Back to Journals » Infection and Drug Resistance » Volume 14
Molecular Mechanisms and Epidemiology of Carbapenem-Resistant Enterobacter cloacae Complex Isolated from Chinese Patients During 2004–2018
Authors Liu S, Huang N, Zhou C, Lin Y, Zhang Y, Wang L, Zheng X, Zhou T , Wang Z
Received 2 July 2021
Accepted for publication 2 September 2021
Published 7 September 2021 Volume 2021:14 Pages 3647—3658
DOI https://doi.org/10.2147/IDR.S327595
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Shixing Liu,1 Na Huang,1 Cui Zhou,1 Yishuai Lin,2 Ying Zhang,2 Lingbo Wang,1 Xiangkuo Zheng,1 Tieli Zhou,1 Zhongyong Wang1
1Department of Clinical Laboratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang Province, People’s Republic of China; 2School of Laboratory Medicine and Life Science, Wenzhou Medical University, Wenzhou, Zhejiang Province, People’s Republic of China
Correspondence: Zhongyong Wang; Tieli Zhou
Department of Clinical Laboratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang Province, People’s Republic of China
Tel +86-577-5557-9725
; +86-577-8668-9885
Email [email protected]; [email protected]
Background: The emergence and spread of carbapenem-resistant Enterobacter cloacae complex (ECC) have posed a serious threat to human health worldwide. This study aimed to investigate the molecular mechanism of carbapenem resistance and its prevalence among ECC in China.
Methods: A total of 1314 ECC clinical isolates were collected from the First Affiliated Hospital of Wenzhou Medical University from 2004 to 2018. Sensitivity to antibiotics was determined using the agar dilution method. The production of carbapenemases and the prevalence of resistance-associated genes were investigated using PCR. The expression of outer membrane porin (OMP) genes (ompC/ompF) and cephalosporinase gene ampC was analyzed by quantitative real-time PCR. The effect of efflux pump mechanism on carbapenem resistance was tested. ECC was typed by multilocus sequence typing (MLST).
Results: In this study, 113 carbapenem-nonsusceptible ECC strains were identified. The prevalence rates of carbapenemase genes blaKPC-2 and blaNDM were 12.4% (14/113) and 17.7% (20/113), and that of the extended-spectrum β-lactamase (ESBL) genes blaCTX-M, blaTEM, and blaSHV were 28.3% (32/113), 27.4% (31/113), and 14.2% (16/113), respectively. Among 67 carbapenem-nonsusceptible ECC isolates producing non-carbapenemase, low expression of ompC/ompF and overexpression of ampC were found in 46 and 40 strains, respectively. In addition, the carbapenem resistance was related to the overexpression of the efflux pump in the study. Finally, the 113 carbapenem-nonsusceptible ECC strains were categorized into 39 different sequence types using MLST.
Conclusion: Carbapenem-nonsusceptible ECC strains producing non-carbapenemase were predominant. The low expression of OMP with the overexpression of cephalosporinase or production of ESBLs and overexpression of efflux pump might contribute to the resistance to carbapenem for carbapenem-nonsusceptible ECC strains producing non-carbapenemase. The blaNDM and blaKPC comprised the principal resistance mechanism of carbapenemase-producing ECC in the hospital, causing a threat to public health. Therefore, monitoring programs to prevent the emergence and further spread of antibiotic resistance are urgently needed.
