Back to Journals » Infection and Drug Resistance » Volume 14
Molecular Epidemiology of mcr-1, blaKPC-2, and blaNDM-1 Harboring Clinically Isolated Escherichia coli from Pakistan
Authors Bilal H , Rehman TU, Khan MA, Hameed F, Jian ZG, Han J, Yang X
Received 20 January 2021
Accepted for publication 11 March 2021
Published 16 April 2021 Volume 2021:14 Pages 1467—1479
DOI https://doi.org/10.2147/IDR.S302687
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Hazrat Bilal,1 Tayyab Ur Rehman,2 Muhammad Asif Khan,2 Fareeha Hameed,2 Zhang Gao Jian,1 Jianxiong Han,1 Xingyuan Yang1
1Faculty of Health Sciences, Institute of Physical Sciences and Information Technology, Anhui University, Hefei, People’s Republic of China; 2Institute of Basic Medical Sciences, Khyber Medical University, Peshawar, Pakistan
Correspondence: Xingyuan Yang
Faculty of Health Sciences, Institute of Physical Science and Information Technology, Anhui University, No, 111 Jiulong Road, Hefei, Anhui, 230601, People’s Republic of China
Email [email protected]
Purpose: The multiple-drug resistant Escherichia coli are among the deadliest pathogens causing life-threatening infections. This study was planned to determine the molecular epidemiology of mcr-1, blaKPC-2, and blaNDM-1 harboring clinically isolated E. coli from Pakistan.
Methods: In total, 545 strains of E. coli from clinical samples were collected from June 2018 to September 2019. All the isolates were screened for colistin-resistance, extended-spectrum-β-lactamases (ESBL), and carbapenemases through the micro-dilution method, Double-Disk-Synergy-Test (DDST), and Modified-Hodge-Test (MHT). The detection, sequence-typing, conjugal transfer, S1-PFGE, plasmid-replicon-typing, and southern-blotting for mcr, ESBL, and carbapenemase-encoding genes were performed.
Findings: A total of four (0.73%) colistin-resistant strains carrying alongside mcr-1 and blaCTX-M-15 genes, three of these strains also had the blaTEM-1 gene. The presence of ESBL genes was detected in 139 (25.5%) isolates harboring blaCTXM-15 (74.82%), blaTEM (34.53%), blaSHV (28.06%) and blaOXA-1 (28.78%). In 129 carbapenemase-producers, 35.83% possessed blaNDM-1, 26.67% blaKPC-2, 8.3% blaOXA-48, 25% blaVIM-1, and 20.83% blaIMP-1 genes. The sequence typing revealed that mcr-1 harboring isolates belonged to ST405, ST117, and ST156. Fifty percent of blaKPC-2 and 48.83% of blaNDM-1 were found on ST131 and ST1196, respectively. Two rare types of STs, ST7584, and ST8671 were also identified in this study. The mcr-1 gene was located on Incl2 (60-kb) plasmid. The blaKPC-2 was present on (140-kb) IncH12, (100-kb) IncN, (90-kb) Incl1, while blaNDM-1 was located on (70-kb) IncFIIK, (140-kb) IncH12, (100-kb) IncN, (60-kb) IncA/C, and (45-kb) IncFII plasmids, which were successfully trans-conjugated. Among the plasmid types, the Incl1 carrying blaKPC-2, IncH12 harboring blaKPC-2 and blaNDM-1, and IncFIIK carrying blaNDM-1 were for the first time detected in Pakistan.
Conclusion: The mcr-1, blaKPC-2, and blaNDM-1 genes finding in various clonal and plasmids types indicate that a substantial selection of the resistance genes had occurred in our clinical strains.
Keywords: colistin-resistance, extended-spectrum-β-lactamases, carbapenemases, mcr1, blaKPC-2, blaNDM-1, plasmids, E. coli
Introduction
Antimicrobial resistance is a grave concern globally, where the situation is continuously getting worse due to overuse and misuse of antibiotics. Various mechanisms of antimicrobial resistance have been established by pathogens, of which extended-spectrum-β-lactamases (ESBL) enzymes production is the most prominent one.
