Back to Journals » Infection and Drug Resistance » Volume 13
Molecular Mechanisms and Epidemiology of Carbapenem-Resistant Escherichia coli Isolated from Chinese Patients During 2002–2017
Authors Tian X, Zheng X, Sun Y, Fang R, Zhang S, Zhang X, Lin J, Cao J, Zhou T
Received 21 September 2019
Accepted for publication 30 January 2020
Published 17 February 2020 Volume 2020:13 Pages 501—512
DOI https://doi.org/10.2147/IDR.S232010
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Xuebin Tian, 1, 2 Xiangkuo Zheng, 2 Yao Sun, 1 Renchi Fang, 1 Siqin Zhang, 1 Xiucai Zhang, 1 Jie Lin, 1 Jianming Cao, 2 Tieli Zhou 1
1Department of Clinical Laboratory, First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang Province, People’s Republic of China; 2School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, Zhejiang Province, People’s Republic of China
Correspondence: Tieli Zhou
Department of Clinical Laboratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, People’s Republic of China
Tel +86-0577-86699370
Email [email protected]
Jianming Cao
School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, People’s Republic of China
Tel +86-0577-88069595
Email [email protected]
Background: The emergence and spread of carbapenem-resistant Escherichia coli (E. coli) pose a serious threat to human health worldwide. This study aimed to investigate the molecular mechanisms underlying carbapenem resistance and their prevalence among E. coli in China.
Methods: A collection of 5796 E. coli clinical isolates were collected from the First Affiliated Hospital of Wenzhou Medical University from 2002 to 2017. Sensitivity to antibiotics was determined using the agar dilution method. The detection of carbapenemases production and the prevalence of resistance-associated genes were investigated through modified carbapenem inactivation method (mCIM), PCR and sequencing. The mutations in outer membrane porins genes (ompC and ompF) were also analyzed by PCR and sequencing assays. The effect of efflux pump mechanism on carbapenem resistance was also tested. E. coli were typed by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST).
Results: A total of 58 strains (1.0%) of carbapenem-resistant E. coli were identified. The strains carrying blaKPC-2 and blaNDM accounted for 22.4% (13/58) and 51.7% (30/58), respectively. Among blaNDM- positive strains, 27 blaNDM genes were assigned to blaNDM-5, while the remaining three strains were blaNDM-1, whereas blaVIM, blaIMP, blaOXA-48, and blaSHV were not found. The CTX-M-type β-lactamase genes accounted for 96.6% (56/58). In addition, blaTEM-1 genes were identified in 58.6% of tested strains. In carbapenem-resistant isolates, mutations in OmpC (the majority of mutated sites were D192G and Q104_F141del, accounting for 54.5%) and OmpF (large deletions S75_V127del, W83_D135del and Q88_D135del) were detected. Of note, the antibiotic resistance was not associated with overexpression of efflux pump. Moreover, MLST categorized the 58 carbapenem-resistant isolates into 19 different sequence types. PFGE analysis revealed that homology among the carbapenem-resistant isolates was low and sporadic.
Conclusion: The blaNDM was the principal resistance mechanism of carbapenem-resistant E. coli in the hospital. blaNDM-5 is becoming a new threat to public health and the alteration of outer membrane porins might help further increase the MIC of carbapenem.