Keywords: carbapenemase, carbapenem-resistant mechanism, Enterobacter cloacae complex, epidemiology, non-carbapenemase
Introduction
The Enterobacter cloacae complex (ECC) is one of the common microorganisms isolated in clinical specimens causing all kinds of infections, like pneumonia, urinary tract infections, and sepsis, in the last few decades.1,2 Multidrug-resistant (MDR) ECC isolates have emerged and spread worldwide with the widespread use of antibiotics.3 With the increase in resistance rates to aminoglycosides, fluoroquinolones, and third-generation cephalosporins, carbapenems, as the last-resort antibiotic, have gradually been used for treating MDR ECC infections.1,4,5 However, the increasing resistance rates of carbapenems have gained special clinical attention.6–8
The mechanisms of carbapenem resistance in ECC are realized by either the acquisition of plasmid-encoded carbapenemase genes and the overexpression of efflux pumps, or, more commonly, the constitutive overexpression of AmpC or production of extended-spectrum β-lactamase (ESBL) combined with disrupted membrane permeability (the decrease in or loss of the outer membrane protein).3,9 ECC is inherently resistant to first- and second-generation cephalosporins because the overexpression of an inducible AmpC β-lactamase is encoded by the chromosome gene ampC.3,10 Moreover, the acquisition of plasmid-mediated ESBL genes, such as blaTEM, blaSHV, blaCTX-M, and so forth, makes ECC resistant to most β-lactam drugs, thus increasing the difficulty in clinical treatment.11–15 Two major categories of carbapenem enzymes, carbapenem-hydrolyzing serine β-lactamases and metallo-β-lactamases, such as KPC, NmcA, IMI, FRI, GES, OXA, VIM, IMP, and NDM, have been identified in carbapenem-resistant Enterobacteriaceae.3 The most common description of KPC and NDM-1 was in ECC isolates.16
Considering the increasing prevalence of carbapenem-nonsusceptible ECC isolates worldwide, longitudinal epidemiological surveillance and resistance mechanism study on the carbapenem-nonsusceptible ECC should be performed to control and prevent the distribution and spread of resistance, which is key to clinical significance in guiding antimicrobial therapy. However, relevant data on the long-term evolution of carbapenem-nonsusceptible ECC are lacking in China. In this study, the epidemiology prevalence and the molecular mechanisms of 113 ECC clinical isolates were characterized for carbapenem resistance during large-scale surveillance in the southeast of China. This study was novel in reporting ECC nonsusceptible to carbapenem antibiotics on a large scale in China.
Materials and Methods
Bacterial Isolates
A total of 1314 ECC clinical isolates were collected from the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) from 2004 to 2018. Antimicrobial susceptibility testing and identification of all isolates were performed using a VITEK ®2 system (bioMérieux, Marcy-l’Étoile, France). The isolates were stored in 30% glycerol at –80°C prior to further analysis. All investigation protocols in this study were approved by the ethics committee of the First Affiliated Hospital of Wenzhou Medical University. Informed consent was waived because this study with observational nature focused mainly on bacteria and involved no interventions to patients.
Minimum Inhibitory Concentration Determination
According to the latest guidelines recommended by the Institute of Clinical and Laboratory Standards (CLSI, 2020), the minimum inhibitory concentrations (MICs) of 15 antimicrobial agents, including aztreonam (ATM), ciprofloxacin (CIP), levofloxacin (LVX), ceftriaxone (CRO), cefepime (FEP), ceftazidime (CAZ), meropenem (MEM), imipenem (IPM), ertapenem (ETP), gentamicin (GEN), tobramycin (TOB), amikacin (AMK), trimethoprim-sulfamethoxazole (SXT), nitrofurantoin (NIT), and colistin (COL), were determined by the agar dilution method. Briefly, the bacteria were suspended in saline to one-tenth the turbidity of the 0.5 McFarland standard. The bacterial suspension was inoculated on the Mueller–Hinton (MH) agar plate containing different drug concentrations using the nail plate. The results were quantified by observing bacterial growth after incubation at 37°C for 16–20 h.17 The MIC determination of colistin was explained by the recommendation of the European Committee on Antimicrobial Susceptibility Testing. Escherichia coli ATCC 25922 was used as the quality control strain for antimicrobial susceptibility testing. All experiments were performed in triplicate.
Detection of Extended-Spectrum Beta-Lactamases
The American Clinical Laboratory Standardization Institute (CLSI) recommended ESBL confirmation test was performed.1 Briefly, a lawn of test bacteria suspension equivalent to 0.5 McFarland turbidity standard solution was swabbed on the surfaces of MHA plates, and then ceftazidime (30 µg) and cefotaxime (30 µg) disks (Kanvax, China) with and without clavulanic acid (10 µg) were seeded within 15 min. All plates were then incubated aerobically at 37°C for 18 h. An isolate was phenotypically confirmed as an ESBL producer when a zone diameter difference of ≥5 mm was observed between both antibiotic disks with clavulanic acid and a similar agent without clavulanic acid.18 The E. coli ATCC 25922 strain was used as the negative control, and the Klebsiella pneumoniae ATCC 700603 strain was used as the positive control.