Carbapenems are considered effective drugs against most ESBL producing pathogens, but due to their extensive practice, various resistance mechanisms have been developed in which carbapenemase production is most threatening.
Colistin is considered the last optional therapy against multiple drug-resistant (MDR) pathogens.
To combat multiple-drugs-resistant superbugs, continuous surveillance studies and molecular typing of bacterial strains, and plasmid typing are required for the current depiction of resistance epidemiology. Aiming this, we in the current study screened 545 E. coli to determine the prevalence of colistin resistance mcr, ESBL, and carbapenemases-encoding genes in Pakistan. We performed antimicrobial susceptibility testing for all isolates. Sequence and plasmid replicon typing, S1-PFGE, and southern blotting were carried out for mcr-1, blaKPC-2, and blaNDM-1 harboring isolates.
Methods
Samples Collection and Species Identification
A total of 545 clinically isolated E. coli were obtained from the microbiology laboratory of the Pakistan Institute of Medical Sciences (PIMS), located in Islamabad, the capital city of Pakistan. PIMS is a government-assisted tertiary care hospital, and Pakistan is a developing country with half of the population living below the poverty line. Therefore, patients approached to PIMS hospital is comparatively high due to their low-cost treatment services, indicating the locality’s health status.
The inclusion criteria for sampling were the entire routine processing samples having E. coli as a bacterial pathogen collected from June 2018 to September 2019 in the microbiology laboratory of PIMS Islamabad, Pakistan. According to laboratory records, the strains were isolated from stool (n=18), wound (n=73), blood (n=109), and urine (n=345) as per standard microbiological and biochemical procedures such as colony morphology, gram staining, and API 20E strip test (BioMérieux).
Antibiotics Sensitivity Testing, Carbapenemase, MBL, and ESBL Detection
The broth microdilution method for colistin, amikacin ampicillin, cefotaxime, aztreonam, chloramphenicol, gentamycin, ciprofloxacin, cefoxitin, fosfomycin, tetracycline, and meropenem, was performed to assist antibiotic sensitivity of all collected strains. The minimum inhibitory concentrations for fosfomycin-resistant isolates were confirmed via the agar dilution method according to CLSI guidelines. The carbapenemases and ESBLs producing ability of all the isolates were determined by modified-Hodge-test (MHT) and double-desk-synergy-test (DDST). All of the MHT positive isolates were subjected to Metallo-β-lactamases (MBL) detection via a combined-disc-test (CDT). CLSI guidelines were used for results interpretations.
Antibiotic Resistance Genes
The genomic DNA from all isolates was extracted using the conventional boiling method.
Molecular Typing
To find out the sequence type (ST) and ST complex of mcr-1, blaKPC-2, and blaNDM-1 harboring isolates, multi-locus-sequence-typing (MLST) were carried out. The seven alleles, adk, fumC, gyrB, icd, mdh, purA, and recA, were amplified and sequenced. The sequencing results were analysed on the MLST database to find out the sequence types of our isolates (http://enterobase.warwick.ac.uk/species/index/ecoli). The phylogenetic analysis based on MLST sequence types and alleles was performed on BioNumerics (Applied math, Belgium).
Trans-Conjugation and Plasmid Replicon Typing
To identify the conjugation-ability of mcr-1, blaKPC-2, and blaNDM-1 bearing strains, trans-conjugation was carried out. The E. coli EC 600 (NalR, RifR) was selected as recipients, and mcr-1, blaKPC-2, and blaNDM-1 bearing strains were taken as donors. The sensitivity of all the donor strains was checked for rifampicin. Colistin (2 mg/L) and rifampicin (600 mg/L) were used to select mcr-1 carrying conjugants. Similarly, meropenem (4 mg/L) and rifampicin (600 mg/L) were used to select blaKPC-2 and blaNDM-1 bearing conjugants. To run the experiment, a protocol already evaluated and published has been used.