Keywords: Escherichia coli, carbapenem-resistant, carbapenemase, outer membrane porin, epidemiology
Introduction
Escherichia coli is one of the most commonly isolated microorganisms in clinical specimens. Multidrug resistance in E. coli has become an upsetting issue observed in humans1 and has been recognized as a contributor to the dissemination of antibiotic-resistance genes.2 Controlling the dissemination of multidrug-resistant (MDR) strains is problematic due to very few new antibiotics available.3,4 Because of increasing resistance to third-generation cephalosporins, fluoroquinolones and aminoglycosides, carbapenems have gradually become the last resort for life-threatening MDR E. coli infections because of their broad-spectrum antimicrobial agents.5,6 Nevertheless, with an increasing consumption of carbapenems, the emergence of carbapenems resistant E. coli has become a serious public health concern worldwide.7,8
The mechanisms of carbapenem resistance are strongly associated with carbapenemase production (acquisition of carbapenemase genes), combination of porin loss with extended-spectrum β-lactamases (ESBLs) and the overexpression of efflux pumps.9,10 Several studies have reported that acquired carbapenemase isolates might cause hospital outbreaks and become endemic in healthcare settings.11,12 Globally predominant carbapenemases include KPC, NDM, VIM, IMP, and OXA, which are encoded by blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA genes present in both the plasmid and the chromosome.13,14 In addition, the carbapenemase genes could co-exist with ESBLs and other resistance genes on plasmids, which further limit the treatment options. Moreover, previous studies have reported that the outer membrane porins of E. coli are involved in the MDR phenotype.15,16 Choi et al have constructed mutants of porins (ompC and ompF mutations) in E. coli and discovered that porins have a distinct role in antibiotic resistance and membrane integrity.17 With the increase in the prevalence of carbapenem-resistant E. coli strains worldwide,18,19 longitudinal epidemiological surveillance and mechanisms research on the carbapenem-resistant E. coli are of great clinical significance for the global control and prevention of the distribution and spread of resistance, as well as the guidance on antibacterial treatment. Nonetheless, there is still a lack of data on the long-term evolution of carbapenem-resistant E. coli in China. In the present study, we characterized the epidemiology prevalence and molecular mechanisms of 58 E. coli clinical isolates during large-scale surveillance for carbapenem resistance in the southeast of China.
Materials and Methods
Bacterial Isolates
A total of 5796 E. coli clinical isolates were collected from the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) between 2002 and 2017. Identification of all isolates was performed using a VITEK®2 system (bioMérieux, Marcy-l’Étoile, France). After collection, isolates were stored in 30% glycerol at –80°C. Relevant clinical data were collected from the medical records. We collected the information about isolation date, age, gender, sample, and ward.
Minimum Inhibitory Concentration Determination
MICs of 12 antimicrobial agents, including imipenem, meropenem, ertapenem, ampicillin, ceftriaxone, ceftazidime, ciprofloxacin, levofloxacin, gentamicin, tobramycin, amikacin, and fosfomycin, were determined by the agar dilution method according to the guidelines recommended by the latest Clinical and Laboratory Standards Institute (CLSI).20 Colistin MIC determination was performed with broth microdilution and interpreted by the recommendation of the European Committee on Antimicrobial Susceptibility Testing clinical breakpoints (http://www.eucast.org/). E. coli ATCC 25922 was used as the control strain for antimicrobial susceptibility testing.
Detection of Carbapenemases and Antibiotic Resistance Determinants
The modified carbapenem inactivation method (mCIM) was used to screen isolates for the production of carbapenemases, according to CLSI guidelines. The presence of resistant mechanisms, including carbapenem resistance genes (blaKPC-2, blaNDM, blaIMP, blaVIM, and blaOXA-48), ESBLs genes (blaSHV, blaTEM, blaCTX-M-1, and blaCTX-M-9), outer membrane porins genes (ompC and ompF), fosfomycin resistance genes (fosA3 and fosA) and colistin resistance genes (mcr-1 and mcr-3) were identified by polymerase chain reaction (PCR) and sequencing. Each isolate DNA was extracted from fresh bacterial colonies using a Biospin Bacterial Genomic DNA Extraction kit (Bioer Technology, Hangzhou, China). The primers used for amplification and sequencing were listed in Table S1. Positive amplification products were sent to Shanghai BGI Technology Co. (Shanghai, China) for sequencing. Nucleotide sequences were compared by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The online PROVEAN platform (http://provean.jcvi.org/seq_submit.php) was used to predict alterations in the biological function of the proteins.
Effect of Efflux Pump Mechanism on Carbapenem-Resistance in E. coli
Carbonyl cyanide m-chlorophenylhydrazone (CCCP) is an energy uncoupler, has been identified as a compound to reverse MDR in E. coli over-expressing efflux pumps.21 CCCP (Sigma, St Louis, MO) was used to measure the activity of efflux pumps in carbapenem-resistant E. coli isolates. The change in the MICs of carbapenems was determined by the agar dilution method in the absence or presence of 10 μg/mL CCCP. A phenotype for positive efflux was defined as a ≧4-fold reduction of the carbapenem MIC in the presence of CCCP.22
Molecular Epidemiology Analysis
MLST analyses of the carbapenem-resistant isolates were carried out by amplifying eight housekeeping genes (dinB, icdA, pabB, polB, putP, trpA, trpB, and uidA). Sequence types were assigned by querying against the database available at the Institut Pasteur’s E. coli MLST website (http://bigsdb.web.pasteur.fr/ecoli/ecoli.html).