Detection of Antibiotic Resistance Determinants
The beta-lactamase genes, including carbapenemase genes (blaKPC, blaIMP, blaNDM, blaSPM, blaIMI, blaVIM, blaOXA-23, blaOXA-24, blaOXA-48, blaOXA-58, blaNmc-A, blaFRI-1, blaSME, blaGIM, blaBIC, blaDIM, blaAIM, blaGES, and blaSIM) and extended-spectrum β-lactamase genes (blaTEM, blaSHV, blaCTX-M-1, blaCTX-M-9, and blaCTX-M-14), were identified by polymerase chain reaction (PCR). The primers of all genes are summarized in Table S1. The positive products of PCR amplification were sequenced by Shanghai Genomics Institute Technology Co. Ltd. (Shanghai, China). All sequencing results of the products were analyzed using BLAST searches against the NCBI database (www.ncbi.nlm.nih.gov/BLAST).
Efflux Pump Inhibition Assay
The efflux pump activity of carbapenem-insensitive ECC strains was determined using the efflux pump inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (Sigma, MO, USA). The MICs of ertapenem in the presence or absence of 8 μg/mL CCCP, which did not inhibit the growth of ECC bacteria, were determined by the agar dilution method. In the presence of CCCP, a reduction in carbapenem MIC ≥4 times was defined as the positive phenotype of the efflux pump downregulation.
Quantitative Real-Time PCR for Cephalosporinase AmpC and Outer Membrane Protein
The total RNA of 67 noncarbapenemase-producing ECC isolates was extracted. Then, 500 ng RNA was mixed with the reverse transcription system, and 10 μL of cDNA was obtained using a PrimeScript™ RT Kit (TaKaRa, Japan). Using a CFX-96 touch real-time PCR system, qPCR (Bio-Rad, CA, USA) was performed. Then, 100 ng cDNA, TB Green Premix Ex Taq II (Tli RNaseH Plus) (2×) (TaKaRa), and specific primers (ompC f: 5′-GCGACCAGACCTACATGCGT-3′, r: 5′-TTCGTTCTCACCAGAGTTACCCT-3′, ompF f: 5′-TCCCTGCCCTGCTGGTAG-3′, r: 5′-TAAGTGTTGTCGCCATCGTTG-3′, ampC f: 5′-GCATGGCGGTGGCCGTTAT-′ r: 5′-CTGCTTGCCCGTCAGCTGT-3′) were added to each sample. The cycling conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. The expression levels of outer membrane genes ompC and ompF and cephalosporinase gene ampC were detected by RT-qPCR; the rpoB gene was used as the internal gene. Compared with carbapenem-sensitive Enterobacter cloacae ATCC700323, the target genes were quantified using the comparative threshold cycle 2−ΔΔCt method. All experiments were repeated three times independently and averaged in the calculation of relative expression levels.
Multilocus Sequence Typing
The carbapenem-nonsusceptible ECC isolates were analyzed by amplifying seven housekeeping genes (dnaA, fusA, gyrB, leuS, pyrG, rplB, and rpoB). Sequence types (STs) were assigned by querying against the database available at the Institut Pasteur’s Enterobacter cloacae MLST website (https://pubmlst.org/bigsdb?db=pubmlst_ecloacae_seqdef). Following the genetic similarity diagram using the goeBURST program, the clonal complexes were analyzed to identify the molecular epidemiological relationships.
Results
Bacterial Isolates and Antimicrobial Susceptibility Test
A total of 113/1314 (8.6%) carbapenem-nonsusceptible ECC isolates were determined with imipenem, meropenem, and ertapenem; 99 (7.5%) carbapenem-resistant strains were determined. The carbapenem-resistant ECC isolate was first detected in the hospital in 2010; the resistance rate increased from 2.5% in 2010 to 11.9% in 2018 (Table 1). The results of the antimicrobial susceptibility test of all 113 carbapenem-nonsusceptible isolates are listed in Table 2, which suggested higher resistance to fluoroquinolones, cephalosporins, and monobactams. Further, 63 (55.8%) and 45 (39.8%) ECC isolates were resistant to ciprofloxacin and levofloxacin, respectively; 30 (26.5%) and 35 (31.0%) isolates were resistant to gentamicin and tobramycin, respectively. Of note, 112 (99%) ECC strains were resistant to ceftriaxone. Nevertheless, 7 (6.2%) and 20 (17.7%) isolates were resistant to amikacin and colistin, respectively.