To explore the incompatibility types of plasmids bearing mcr-1, blaKPC-2, and blaNDM-1, PCR-based replicon typing (PBRT) was performed. For this experiment, plasmid DNA from successful transconjugants was obtained by alkaline-lysis procedure.
S1-PFGE and Southern Hybridization
To determine the sizes of plasmid, S1 Pulse Field Gel Electrophoresis (PFGE) of the trans-conjugants were performed following the protocol.
Genetic Context of mcr-1, PCR Mapping
The genetic context of mcr-1 was determined by PCR mapping following the published data of pHNSHP45 and mcr-1 harboring Incl2 plasmid detected in healthy broiler from Pakistan.
Results
Bacterial Isolation and Antibiotic Susceptibility Profile
A total of 545 E. coli strains were confirmed via colony morphology and 16S rDNA sequence blast. The antibiotic susceptibility testing of isolates for twelve antibiotics was performed through microdilution (Figure 1). Colistin sulphate and fosfomycin were the most susceptible drugs, with 99.27% and 93.03% susceptibility. Only four (0.73%) isolates showed resistance to colistin.
 |
Figure 1 Antibiotic resistance profile of all tested E. coli isolates. |
ESBL and Carbapenemases Producing Isolates
The DDST and MHT were performed for all isolates to determine the prevalence of ESBL and carbapenemases-producing strains. Among 545 tested isolates, 139 (25.5%) and 120 (22.02%) isolates were ESBL positive and carbapenemases producers, respectively, based on DDST and MHT results from interpretation. Furthermore, to determine the prevalence of MBL among MHT positive isolates, the CDT was carried out. The results revealed that 80 (66.67%) out of 120 carbapenemases producing strains were also MBL positive. The DDST, MHT, and CDT revealed that four colistin-resistant isolates were ESBL positive but not carbapenemases producers.
Antibiotic-Resistant Genes
Upon molecular confirmation of colistin resistance through PCR, a total of four (0.73%) colistin-resistant strains carrying alongside mcr-1 and blaCTX-M-15 genes, three of these strains also had the blaTEM-1 gene (Table 1).
 |
Table 1 Antimicrobial Susceptibility Profile of mcr-1 Harbored Isolates |
Among 139 ESBL producer strains, 74.82% (104 of 139) harbored blaCTXM-15 gene, 34.53% (48 of 139) carried blaTEM gene (n=29 blaTEM-1, n=19 blaTEM-116), 28.78% (40 of 139) contained blaOXA-1 gene and 5.75% (8 of 139) harbored blaSHV-75 gene. The blaSHV-11, a non ESBL variant of SHV was identified in 31 isolates. In 129 carbapenemase-producers, 35.83% (43 of 129) possessed blaNDM-1, 26.67% (32 of 129) blaKPC-2, 8.3% (10 of 129) blaOXA-48, 25% (30 of 129) blaVIM-1, and 20.83% (25 of 129) possessed blaIMP-1 gene. The 51.07% (71 0f 139) ESBL positive isolates were also positive for carbapenemases. Co-harboring ESBL and carbapenem-resistant genes were detected in these isolates. The prevalence of the co-occurrence of blaNDM-1, blaCTXM-15, and blaOXA-1 were 16.9% (12 of 71), blaNDM-1 and blaCTXM-15 were 15.49% (11 of 71), blaIMP-1, blaCTXM-15, and blaTEM-1 were 11.27% (8 of 71), blaIMP-1 and blaOXA-1 were 11.27% (8 of 71), blaKPC-2, blaCTXM-15, and blaTEM were 9.86% (7 of 71), blaVIM-1 and blaSHV were 8.45% (6 of 71), blaKPC-2 and blaCTXM-15 were 7.04% (5 of 71), blaKPC-2, blaTEM, and blaSHV were 5.63% (4 of 71), blaOXA-48 and blaSHV were 5.63% (4 of 71), blaVIM-1 and blaCTXM-15 were 2.82% (2 of 71), blaKPC-2, blaVIM-1 and blaTEM were 2.82% (2 of 71), blaNDM-1, blaOXA-48, and blaTEM were 2.82% (2 of 71%) (Table 2).