To further identify potential clonal spread, PFGE was performed using a CHEF-Mapper XA PFGE system (Bio-Rad, Hercules, CA). Briefly, genomic DNA was extracted from all tested isolates, followed by Xba I restriction enzyme (Takara Bio, Inc., Kusatsu, Japan) digestion. Electrophoresis was then performed under the following conditions: temperature, 14°C; voltage, 6 V/cm; pulse angle 120°; and pulse duration, 2.16–54.17 s for 18.5 hrs. The universal standard strain Salmonella enterica serotype H9812 was used as a molecular marker.23 Band patterns were analyzed and interpreted according to the criteria proposed by Tenover et al.24
Results
Bacterial Strains and Antimicrobial Susceptibility Testing
A total of 58 (1.0%) carbapenem-resistant E. coli isolates were identified with carbapenems (including imipenem, meropenem, and ertapenem), MICs ranging from 2 μg/mL to ≧16 μg/mL. Carbapenem-resistant E. coli isolates at our hospital were first detected in 2012; after that, the resistance rate has increased from 0.85% to 1.85% as was detected in 2017 (Table 1). Table 2 summarized the patient characteristics and species distribution. Overall, the carbapenem-resistant organisms were mainly from urine samples (31.0%, 18/58), followed by blood (27.6%, 16/58) and drainage (19.0%, 11/58). There were more isolates from males than females (62.1% vs 37.9%, respectively). Isolates were cultured from patients aged 19 to 91 years (average age 62.5 years). The majority of the isolates were from patients in the intensive care unit (ICU) (31.0%, 18/58), Hepatobiliary Surgery (10.3%, 6/58). The antimicrobial resistance profiles of the 58 carbapenem-resistant isolates were summarized in Table 3. According to the results of antimicrobial susceptibility testing, all 58 isolates showed higher resistance rates to cephalosporins, fluoroquinolones, and aminoglycosides. Thereinto, 55 (94.8%) isolates were resistant to fluoroquinolones, including levofloxacin and ciprofloxacin; 49 (84.5%) isolates were resistant to aminoglycosides, including gentamicin, tobramycin and amikacin. Furthermore, 58 (100%) isolates were resistant to ampicillin, while 18 (31.0%) isolates were resistant to fosfomycin and 2 (3.4%) to colistin.
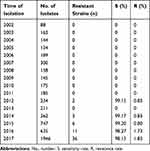 |
Table 1 Carbapenems Susceptibility of E. coli Clinical Isolates |
 |
Table 2 Patient’s Clinical Data and Characteristics of Analyzed Strains |
 |
Table 3 Minimum Inhibitory Concentrations (MICs) of 58 Carbapenem-Resistant E. coli Isolates |
Prevalence of β-Lactamase Genes
Forty-three carbapenem-resistant E. coli isolates produced carbapenemases (Figure S1 and Table S2). The prevalence rates of blaKPC-2 and blaNDM in carbapenem-resistant isolates were 22.4% and 51.7%, respectively (Figure 1), while blaIMP, blaVIM and blaOXA-48 were not detected. In addition, the number of isolates harbored blaNDM-1 or blaNDM-5 were 3 (5.2%) and 27 (46.6%), respectively. Moreover, the most prevalent CTX-M-type among analyzed strains was blaCTX-M-1 (75.9%, 44/58), followed by blaCTX-M-9 (65.5%, 38/58). In general, the CTX-M-type β-lactamase genes accounted for 96.6% (56/58). In addition to blaCTX-M genes, blaTEM-1 genes were also identified in 58.6% of tested strains, blaSHV was not detected.