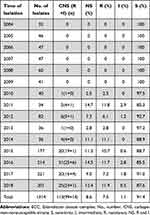 |
Table 1 Carbapenem Susceptibility of ECC Clinical Isolates |
 |  |  |  |
Table 2 Minimum Inhibitory Concentrations (MICs) of 113 Carbapenem-Nonsusceptible ECC Isolates |
Frequency of β-Lactamase Genes
A total of 46 carbapenem-nonsusceptible ECC isolates carried carbapenemase genes (Figure 1). The prevalence rate of blaKPC-2, blaNDM, blaIMP, and blaOXA-23 in carbapenem-nonsusceptible strains was 12.4% (14/113), 17.7% (20/113), 8.0% (9/113), and 3.5% (4/113), respectively, including one isolate carrying blaKPC-2 and blaNDM-1 specially (Figure 1), while blaNmc-A, blaBIC, blaGES, blaAIM, blaGIM, blaDIM, blaSIM, blaOXA-24, blaOXA-48, blaFRI-1, blaOXA-58, blaSME, and blaAIM were not detected in all isolates. Moreover, 49 ECC strains were positive for the ESBL phenotypic test in 113 strains, and the ESBLs genes blaTEM (27.4%, 31/113), blaCTX-M-14 (19.5%, 22/113), blaCTX-M-1 (15.9%, 18/113), blaCTX-M-9 (14.2%, 16/113), and blaSHV (14.2%, 16/113) among analyzed strains were also determined. In general, the harboring of carbapenemase genes and ESBLs accounted for 40.7% (46/113) and 58.4% (66/113), respectively.
Overexpression of the Efflux Pump
The efflux pump inhibition test was performed to explore the effect of the efflux pump on carbapenem resistance. The results showed that 36 strains had a ≥4 times reduction in ertapenem MICs in the presence of 8 μg/mL CCCP, suggesting that the efflux pump had a significant effect on carbapenem resistance.
Outer Membrane Protein Gene Expressions
This study investigated the relationship between resistance to carbapenems and the expression level of outer membrane genes ompC and ompF and cephalosporinase gene ampC. Further, 67 ECC noncarbapenemase-producing isolates and Enterobacter cloacae ATCC700323 as the control strain were used. The results of RT-qPCR showed that the expression level of ompC in 29 ECC isolates decreased compared with that in ATCC 700323, and the decreased expression of ompF was found in 42 ECC strains. In addition, 24 ECC isolates had low expression of both ompC and ompF. The overexpression of cephalosporinase gene ampC was found in 40 ECC strains, with the highest level as 26-fold (Figures 1 and S1). Further, 24 ECC isolates had the overexpression of cephalosporinase gene ampC and low expression of ompC or/and ompF. ESBLs and low-level expression of ompC or/and ompF were detected in 33 ECC isolates (Figure 1).
Multilocus Sequence Typing Analysis
The 113 carbapenem-nonsusceptible ECC isolates were assigned to 39 different STs, including the most prevalent ST190 (14.2%, 16/113), followed by ST114 (4.4%, 5/113), ST93 (4.4%, 5/113), ST97 (3.5%, 4/113), ST78 (3.5%, 4/113), ST32 (2.7%, 3/113), ST46 (1.8%, 2/113), ST120 (1.8%, 2/113), ST523 (1.8%, 2/113), ST557 (1.8%, 2/113), ST1411 (1.8%, 2/113), and ST1439 (1.8%, 2/113); the remaining STs contained 1 strain for each, except 34 novel STs (marked as NEW in Figure 1, currently not registered in the MLST database) as detected by MLST analysis (Figure 1). Also, 39 STs were analyzed for the molecular epidemiological relationships using goeBURST. The result showed that all 39 STs belonged to singletons with no CCs (Figure 2). In addition, of 16 ST190 strains, 4 isolates produced KPC-2, 1 produced NDM-1, 1 produced OXA-23, 1 produced IMP, and other produced ESBLs. All ST114 isolates produced NDM, including three (75%) strains with blaNDM-1, and two produced NDM-5. All ST97 strains produced carbapenemase; three strains carried blaNDM-1 and one blaIMP.