 |
Table 2 Prevalence of Solo or in Co-Existence of Antibiotic Resistance Genes Detected in This Study |
Molecular Typing
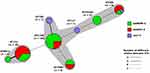
The MLST was performed according to Warwick MLST Database to find out the sequence types of mcr-1, blaKPC-2, and blaNDM-1 harboring isolates. The clonality of mcr-1, blaKPC-2, and blaNDM-1 carrying isolates was assessed as a minimum spanning tree based on MLST sequence types and alleles (Figure 2). According to MLST results, the sequence types for mcr-1 harboring isolates were ST156 (n=2), ST117 (n=1), and ST405 (n=1). The sequence types of blaKPC-2 harboring isolates were ST131 (n=16), ST1196 (n=7), ST405 (n=5), and ST7584 (n=4). Likewise, the sequence types of blaNDM-1 harboring isolates were ST1196 (n=21), ST131 (n=9), ST10184 (n=6), ST7584 (n=4), ST405 (n=2) ST8671 (n=1) (Table S2, supplementary data). Overall, the MLST result indicates the diversity in clonal type for mcr-1, blaKPC-2, and blaNDM-1 harboring isolates.
Transconjugation and PBRT
The transconjugation experiment was performed to confirm the transferability and localization of mcr-1, blaKPC-2, and blaNDM-1 genes on plasmids. The conjugation experiments succeeded for all isolates except for five ST131 strains carrying blaKPC-2. The transconjugants were evaluated for antibiotic sensitivity testing and detection of relevant resistant genes via PCR. In mcr-1 harboring transconjugants, only the mcr-1 gene was detected; however, PCR did not detect the ESBL genes. Similarly, the transconjugants of blaKPC-2, and blaNDM-1 showed two to four-fold increases in meropenem MIC than donor strains, though remaining resistance profiles were similar to that of the donor strains (Table S3, supplementary data).
To find out the plasmids type for blaKPC-2, and blaNDM-1, and mcr-1, the PBRT was carried out from the trans-conjugants strains. The Incl2 type plasmid was detected in all the mcr-1 harboring isolates. The plasmids incompatibility types for blaKPC-2 harboring isolates were IncH12 (n=15), IncN (n=8), and Incl1 (n=4). For blaNDM-1 harboring isolates were IncFIIK (n=17), IncH12 (n=14), IncN (n=5), IncA/C (n=4), and IncFII (n=1) (Table 3). Among the plasmid types, the Incl1 carrying blaKPC-2, IncH12 harboring blaKPC-2, and blaNDM-1, and IncFIIK carrying blaNDM-1 were for the first time detected in Pakistan.
 |
Table 3 Number of mcr-1, blaKPC-2, and blaNDM-1 Harboring Plasmids Incompatibility Type Prevalent on Various Sequence Types |
S1 PFGE and Southern Hybridization
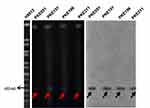
S1 PFGE and southern- hybridization were performed for four mcr-1 transconjugants, five blaKPC-2 transconjugants (two having Incl1 plasmid, two having IncN plasmid, and one having IncH12 plasmid), and five blaNDM-1 (one from each type of plasmid). The S1 PFGE and southern-blotting results revealed that all the mcr-1 harboring transconjugants have a plasmid size near 60-kb (Figure 3). Similarly, plasmids sizes near to 90-kb (Incl1), 100-kb (IncN) and 140-kb (IncH12) were detected for blaKPC-2 (Figure 4). The plasmid sizes for blaNDM-1 were 70-kb (IncFIIK), 140-kb (IncH12), 100-kb (IncN), 60-kb (IncA/C), and 45-kb (IncFII) (Figure 5).
mcr-1 Genetic Context, PCR Mapping
For PCR mapping of mcr-1 genetic background, seven genes closed to mcr-1 were amplified and sequenced. Among these, six genes vird4, pilN, hp, mcr-1, nikB, and ParA showed 100% similarity with pHNSHP45 upon sequence alignment. Though, the amplicon size of the tnpA gene was unexpected (Figure 6). New primers for tnpA (having no IsApl1 loci) were designed to clear this ambiguity. The PCR result and sequencing for new tnpA primers revealed that IsApl1 is absent in our isolates.