Detection of Mutations in ompC and ompF
Mutations in ompC and ompF genes were detected in carbapenem-resistant isolates, including amino acid substitutions and deletions. Deleterious mutations of OmpC and OmpF occurred in 22 and 21 isolates, respectively. Moreover, four carbapenem-resistant isolates had mutations in both OmpC and OmpF. The majority of mutation sites in ompC were D192G followed by Q104_F141del. Notably, several large deletions (S75_V127del, W83_D135del and Q88_D135del) of an amino acid sequence encoded by the ompF gene were also detected. Amino acid substitutions in ompC and ompF were considered deleterious by PROVEAN (Tables 4 and 5).
 |
Table 4 Mutations in Carbapenem-Resistant E. coli Isolates |
 |
Table 5 Analysis of Mutations in ompC and ompF |
Phenotypic Detection of the Efflux Pump Overexpression
The effect of efflux pumps on the antibiotic resistance profiles of isolates was examined using the efflux pump inhibitor CCCP. When exposed to 10 μg/mL CCCP, none of the isolates showed a ≧4-fold decrease in the carbapenem MIC, suggesting that the antibiotic resistance was not associated with overexpression of efflux pump in this study.
Epidemiological Characterization
MLST analysis assigned the 58 carbapenem-resistant isolates into 19 different sequence types (STs) (Figure 2). ST8 was the predominant ST, accounting for 29.3% (17/58), followed by ST19 (12.1%, 7/58) and ST692 (12.1%, 7/58). Moreover, there were two novel STs (labelled as “New” in Figure 2; currently not registered in the MLST database). PFGE analysis revealed that homology among the resistant isolates was low and sporadic, suggesting a very low likelihood of clonal spread (Figure 2).
Discussion
Carbapenems are extensively applied in clinical settings for the therapeutic management of MDR Gram-negative bacterial infections due to their broad spectrum of antimicrobial activity.25 Yet, several surveillance programs have reported a highly increasing carbapenem resistance, making clinical treatment more challenging.26,27 In the current study, 58 of 5796 E. coli isolates exhibited an increasing carbapenem-resistant rate from 2002 to 2017. The relatively higher incidence revealed that the ongoing surveillance is urgently warranted in China.
From the clinical perspective, there have been reports of transmission of E. coli in the ICU,28,29 and clinicians should be vigilant about the potential presence of this species. Our study also confirmed that carbapenem-resistant strains were most commonly isolated from patients aged >65 years who were treated in the ICU. The KPC-type enzyme was first reported in Klebsiella pneumoniae from the southern United States in 200130 and now endemic all over the world.31,32 In China, dissemination of KPC-producing Enterobacteriaceae spp. has been confirmed in Shandong, Zhejiang, Taiwan, and other provinces.33–35 KPC-2 was the most important in K. pneumoniae, whereas NDM-1 was the most important in E. coli. Notably, in previous studies in China, a few strains of E. coli with KPC-2 were detected.36 However, in our study, 22.4% (13/58) of the strains were detected with KPC-2. This finding suggested that more attention should be paid to the spread of KPC-2 in this region. The IMP and VIM genes were reported in several regions, OXA-48 was more common in Europe but had not been found in our study.37 New Delhi metallo-β-lactamase (NDM), which was first reported in Sweden in 2009 in a patient who developed an infection while travelling in India,38 could confer resistance to most β-lactams. Over the recent years, a high prevalence of NDM-1 has been observed in China and India.39,40 In addition, the rapid global spread of NDM-producing isolates via MDR plasmids has led many into thinking that common infections with such strains may soon be untreatable.41 Selective pressure caused by increased use of antibiotics may drive the evolution of NDM-1, thus resulting in the emergence of its variants. In the current study, the emergence of NDM-5 reflected a new prevalence since 2017. M154L amino acid substitution in NDM-5 was the most common substitution in all NDMs variants,42 responsible for increased carbapenemase activity. Moreover, NDM-5 has an extra V88L substitution; the emergence of V88L may contribute to lower catalytic activity on imipenem and meropenem.43,44 Although NDM-5 made anti-infective treatment more difficult,45 the lower hydrolytic activity of imipenem and meropenem implied these were still the first choice for MDR E. coli isolates. Our study indicated an increased number of carbapenemases-producing E. coli isolates over the last few years. It also revealed the high incidence of blaNDM since it was first discovered at the hospital between 2015 and 2017. Interestingly, our results revealed that NDM-5 may even replace the NDM-1 in carbapenem-resistant E. coli isolates from 2017 in China. To date, several studies showed that blaNDM-5 was carried by IncX3 plasmids in China,46,47 India,48 Denmark49 and Australia.50 The fact that IncX-type plasmids have been shown to be conjugatable in most studies could explain the rapid spread of blaNDM-5-carrying isolates. Therefore, it is imperative that feasible and effective measures are taken immediately.