Discussion
Carbapenem is widely used in the treatment and management of MDR Gram-negative bacterial infections in the clinical environment due to its broad-spectrum antibacterial activity.19 However, some surveillance programs reported a significant increase in the resistance of carbapenems, making clinical treatment a great challenge.20,21 In the present study, we collected 1314 ECC clinical isolates from the First Affiliated Hospital of Wenzhou Medical University from 2004 to 2018, and 113/1314 (8.6%) carbapenem-nonsusceptible ECC isolates were determined. The resistance of ECC strains to carbapenems showed a fluctuating upward trend from 2004 to 2018 (Table 1). In addition, the susceptibility of these 113 carbapenem-nonsusceptible ECC strains to commonly used antibacterial drugs showed that the 113 ECC strains showed a trend of multi-drug resistance as a whole. The high resistance to fluoroquinolones, cephalosporins, monobactams, and colistin, but low to amikacin, was found in our study, which corresponded to previous findings.22,23 It is worth noting that colistin resistance rate was as high as 17.7%, which was related to inherent colistin resistance in the genogroups of the ECC.23 Therefore, exploring the mechanism of these strains resistant to carbapenem is important so as to better prevent resistance. As far as I know, it is first time to report the analysis of molecular mechanisms and epidemiology of carbapenem-nonsusceptible ECC isolates with a longer period and large number of strains. The findings might provide a reference for the monitoring and control of carbapenem-nonsusceptible ECC isolates.
The production of carbapenemase is one of the carbapenem resistance mechanisms of ECC.9 New Delhi metallo-β-lactamase (NDM), which is encoded by the gene blaNDM, can lead to resistance to most β-lactam antibiotics, which was first found in New Delhi, India, in 2009.24 blaNDM-1 is prevalent in the Indian region, and multiple blaNDM alleles have been detected in hospitals in eastern China and Czech.6,25,26 The present study also showed the emergence and spread of NDM-1, reflecting a high prevalence since 2012. The KPC enzyme was first discovered in ECC in 2005 and is extremely endemic worldwide now.21,27,28 The dissemination of the ECC that produces KPC has been verified in China.29,30 In addition, a previous study reported that 8.6% KPC-2-producing strains were detected in Chongqing.31 However, our study showed that 12.4% (14/113) of the KPC-2-producing strains were found. VIM enzyme, which was reported in many regions, and OXA-48 enzyme, which was more common in Europe, were not found in the present study.32,33 The study also indicated an increased number of carbapenemase-producing ECC isolates over the past few years, implying that the prevalence of carbapenem-nonsusceptible ECC was dependent on geographic regions and differences in drug use, even within a single country. The blaNDM and blaKPC were prevalent in carbapenemase-producing ECC in the hospital. Therefore, effective measures need to be taken to prevent the incidence.
In addition, ESBL genes were related to increased MIC to carbapenem, which were first identified in ECC in 1989.34,35 Since then, the prevalence of ECC carrying ESBL has increased in hospital settings and in patients previously exposed to antibiotics.36–38 Compared with non-ESBL-producing strains, ESBL-producing ECC showed a higher health risk associated with hospital-acquired infections.39 In this study, 58.6% (66/113) ECC strains produced ESBLs; most of them had more than one ESBL gene. Moreover, the ESBLs gene blaTEM was popular in this study, which was totally different from the predominant blaCTX-M-15 in Bulgaria.40 We found that the carrying rates of ESBLs were higher in carbapenem-resistant ECC than in carbapenem-sensitive ECC, compared with other reports.41
The production of ESBLs or the overexpression of AmpC combined with disrupted membrane permeability (outer membrane protein decreased or loss) was another reason for carbapenem resistance.3,9,42 In this study, 40 of 67 strains producing non-carbapenemase had the overexpression of AmpC or produced ESBLs combined with a decrease in the outer membrane proteins (Figure 1 and Table S2). Of the remaining 27 strains, 10 strains including 4 isolates with only decreased expression of ompC or/and ompF, 3 isolates with only overexpression of ampC, 2 isolates only producing ESBLs, and 1 isolate with ESBL and overexpression of ampC were intermediate to carbapenem, which correspond to a previous report that the overexpression of ampC genes encoding cephalosporinase, leading to ECC strains resistant to first- and second-generation cephalosporins, and the disrupted membrane permeability (outer membrane protein decreased or loss) slightly increased MIC to carbapenem but did not lead to resistance.35 However, 15 other ertapenem-resistant strains were also found in the aforementioned mechanism combinations. We suspected that these combinations might increase the MIC of these strains to carbapenem drugs and even reach the level of resistance (Table S2), which still requires further exploration.