Discussion
Antimicrobial-resistance is a global matter of concern, but developing countries like Pakistan, where there are poor hygienic conditions and deprived clinical structures, are at risk. The present study was designed to determine the prevalence of plasmid-mediated colistin resistance, ESBL, and carbapenemases producing genes in clinically isolated E. coli from Pakistan. The molecular epidemiology of mcr-1, blaKPC-2, and blaNDM-1 was determined.
In the present study, the prevalence of ESBL was 25%, which agreed with previous studies from Pakistan that reported 23.56% and 23.91% ESBL positive isolates.
The cumulative prevalence of carbapenemases producers was 22.02%, of which 13.03% were also ESBL positive, much higher than a formerly reported 1.9–2.4% prevalence of carbapenem resistance in Asian countries.
A very little data about the clonal diversity of carbapenemases producing isolates are available from Pakistan. Therefore, we performed the MLST of blaKPC-2, and blaNDM-1 harboring isolates. The ST131 and ST1196 are the most prevalent sequence types for blaKPC-2, and blaNDM-1 detected in the current study. ST131 exhibits multiple virulences and antibiotic resistance genes considering public health threats globally.
All the blaKPC-2 and blaNDM-1 harboring isolates have successfully trans-conjugated except five ST131 blaKPC-2 carrying strains. The failure of blaKPC-2 conjugation in these strains might be due to its localization on the chromosome. A study from China recently determined the occurrence of blaKPC-2 on the chromosomal DNA of ST131 E. coli.
This study identified the first mcr-1 bearing Incl2 plasmid in clinically isolated E. coli from Islamabad, Pakistan. Four (0.73%) isolates harbored the mcr-1 gene out of 545 tested isolates in our study. To date, only two studies reported mcr-1 harboring clinically isolated E. coli From Pakistan. In Peshawar, Pakistan, a study reported 5% mcr-1 out of 120 multiple drug-resistant E. coli.
Diverse sequence types of mcr-1 harboring E. coli ST156 (n=2), ST405 (n=1), and ST117 (n=1) are detected in this study. The sequence type’s diversity suggests multiclonal dissemination of colistin resistance mcr-1 from 2018 to 2019 in Pakistan. The ST156 and ST405 having mcr-1 are for the first time reported in the present study from Pakistan. The ST156 and ST405 having blaNDM-1 are previously detected from chicken meat and clinical isolates.
In this study, the colistin resistance isolates harbor a 60-kb Incl2 plasmid, having transferability. The PCR results of mcr-1 transconjugants revealed that the Incl2 plasmids harbor only the mcr-1 gene, and the ESBLs genes (blaCTXM15 and blaTEM) were not detected in the transconjugants. Another study from Pakistan reported a similar scenario for Incl2 plasmid having only mcr-1 gene in ST155 isolated from a healthy broiler. The previously reported ST155 incl2 plasmid from a healthy broiler had a similar genetic context to pHNSHP45, except for the absence of IsApl1 insertion sequence.
The detection of mcr-1 on ST117 and similar plasmid features with a previously reported plasmid from healthy broilers suggest that poultry-origin mcr-1 harboring Incl2 plasmid is circulating in Pakistan.
Conclusion
In this study, we report the prevalence of mcr-1, ESBL, and carbapenemase-encoding genes in clinically isolated E. coli collected from PIMS hospital Islamabad, Pakistan. The isolates were belonging to diverse clonal and plasmid types. The rare clonal types were identified for blaNDM-1 and blaKPC-2. This indicates that strong selection regarding the resistance genes had occurred in our clinical strains. Moreover, the mcr-1 gene is of the avian pathogenic E. coli lineage. Sequence typing and plasmid analysis of mcr-1 suggests its dissemination via horizontal transfer and food chain. Implementation of antibiotics guidelines for animal farming and human well-being are must, to tackle antibiotic resistance at appropriate levels.