ESBL-producing E. coli showed higher health risks related to hospital-acquired infections compared to non-ESBL-producing isolates.51 CTX-M β-lactamases are the most widespread types of ESBLs, which have been identified in the mid-2000s in clinical E. coli isolates.52 In this study, 96.6% of ESBL genes were classified as blaCTX-M. Several reports have indicated that the transfer of CTX-M mobile plasmids could be frequently accompanied by the acquisition of fosfomycin resistance genes.53,54 In our study, 17 carbapenem-resistant strains harboring CTX-M plasmids were positive for the fosA3 gene. Colistin resistance represents another health concern. Two colistin-resistant E. coli strains detected in our study carried mcr-1 gene. Moreover, co-harboring of blaNDM-1, fosA3, and mcr-1 were detected in DC-3737, like a reservoir, which posed serious concern on public health.
It has been reported that resistance to carbapenems could be mediated by non-specific outer membrane porins OmpC and OmpF in E. coli.17 In the current study, the deleterious mutations were detected in 39 isolates, whereas OmpC and OmpF alteration occurred in 22 and 21 isolates, respectively. Mutation prediction showed that the amino acid substitutions in ompC, such as D192G might be the key factor driving resistance to carbapenems, while amino acid deletions could make an important impact in ompF mutations. The mutations in OmpF and OmpC were the important mechanisms contributing to the elevated MICs to carbapenems.
All of the isolates (100%, 58/58) were ertapenem non-susceptible; however, the abundance of imipenem-resistant strains was relatively smaller, promising the suitability of imipenem as the choice of treatment for infections caused by ertapenem-non-susceptible E. coli isolates. Furthermore, the alteration of outer membrane porins combined with carbapenemase production were found in 39 isolates, which further decreased the sensitivity of imipenem and meropenem. Otherwise, it is worth noting that the carbapenem resistance mechanism of DC-38 still remains unclear, and needs to be further researched in the future.
So far, few studies have reported the effect of efflux pump on carbapenems resistance in Enterobacter spp.55–57 The current study showed that the efflux pump inhibitor CCCP was not able to restore the susceptibility of carbapenem-resistant E. coli, indicating that efflux pump was not involved in the carbapenem resistance in our study.
Our analysis showed that the majority of carbapenem-resistant clinical E. coli isolates showed different PFGE patterns, suggesting that they were genetically unrelated. The results of MLST demonstrated that these carbapenem-resistant isolates were polyclonal without a clonal dissemination. We speculated that carbapenem-resistant E. coli isolates might originate from different lineages and sources, instead of expansion of a single clonal lineage, which is in line with previous reports.58 Among them, ST8 was the main clone type (29.3%, 17/58). Interestingly, 76.9% (10/13) KPC-2-producing E. coli isolates belonged to ST8 in our study, indicated that a high prevalence of blaKPC-2 was linked with ST8. We hypothesized that ST8 had a better ability to capture or accumulate blaKPC compared with the other types. Furthermore, both STs ST19 and ST692 were present in association with the blaNDM-5 gene, which was firstly reported to be linked with NDM-5- producing isolates.
In summary, we described the resistance mechanisms and the molecular epidemiology of carbapenem-resistant E. coli isolates at the First Affiliated Hospital of Wenzhou Medical University between 2002 and 2017. To best of our knowledge, this is the first report on the long duration and large scale of carbapenem-resistant E. coli isolates in China. Due to the limited treatment options, the rising resistance rate has further exacerbated the threat to public health. The prevalence of variant blaNDM-5 represents a new threat. Moreover, ESBLs genes have shown to have a significant role in the carbapenem-resistant E. coli isolates, among which, CTX-M-type ESBLs were prevalent. As carbapenems are becoming ever more used as an effective therapeutic option, monitoring programs are urgently required to prevent the emergence and further spread of its resistance.
Ethical Statement
No samples were collected specifically for this research; only anonymized clinical residual samples collected during routine hospital procedures were used for this study.