Several studies reported the effect of the efflux pump on the carbapenem resistance of ertapenem-insensitive ECC strains.9,42 The present study indicated that CCCP, an efflux pump inhibitor, decreased the MIC of 36 ECC strains to carbapenem, suggesting that efflux pumps played an important role in carbapenem resistance. However, the expression of the efflux pump acrB revealed no difference in ertapenem-resistant ECC strains, suggesting the presence of an additional unknown efflux pump influencing ertapenem resistance.9
The results of MLST analysis suggested that the distribution of STs of all carbapenem-nonsusceptible ECC isolates was diversified (Figure 2). ST190 was the most prevalent isolate in the hospital; a small-scale explosion in 2016 (Figures 1 and 2), different from previous studies, revealed that ST66, ST78, ST108, and ST114 were the most prevalent and widespread ECC STs.35,43 The present study found that ST190 isolates producing carbapenemase and various other β-lactamase profiles with a higher risk might cause severe drug-resistant outbreaks in the hospital. ST78 and ST114 producing carbapenemase were major international clones, which were worth noting and reminded us of the spread of these strains.
In conclusion, this study summarized the resistance mechanisms and molecular epidemiology of carbapenem-nonsusceptible ECC strains in the hospital from 2004 to 2018. This was the first time that ECC nonsusceptible to carbapenem antibiotics was reported on a large scale in China. The increasing rates of resistance to antibiotics have further aggravated the threat to human health because of limited treatment options. ECC isolates that do not produce carbapenemase are predominant in carbapenem-nonsusceptible ECC isolates. Carbapenem resistance is mediated by the overexpression of efflux pumps, or, more commonly, through the acquisition of constitutive overexpression of AmpC or ESBL combined with a decrease in the outer membrane proteins. The resistance of carbapenemase-producing ECC isolates is conferred through the acquisition of carbapenemase genes and the overexpression of efflux pumps. As carbapenem antibiotics are gradually applied as an effective treatment option, monitoring programs to prevent the emergence and further spread of antibiotic resistance are urgently needed.
Ethics Approval
The need for ethics approval and consent was deemed unnecessary in this study according to the ethics committee of the First Affiliated Hospital of Wenzhou Medical University.
Acknowledgments
We thank the Health Department of Zhejiang Province of the People’s Republic of China (no. 2011KYA106) for providing financial funding.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the manuscript; gave final approval of the version to be published; have agreed on the journal to which the manuscript has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest.
References
1. Sanders WE
2. Davin-Regli A, Pages JM. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol. 2015;6:392. doi:10.3389/fmicb.2015.00392
3. Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front Microbiol. 2019;10:44. doi:10.3389/fmicb.2019.00044
4. Huang S, Dai W, Sun S, et al. Prevalence of plasmid-mediated quinolone resistance and aminoglycoside resistance determinants among carbapenem non-susceptible Enterobacter cloacae. PLoS One. 2012;7(10):e47636. doi:10.1371/journal.pone.0047636
5. Corkill JE, Anson JJ, Hart CA. High prevalence of the plasmid-mediated quinolone resistance determinant qnrA in multidrug-resistant Enterobacteriaceae from blood cultures in Liverpool, UK. J Antimicrob Chemother. 2005;56(6):1115–1117. doi:10.1093/jac/dki388
6. Jin C, Zhang J, Wang Q, et al. Molecular characterization of carbapenem-resistant Enterobacter cloacae in 11 Chinese cities. Front Microbiol. 2018;9:1597. doi:10.3389/fmicb.2018.01597
7. Raimondi A, Traverso A, Nikaido H. Imipenem- and meropenem-resistant mutants of Enterobacter cloacae and proteus rettgeri lack porins. Antimicrob Agents Chemother. 1991;35(6):1174–1180. doi:10.1128/aac.35.6.1174