Ethical Approval
This study was approved by Institutional review board of Anhui University; the ethical approval number is 2020KYNO. 19.
Acknowledgment
The authors are thankful to the Institute of Basic Medical Sciences, Khyber Medical University Peshawar, Pakistan and Institute of Health sciences, Anhui University, China for supporting and facilitating this work.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, took part in drafting, revising, gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by grants from Natural Science Foundation of China (number 31771310 to Xingyuan Yang) and Anhui Province Natural Science Foundation (number 1708085MC67 to Xingyuan Yang).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Theuretzbacher U, Bush K, Harbarth S. Critical analysis of antibacterial agents in clinical development. Nat Rev Microbiol. 2020;18(5):286–298. doi:10.1038/s41579-020-0340-0
2. Shafiq M, Huang J, Ur Rahman S, et al. High incidence of multidrug-resistant Escherichia coli coharboring mcr-1 and bla (CTX-M-15) recovered from pigs. Infect Drug Resist. 2019;12:2135–2149. doi:10.2147/IDR.S209473
3. Woerther PL, Burdet C, Chachaty E, Andremont A. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev. 2013;26(4):744–758. doi:10.1128/CMR.00023-13
4. Abrar S, Hussain S, Khan RA, Ul Ain N, Haider H, Riaz S. Prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae: first systematic meta-analysis report from Pakistan. Antimicrob Resist Infect Control. 2018;7(1):26. doi:10.1186/s13756-018-0309-1
5. Codjoe FS, Donkor ES. Carbapenem Resistance: a Review. Med Sci (Basel). 2017;6(1). doi:10.3390/medsci6010001
6. Suay-García B, Pérez-Gracia MT. Present and Future of Carbapenem-resistant Enterobacteriaceae (CRE) Infections. Antibiotics (Basel). 2019;8(3). doi:10.3390/antibiotics8030122
7. Ain N, Abrar S, Sherwani RAK, Hannan A, Imran N, Riaz S. Systematic surveillance and meta-analysis on the prevalence of metallo-β-lactamase producers among carbapenem resistant clinical isolates in Pakistan. J Glob Antimicrob Resist. 2020;23:55–63. doi:10.1016/j.jgar.2020.07.024
8. Karaiskos I, Lagou S, Pontikis K, Rapti V, Poulakou G. The “Old” and the “New” antibiotics for MDR gram-negative pathogens: For Whom, When, and How. Front Public Health. 2019;7:151.
9. Zhai YJ, Sun HR, Luo XW, et al. CpxR regulates the colistin susceptibility of Salmonella typhimurium by a multitarget mechanism. J Antimicrob Chemother. 2020;75(10):2780–2786. doi:10.1093/jac/dkaa233
10. Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
11. Nang SC, Li J, Velkov T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit Rev Microbiol. 2019;45(2):131–161. doi:10.1080/1040841X.2018.1492902
12. Mohsin M, Azam M, Ur Rahman S, et al. Genomic background of a colistin-resistant and highly virulent MCR-1-positive Escherichia coli ST6395 from a broiler chicken in Pakistan. Pathog Dis. 2019;77(7). doi:10.1093/femspd/ftz064.