Acknowledgment
We thank the Planned Science and Technology Project of Wenzhou (no. Y20170204) for providing financial funding.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gajdacs M, Abrok M, Lazar A, et al. Comparative epidemiology and resistance trends of common urinary pathogens in a tertiary-care hospital: a 10-year surveillance study. Medicina (Kaunas). 2019;55. doi:10.3390/medicina55070356
2. Kappell AD, DeNies MS, Ahuja NH, et al. Detection of multi-drug resistant Escherichia coli in the urban waterways of Milwaukee, WI. Front Microbiol. 2015;6:336. doi:10.3389/fmicb.2015.00336
3. Gajdacs M. The concept of an ideal antibiotic: implications for drug design. Molecules. 2019;24:892. doi:10.3390/molecules24050892
4. Gajdacs M, Albericio F. Antibiotic resistance: from the bench to patients. Antibiotics (Basel). 2019;8. doi:10.3390/antibiotics8030129
5. Tamma PD, Han JH, Rock C, et al. Carbapenem therapy is associated with improved survival compared with piperacillin-tazobactam for patients with extended-spectrum beta-lactamase bacteremia. Clin Infect Dis. 2015;60:1319–1325. doi:10.1093/cid/civ003
6. Ji X, Zheng B, Berglund B, et al. Dissemination of extended-spectrum beta-lactamase-producing Escherichia coli carrying mcr-1 among multiple environmental sources in rural China and associated risk to human health. Environ Pollut. 2019;251:619–627. doi:10.1016/j.envpol.2019.05.002
7. De Gheldre Y, Maes N, Rost F, et al. Molecular epidemiology of an outbreak of multidrug-resistant Enterobacter aerogenes infections and in vivo emergence of imipenem resistance. J Clin Microbiol. 1997;35:152–160. doi:10.1128/JCM.35.1.152-160.1997
8. Ehrhardt AF, Sanders CC, Thomson KS, et al. Emergence of resistance to imipenem in Enterobacter isolates masquerading as Klebsiella pneumoniae during therapy with imipenem/cilastatin. Clin Infect Dis. 1993;17:120–122. doi:10.1093/clinids/17.1.120
9. Cuzon G, Naas T, Guibert M, et al. In vivo selection of imipenem-resistant Klebsiella pneumoniae producing extended-spectrum beta-lactamase CTX-M-15 and plasmid-encoded DHA-1 cephalosporinase. Int J Antimicrob Agents. 2010;35:265–268. doi:10.1016/j.ijantimicag.2009.10.021
10. Fuste E, Lopez-Jimenez L, Segura C, et al. Carbapenem-resistance mechanisms of multidrug-resistant Pseudomonas aeruginosa. J Med Microbiol. 2013;62:1317–1325. doi:10.1099/jmm.0.058354-0
11. Muscarella LF. Risk of transmission of carbapenem-resistant Enterobacteriaceae and related “superbugs” during gastrointestinal endoscopy. World J Gastrointest Endosc. 2014;6:457–474. doi:10.4253/wjge.v6.i10.457
12. Logan LK, Weinstein RA. the epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215:S28–S36. doi:10.1093/infdis/jiw282
13. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–1798. doi:10.3201/eid1710.110655
14. Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi:10.1128/CMR.00001-07
15. Mach T, Neves P, Spiga E, et al. Facilitated permeation of antibiotics across membrane channels–interaction of the quinolone moxifloxacin with the OmpF channel. J Am Chem Soc. 2008;130:13301–13309. doi:10.1021/ja803188c
16. Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009;1794:808–816. doi:10.1016/j.bbapap.2008.11.005
17. Choi U, Lee CR. Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front Microbiol. 2019;10:953. doi:10.3389/fmicb.2019.00953
18. Satlin MJ, Chen L, Patel G, et al. Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant Enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob Agents Chemother. 2017;61. doi:10.1128/AAC.02349-16
