Infect Dis. 2002;34(1):39–45. doi:10.1086/324623
12. Kai M, Fafutis-Morris M, Miyamoto Y, et al. Mutations in the drug resistance-determining region of Mycobacterium lepromatosis isolated from leprosy patients in Mexico. J Dermatol. 2016;43(11):1345–1349. doi:10.1111/1346-8138.13483
13. Comas I, Borrell S, Roetzer A, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44(1):106–110. doi:10.1038/ng.1038
14. Benjak A, Avanzi C, Singh P, et al. Phylogenomics and antimicrobial resistance of the leprosy bacillus Mycobacterium leprae. Nat Commun. 2018;9(1):352. doi:10.1038/s41467-017-02576-z
15. Malik S, Willby M, Sikes D, Tsodikov OV, Posey JE. New insights into fluoroquinolone resistance in mycobacterium tuberculosis: functional genetic analysis of gyrA and gyrB mutations. PLoS One. 2012;7(6):e39754. doi:10.1371/journal.pone.0039754
16. Wu L, Shen Y, Yao Q, et al. Temporal-spatial distribution characteristics of leprosy: a new challenge for leprosy prevention and control in Zhejiang, China. PLoS Negl Trop Dis. 2021;15(1):e0008956. doi:10.1371/journal.pntd.0008956
17. Gelber RH, Grosset J. The chemotherapy of leprosy: an interpretive history. Lepr Rev. 2012;83(3):221–240. doi:10.47276/lr.83.3.221
18. Faust L, Klowak M, MacRae C, Kopalakrishnan S, Showler AJ, Boggild AK. Ofloxacin-containing multidrug therapy in ambulatory leprosy patients: a case series. J Cutan Med Surg. 2021;25(1):45–52. doi:10.1177/1203475420952437
19. Balagon MF, Cellona RV, Abalos RM, Gelber RH, Saunderson PR. The efficacy of a four-week, ofloxacin-containing regimen compared with standard WHO-MDT in PB leprosy. Lepr Rev. 2010;81(1):27–33. doi:10.47276/lr.81.1.27
20. Lee SB, Kim SK, Kang TJ, et al. The prevalence of folP1 mutations associated with clinical resistance to dapsone, in Mycobacterium leprae isolates from South Korea. Ann Trop Med Parasitol. 2001;95(4):429–432. doi:10.1080/00034983.2001.11813656
21. Chokkakula S, Chen Z, Wang L, et al. Molecular surveillance of antimicrobial resistance and transmission pattern of Mycobacterium leprae in Chinese leprosy patients. Emerg Microbes Infect. 2019;8(1):1479–1489. doi:10.1080/22221751.2019.1677177
22. Chauffour A, Morel F, Reibel F, et al. A systematic review of Mycobacterium leprae DNA gyrase mutations and their impact on fluoroquinolone resistance. Clin Microbiol Infect. 2021;27(11):1601–1612. doi:10.1016/j.cmi.2021.07.007
23. Jian-Ping S, Gupte MD, Cheng J, Manickam P, Meiwen Y, Zhong LW. Trends of case detection and other indicators of leprosy in China during 1985–2002. Chin Med Sci J Chung-Kuo Hsueh Ko Hsueh Tsa Chih. 2005;20(2):77–82.
24. Jing Z, Zhang R, Zhou D, Chen J. Twenty five years follow up of MB leprosy patients retreated with a modified MDT regimen after a full course of dapsone mono-therapy. Lepr Rev. 2009;80(2):170–176.
25. Avanzi C, Maia RC, Benjak A, et al. Emergence of mycobacterium leprae rifampin resistance evaluated by whole-genome sequencing after 48 years of irregular treatment. Antimicrob Agents Chemother. 2020;64(7):e00330–20. doi:10.1128/AAC.00330-20
26. Lavania M, Singh I, Turankar RP, et al. Enriched whole genome sequencing identified compensatory mutations in the RNA polymerase gene of rifampicin-resistant Mycobacterium leprae strains. Infect Drug Resist. 2018;11:169–175. doi:10.2147/IDR.S152082
27. Pantel A, Petrella S, Veziris N, et al. Extending the definition of the GyrB quinolone resistance-determining region in Mycobacterium tuberculosis DNA gyrase for assessing fluoroquinolone resistance in M. tuberculosis. Antimicrob Agents Chemother. 2012;56(4):1990–1996. doi:10.1128/AAC.06272-11
28. Zharkov DO. Base excision DNA repair. Cell Mol Life Sci CMLS. 2008;65(10):1544–1565. doi:10.1007/s00018-008-7543-2
29. Moolla N, Goosens VJ, Kana BD, Gordhan BG. The contribution of Nth and Nei DNA glycosylases to mutagenesis in Mycobacterium smegmatis. DNA Repair. 2014;13:32–41. doi:10.1016/j.dnarep.2013.11.003
30. Ford CB, Shah RR, Maeda MK, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45(7):784–790. doi:10.1038/ng.2656
31. Shen J, Yan L, Sun P. Clinical features of relapse after multidrug therapy for leprosy in China. Lepr Rev. 2015;86(2):165–169. doi:10.47276/lr.86.2.165
32. Xiao YH, Wang J, Li Y; MOH National Antimicrobial Resistance Investigation Net. Bacterial resistance surveillance in China: a report from Mohnarin 2004–2005. Eur J Clin Microbiol Infect Dis off Publ Eur Soc Clin Microbiol. 2008;27(8):697–708. doi:10.1007/s10096-008-0494-6
33. Fontana C, Favaro M, Minelli S, et al. New site of modification of 23S rRNA associated with clarithromycin resistance of Helicobacter pylori clinical isolates. Antimicrob Agents Chemother. 2002;46(12):3765–3769. doi:10.1128/AAC.46.12.3765-3769.2002
34. De Francesco V, Zullo A, Giorgio F, et al. Change of point mutations in Helicobacter pylori rRNA associated with clarithromycin resistance in Italy. J Med Microbiol. 2014;63(Pt 3):453–457. doi:10.1099/jmm.0.067942-0
35. Gunawan H, Sasmojo M, Putri HE, Avriyanti E, Hindritiani R, Suwarsa O. Clinical pilot study: clarithromycin efficacy in multibacillary leprosy therapy. Int J Mycobacteriology. 2018;7(2):152–155. doi:10.4103/ijmy.ijmy_58_18
36. Lockwood DN, Reid AJ. The diagnosis of leprosy is delayed in the United Kingdom. QJM Mon J Assoc Physicians. 2001;94(4):207–212. doi:10.1093/qjmed/94.4.207
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