13. Lv J, Mohsin M, Lei S, et al. Discovery of a mcr-1-bearing plasmid in commensal colistin-resistant Escherichia coli from healthy broilers in Faisalabad, Pakistan. Virulence. 2018;9(1):994–999. doi:10.1080/21505594.2018.1462060
14. Azam M, Ehsan I, Sajjad Ur R, Saleemi MK, Javed MR, Mohsin M. Detection of the colistin resistance gene mcr-1 in avian pathogenic Escherichia coli in Pakistan. J Glob Antimicrob Resist. 2017;11:152–153. doi:10.1016/j.jgar.2017.10.012
15. Mohsin M, Raza S, Roschanski N, Guenther S, Ali A, Schierack P. Description of the First Escherichia coli clinical isolate harboring the colistin resistance gene mcr-1 from the Indian Subcontinent. Antimicrob Agents Chemother. 2017;61(1). doi:10.1128/AAC.01945-16
16. Mohsin M, Raza S, Roschanski N, Schaufler K, Guenther S. First description of plasmid-mediated colistin-resistant extended-spectrum β-lactamase-producing Escherichia coli in a wild migratory bird from Asia. Int J Antimicrob Agents. 2016;48(4):463–464. doi:10.1016/j.ijantimicag.2016.07.001
17. Lupindu AM. Isolation and Characterization of Escherichia coli from animals, humans, and environment. In: Samie A, editor. Escherichia Coli - Recent Advances on Physiology, Pathogenesis and Biotechnological Applications. London, United Kingdom: IntechOpen Limited; 2017:187–206.
18. Wayne P. Performance Standards for Antimicrobial Susceptibility Testing. Clinical and laboratory standards institute, CLSI supplement M100; 2020.
19. Dilhari A, Sampath A, Gunasekara C, et al. Evaluation of the impact of six different DNA extraction methods for the representation of the microbial community associated with human chronic wound infections using a gel-based DNA profiling method. AMB Express. 2017;7(1):179. doi:10.1186/s13568-017-0477-z
20. Feliciello I, Chinali G. A modified alkaline lysis method for the preparation of highly purified plasmid DNA from Escherichia coli. Anal Biochem. 1993;212(2):394–401. doi:10.1006/abio.1993.1346
21. Rahman H, Naeem M, Khan I, et al. Molecular prevalence and antibiotics resistance pattern of class A bla CTX-M-1 and bla TEM-1 beta lactamases in uropathogenic Escherichia coli isolates from Pakistan. Turk J Med Sci. 2016;46(3):897–902. doi:10.3906/sag-1502-14
22. Ali F, Niaz Z, Shah PT, Shakeela Q, Uzma B, Ahmed S. Antibiogram of ESBL and MBL producing Pseudomonas aeruginosa among the population of Hazara division, KPK, Pakistan. J Pak Med Assoc. 2020;70(11):1979–1984. doi:10.5455/JPMA.19089
23. Ahmed I, Sajed M, Sultan A, et al. The erratic antibiotic susceptibility patterns of bacterial pathogens causing urinary tract infections. EXCLI J. 2015;14:916–925.
24. Nahid F, Khan AA, Rehman S, Zahra R. Prevalence of metallo-β-lactamase NDM-1-producing multi-drug resistant bacteria at two Pakistani hospitals and implications for public health. J Infect Public Health. 2013;6(6):487–493. doi:10.1016/j.jiph.2013.06.006
25. Abrar S, Vajeeha A, Ul-Ain N, Riaz S. Distribution of CTX-M group I and group III β-lactamases produced by Escherichia coli and klebsiella pneumoniae in Lahore, Pakistan. Microb Pathog. 2017;103:8–12. doi:10.1016/j.micpath.2016.12.004
26. Sid Ahmed MA, Bansal D, Acharya A, et al. Antimicrobial susceptibility and molecular epidemiology of extended-spectrum beta-lactamase-producing Enterobacteriaceae from intensive care units at Hamad Medical Corporation, Qatar. Antimicrob Resist Infect Control. 2016;5(4). doi:10.1186/s13756-016-0103-x.