19. Tangden T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med. 2015;277:501–512. doi:10.1111/joim.12342
20. CLSI. Performance standard for antimicrobial susceptibility testing. In: CLSI Supplement M100.
21. Spengler G, Kincses A, Gajdacs M, et al. New roads leading to old destinations: efflux pumps as targets to reverse multidrug resistance in bacteria. Molecules. 2017;22:468. doi:10.3390/molecules22030468
22. Zhang X, Zhang Y, Wang F, et al. Unravelling mechanisms of nitrofurantoin resistance and epidemiological characteristics among Escherichia coli clinical isolates. Int J Antimicrob Agents. 2018;52:226–232. doi:10.1016/j.ijantimicag.2018.04.021
23. Hunter SB, Vauterin P, Lambert-Fair MA, et al. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. 2005;43:1045–1050. doi:10.1128/JCM.43.3.1045-1050.2005
24. Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi:10.1128/JCM.33.9.2233-2239.1995
25. Nicolau DP, Carmeli Y, Crank CW, et al. Carbapenem stewardship: does ertapenem affect Pseudomonas susceptibility to other carbapenems? A review of the evidence. Int J Antimicrob Agents. 2012;39:11–15. doi:10.1016/j.ijantimicag.2011.08.018
26. Gupta V, Ye G, Olesky M, et al. National prevalence estimates for resistant Enterobacteriaceae and Acinetobacter species in hospitalized patients in the United States. Int J Infect Dis. 2019;85:203–211. doi:10.1016/j.ijid.2019.06.017
27. Khosravi AD, Taee S, Dezfuli AA, et al. Investigation of the prevalence of genes conferring resistance to carbapenems in Pseudomonas aeruginosa isolates from burn patients. Infect Drug Resist. 2019;12:1153–1159. doi:10.2147/IDR.S197752
28. Hoang CQ, Nguyen HD, Vu HQ, et al. Emergence of New Delhi Metallo-Beta-Lactamase (NDM) and Klebsiella pneumoniae Carbapenemase (KPC) Production by Escherichia coli and Klebsiella pneumoniae in Southern Vietnam and Appropriate Methods of Detection: a Cross-Sectional Study. Biomed Res Int. 2019;2019:9757625. doi:10.1155/2019/9757625
29. Gong X, Zhang J, Su S, et al. Molecular characterization and epidemiology of carbapenem non-susceptible Enterobacteriaceae isolated from the Eastern region of Heilongjiang Province, China. BMC Infect Dis. 2018;18:417. doi:10.1186/s12879-018-3294-3
30. Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi:10.1128/AAC.45.4.1151-1161.2001
31. Oteo J, Perez-Vazquez M, Bautista V, et al. The spread of KPC-producing Enterobacteriaceae in Spain: WGS analysis of the emerging high-risk clones of Klebsiella pneumoniae ST11/KPC-2, ST101/KPC-2 and ST512/KPC-3. J Antimicrob Chemother. 2016;71:3392–3399. doi:10.1093/jac/dkw321
32. Sekizuka T, Yatsu K, Inamine Y, et al. Complete genome sequence of a blaKPC-2-positive Klebsiella pneumoniae strain isolated from the effluent of an urban sewage treatment plant in Japan. mSphere. 2018;3.doi. doi:10.1128/mSphere.00314-18
33. Huang J, Ding H, Shi Y, et al. Further spread of a blaKPC-harboring untypeable plasmid in Enterobacteriaceae in China. Front Microbiol. 2018;9:1938. doi:10.3389/fmicb.2018.01938
34. Tseng SP, Wang SF, Ma L, et al. The plasmid-mediated fosfomycin resistance determinants and synergy of fosfomycin and meropenem in carbapenem-resistant Klebsiella pneumoniae isolates in Taiwan. J Microbiol Immunol Infect. 2017;50:653–661. doi:10.1016/j.jmii.2017.03.003
35. Liang WJ, Liu HY, Duan GC, et al. Emergence and mechanism of carbapenem-resistant Escherichia coli in Henan, China, 2014. J Infect Public Health. 2018;11:347–351. doi:10.1016/j.jiph.2017.09.020
36. Chang YT, Siu LK, Wang JT, et al. Resistance mechanisms and molecular epidemiology of carbapenem-nonsusceptible Escherichia coli in Taiwan, 2012–2015. Infect Drug Resist. 2019;12:2113–2123. doi:10.2147/IDR.S208231
37. Albiger B, Glasner C, Struelens MJ, et al. Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill. 2015;20. doi:10.2807/1560-7917.ES.2015.20.45.30062
38. Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. doi:10.1128/AAC.00774-09
39. Dong F, Lu J, Wang Y, et al. A five-year surveillance of carbapenemase-producing Klebsiella pneumoniae in a pediatric hospital in china reveals increased predominance of NDM-1. Biomed Environ Sci. 2017;30:562–569. doi:10.3967/bes2017.075
40. Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi:10.1016/S1473-3099(10)70143-2
41. Walsh TR. Emerging carbapenemases: a global perspective. Int J Antimicrob Agents. 2010;36(Suppl 3):S8–S14. doi:10.1016/S0924-8579(10)70004-2
42. Groundwater PW, Xu S, Lai F, et al. New Delhi metallo-beta-lactamase-1: structure, inhibitors and detection of producers. Future Med Chem. 2016;8:993–1012. doi:10.4155/fmc-2016-0015
43. Nordmann P, Boulanger AE, Poirel L. NDM-4 metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agents Chemother. 2012;56:2184–2186. doi:10.1128/AAC.05961-11
44. Khan S, Ali A, Khan AU. Structural and functional insight of New Delhi Metallo beta-lactamase-1 variants. Future Med Chem. 2018;10:221–229. doi:10.4155/fmc-2017-0143
45. Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011;55:5952–5954. doi:10.1128/AAC.05108-11
46. Zhang F, Xie L, Wang X, et al. Further Spread of blaNDM-5 in Enterobacteriaceae via IncX3 Plasmids in Shanghai, China. Front Microbiol. 2016;7:424. doi:10.3389/fmicb.2016.00424
47. Yang P, Xie Y, Feng P, et al. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother. 2014;58:7548–7552. doi:10.1128/AAC.03911-14
48. Krishnaraju M, Kamatchi C, Jha AK, et al. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J Med Microbiol. 2015;33:30–38. doi:10.4103/0255-0857.148373
49. Hammerum AM, Hansen F, Olesen B, et al. Investigation of a possible outbreak of NDM-5-producing ST16 Klebsiella pneumoniae among patients in Denmark with no history of recent travel using whole-genome sequencing. J Glob Antimicrob Resist. 2015;3:219–221. doi:10.1016/j.jgar.2015.05.003
50. Wailan AM, Paterson DL, Caffery M, et al. Draft Genome Sequence of NDM-5-Producing Escherichia coli Sequence Type 648 and Genetic Context of blaNDM-5 in Australia. Genome Announc. 2015;3. doi:10.1128/genomeA.00194-15
51. Cyoia PS, Koga VL, Nishio EK, et al. Distribution of ExPEC virulence factors, bla CTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front Microbiol. 2018;9:3254. doi:10.3389/fmicb.2018.03254
52. Bush K. Past and Present Perspectives on beta-Lactamases. Antimicrob Agents Chemother. 2018;62. doi:10.1128/AAC.01076-18
53. Sato N, Kawamura K, Nakane K, et al. First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microb Drug Resist. 2013;19:477–482. doi:10.1089/mdr.2013.0061
54. Xie M, Lin D, Chen K, et al. Molecular characterization of Escherichia coli strains isolated from retail meat that harbor blaCTX-M and fosA3 genes. Antimicrob Agents Chemother. 2016;60:2450–2455. doi:10.1128/AAC.03101-15
55. Rosa JF, Rizek C, Marchi AP, et al. Clonality, outer-membrane proteins profile and efflux pump in KPC- producing Enterobacter sp. in Brazil. BMC Microbiol. 2017;17:69. doi:10.1186/s12866-017-0970-1
56. Shi W, Li K, Ji Y, et al. Carbapenem and cefoxitin resistance of Klebsiella pneumoniae strains associated with porin OmpK36 loss and DHA-1 beta-lactamase production. Braz J Microbiol. 2013;44:435–442. doi:10.1590/S1517-83822013000200015
57. Osei Sekyere J, Amoako DG. Carbonyl Cyanide m-Chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front Microbiol. 2017;8:228. doi:10.3389/fmicb.2017.00228
58. Gajdacs M, Urban E. Resistance trends and epidemiology of citrobacter-enterobacter-serratia in urinary tract infections of inpatients and outpatients (RECESUTI): a 10-year survey. Medicina (Kaunas). 2019;55. doi:10.3390/medicina55060285
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