8. Galani I, Souli M, Chryssouli Z, et al. Characterization of a new integron containing bla(VIM-1) and aac(6ʹ)-IIc in an Enterobacter cloacae clinical isolate from Greece. J Antimicrob Chemother. 2005;55(5):634–638. doi:10.1093/jac/dki073
9. Yang FC, Yan JJ, Hung KH, et al. Characterization of ertapenem-resistant Enterobacter cloacae in a Taiwanese university hospital. J Clin Microbiol. 2012;50(2):223–226. doi:10.1128/JCM.01263-11
10. Boyd DA, Mataseje LF, Davidson R, et al. Enterobacter cloacae complex isolates harboring bla NMC-A or bla IMI-type class a carbapenemase genes on novel chromosomal integrative elements and plasmids. Antimicrob Agents Chemother. 2017;61(5). doi:10.1128/AAC.02578-16
11. Van Maerken T, De Brabandere E, Noel A, et al. A recurrent and transesophageal echocardiography-associated outbreak of extended-spectrum beta-lactamase-producing Enterobacter cloacae complex in cardiac surgery patients. Antimicrob Resist Infect Control. 2019;8(1):152. doi:10.1186/s13756-019-0605-4
12. Arpin C, Labia R, Dubois V, et al. TEM-80, a novel inhibitor-resistant β-lactamase in a clinical isolate of Enterobacter cloacae. Antimicrob Agents Chemother. 2002;46(5):1183–1189. doi:10.1128/aac.46.5.1183-1189.2002
13. Bornet C, Davin-Regli A, Bosi C, et al. Imipenem resistance of Enterobacter aerogenes mediated by outer membrane permeability. J Clin Microbiol. 2000;38(3):1048–1052. doi:10.1128/JCM.38.3.1048-1052.2000
14. Pitout JD, Moland ES, Sanders CC, et al. Beta-lactamases and detection of beta-lactam resistance in Enterobacter spp. Antimicrob Agents Chemother. 1997;41(1):35–39. doi:10.1128/AAC.41.1.35
15. Szabo D, Melan MA, Hujer AM, et al. Molecular analysis of the simultaneous production of two SHV-type extended-spectrum beta-lactamases in a clinical isolate of Enterobacter cloacae by using single-nucleotide polymorphism genotyping. Antimicrob Agents Chemother. 2005;49(11):4716–4720. doi:10.1128/AAC.49.11.4716-4720.2005
16. Chavda KD, Chen L, Fouts DE, et al. Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. mBio. 2016;7(6):e02093–e02116. doi:10.1128/mBio.02093-16
17. Fernandes CJ, O’Sullivan MV, Cai Y, et al. Agar dilution method for detection of inducible clindamycin resistance in Staphylococcus spp. J Clin Microbiol. 2007;45(12):4018–4020. doi:10.1128/JCM.01158-07
18. Silago V, Kovacs D, Samson H, et al. Existence of multiple ESBL genes among phenotypically confirmed ESBL producing Klebsiella pneumoniae and Escherichia coli concurrently isolated from clinical, colonization and contamination samples from neonatal units at Bugando Medical Center, Mwanza, Tanzania. Antibiotics. 2021;10:476. doi:10.3390/antibiotics10050476
19. Nicolau DP, Carmeli Y, Crank CW, et al. Carbapenem stewardship: does ertapenem affect Pseudomonas susceptibility to other carbapenems? A review of the evidence. Int J Antimicrob Agents. 2012;39(1):11–15. doi:10.1016/j.ijantimicag.2011.08.018
20. Gupta V, Ye G, Olesky M, et al. National prevalence estimates for resistant Enterobacteriaceae and Acinetobacter species in hospitalized patients in the United States. Int J Infect Dis. 2019;85:203–211. doi:10.1016/j.ijid.2019.06.017
21. Wilson BM, El Chakhtoura NG, Patel S, et al. Carbapenem-resistant Enterobacter cloacae in Patients from the US veterans health administration, 2006–2015. Emerg Infect Dis. 2017;23(5):878–880. doi:10.3201/eid2305.162034
22. Pot M, Reynaud Y, Couvin D, et al. Wide distribution and specific resistance pattern to third-generation cephalosporins of Enterobacter cloacae complex members in humans and in the environment in Guadeloupe (French West Indies). Front Microbiol. 2021;12:628058. doi:10.3389/fmicb.2021.628058
23. Mushtaq S, Reynolds R, Gilmore MC, et al. Inherent colistin resistance in genogroups of the Enterobacter cloacae complex: epidemiological, genetic and biochemical analysis from the BSAC Resistance Surveillance Programme. J Antimicrob Chemother. 2020;75(9):2452–2461. doi:10.1093/jac/dkaa201
24. Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-β-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi:10.1128/AAC.00774-09
25. Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi:10.1093/cid/ciy660
26. Paskova V, Medvecky M, Skalova A, et al. Characterization of NDM-encoding plasmids from Enterobacteriaceae recovered from Czech hospitals. Front Microbiol. 2018;9:1549. doi:10.3389/fmicb.2018.01549
27. Park SO, Liu J, Furuya EY, et al. Carbapenem-resistant Klebsiella pneumoniae infection in Three New York city hospitals trended downwards from 2006 to 2014. Open Forum Infect Dis. 2016;3(4):ofw222. doi:10.1093/ofid/ofw222
28. Bratu S, Mooty M, Nichani S, et al. Emergence of KPC-possessing Klebsiella pneumoniae in Brooklyn, New York: epidemiology and recommendations for detection. Antimicrob Agents Chemother. 2005;49(7):3018–3020. doi:10.1128/AAC.49.7.3018-3020.2005
29. Zhao Y, Zhang J, Fu Y, et al. Molecular characterization of metallo-beta-lactamase- producing carbapenem-resistant Enterobacter cloacae complex isolated in Heilongjiang Province of China. BMC Infect Dis. 2020;20(1):94. doi:10.1186/s12879-020-4768-7
30. Huang J, Ding H, Shi Y, et al. Further spread of a blaKPC-harboring untypeable plasmid in Enterobacteriaceae in China. Front Microbiol. 2018;9:1938. doi:10.3389/fmicb.2018.01938
31. Dai W, Sun S, Yang P, et al. Characterization of carbapenemases, extended spectrum beta-lactamases and molecular epidemiology of carbapenem-non-susceptible Enterobacter cloacae in a Chinese hospital in Chongqing. Infect Genet Evol. 2013;14:1–7. doi:10.1016/j.meegid.2012.10.010
32. Albiger B, Glasner C, Struelens MJ, et al. Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill. 2015;20(45):30062. doi:10.2807/1560-7917.ES.2015.20.45.30062
33. Bitar I, Papagiannitsis CC, Kraftova L, et al. Detection of five mcr-9 –carrying Enterobacterales isolates in four Czech hospitals. mSphere. 2020;5(6):e01008–e01020. doi:10.1128/mSphere.01008-20
34. De Champs C, Sauvant MP, Chanal C, et al. Prospective survey of colonization and infection caused by expanded-spectrum-beta-lactamase-producing members of the family Enterobacteriaceae in an intensive care unit. J Clin Microbiol. 1989;27(12):2887–2890. doi:10.1128/JCM.27.12.2887-2890.1989
35. Cai Y, Chen C, Zhao M, et al. High prevalence of metallo-beta-lactamase-producing Enterobacter cloacae from three tertiary hospitals in China. Front Microbiol. 2019;10:1610. doi:10.3389/fmicb.2019.01610
36. Kluytmans-van den Bergh MF, Rossen JW, Bruijning-Verhagen PC, et al. Whole-genome multilocus sequence typing of extended-spectrum-beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2016;54(12):2919–2927. doi:10.1128/JCM.01648-16
37. Jean SS, Hsueh PR, Group SA-P. Distribution of ESBLs, AmpC beta-lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008–14: results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). J Antimicrob Chemother. 2017;72:166–171. doi:10.1093/jac/dkw398
38. Peirano G, Matsumura Y, Adams MD, et al. Genomic epidemiology of global carbapenemase-producing Enterobacter spp., 2008–2014. Emerg Infect Dis. 2018;24(6):1010–1019. doi:10.3201/eid2406.171648
39. Cyoia PS, Koga VL, Nishio EK, et al. Distribution of ExPEC Virulence Factors, bla CTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front Microbiol. 2018;9:3254. doi:10.3389/fmicb.2018.03254
40. Markovska R, Stoeva T, Dimitrova D, et al. Quinolone resistance mechanisms among third-generation cephalosporin resistant isolates of Enterobacter spp. in a Bulgarian university hospital. Infect Drug Resist. 2019;12:1445–1455. doi:10.2147/IDR.S204199
41. Hoffmann H, Sturenburg E, Heesemann J, et al. Prevalence of extended-spectrum beta-lactamases in isolates of the Enterobacter cloacae complex from German hospitals. Clin Microbiol Infect. 2006;12(4):322–330. doi:10.1111/j.1469-0691.2006.01360.x
42. Szabo D, Silveira F, Hujer AM, et al. Outer membrane protein changes and efflux pump expression together may confer resistance to ertapenem in Enterobacter cloacae. Antimicrob Agents Chemother. 2006;50(8):2833–2835. doi:10.1128/AAC.01591-05
43. Izdebski R, Baraniak A, Herda M, et al. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother. 2015;70(1):48–56. doi:10.1093/jac/dku359
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