27. Xu Y, Gu B, Huang M, et al. Epidemiology of carbapenem resistant Enterobacteriaceae (CRE) during 2000–2012 in Asia. J Thorac Dis. 2015;7(3):376–385. doi:10.3978/j.issn.2072-1439.2014.12.33
28. Ain NU, Iftikhar A, Bukhari SS, et al. High frequency and molecular epidemiology of metallo-β-lactamase-producing gram-negative bacilli in a tertiary care hospital in Lahore, Pakistan. Antimicrob Resist Infect Control. 2018;7(1):128. doi:10.1186/s13756-018-0417-y
29. Safari M, Mozaffari Nejad AS, Bahador A, Jafari R, Alikhani MY. Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU). Saudi J Biol Sci. 2015;22(4):424–429. doi:10.1016/j.sjbs.2015.01.004
30. Maurya N, Jangra M, Tambat R, Nandanwar H. Alliance of Efflux Pumps with β-lactamases in multidrug-resistant Klebsiella pneumoniae Isolates. Microb Drug Resist. 2019;25(8):1155–1163. doi:10.1089/mdr.2018.0414
31. Kim YA, Park YS, Youk T, Lee H, Lee K. Trends in South Korean antimicrobial use and association with changes in Escherichia coli resistance rates: 12-year ecological study using a nationwide surveillance and antimicrobial prescription database. PLoS One. 2018;13(12):e0209580. doi:10.1371/journal.pone.0209580
32. Aung MS, San N, Maw WW, et al. Prevalence of extended-spectrum beta-lactamase and carbapenemase genes in clinical isolates of Escherichia coli in Myanmar: dominance of bla(NDM-5) and emergence of bla(OXA-181). Microb Drug Resist. 2018;24(9):1333–1344. doi:10.1089/mdr.2017.0387
33. Qamar MU, Walsh TR, Toleman MA, et al. Dissemination of genetically diverse NDM-1, −5, −7 producing-Gram-negative pathogens isolated from pediatric patients in Pakistan. Future Microbiol. 2019;14(8):691–704. doi:10.2217/fmb-2019-0012
34. Cai JC, Zhang R, Hu YY, Zhou HW, Chen GX. Emergence of Escherichia coli sequence type 131 isolates producing KPC-2 carbapenemase in China. Antimicrob Agents Chemother. 2014;58(2):1146–1152. doi:10.1128/AAC.00912-13
35. Abd El Ghany M, Sharaf H, Al-Agamy MH, Shibl A, Hill-Cawthorne GA. Genomic characterization of NDM-1 and 5, and OXA-181 carbapenemases in uropathogenic Escherichia coli isolates from Riyadh, Saudi Arabia. PLoS One. 2018;13(8):e0201613. doi:10.1371/journal.pone.0201613
36. Wang D, Mu X, Chen Y, et al. emergence of a clinical escherichia coli sequence Type 131 strain carrying a chromosomal bla (KPC-2) Gene. Front Microbiol. 2020;11:586764. doi:10.3389/fmicb.2020.586764
37. Pérez-Vazquez M, Oteo-Iglesias J, Sola-Campoy PJ. Characterization of carbapenemase-producing Klebsiella oxytoca in Spain, 2016–2017. Antimicrob Agents Chemother. 2019;63(6):e025029–025018. doi:10.1128/AAC.02529-18
38. Mohamed ER, Ali MY, Waly N, Halby HM, El-Baky RMA. The Inc FII plasmid and its contribution in the transmission of bla(NDM-1) and bla(KPC-2) in Klebsiella pneumoniae in Egypt. Antibiotics (Basel). 2019;8(4):266.
39. Sartor AL, Raza MW, Abbasi SA, et al. Molecular epidemiology of NDM-1-producing Enterobacteriaceae and Acinetobacter baumannii isolates from Pakistan. Antimicrob Agents Chemother. 2014;58(9):5589–5593. doi:10.1128/AAC.02425-14
40. Nahid F, Zahra R, Sandegren L. A blaOXA-181-harbouring multi-resistant ST147 Klebsiella pneumoniae isolate from Pakistan that represent an intermediate stage towards pan-drug resistance. PLoS One. 2017;12(12):e0189438. doi:10.1371/journal.pone.0189438
41. Gondal AJ, Saleem S, Jahan S. Novel carbapenem-resistant Klebsiella pneumoniae ST147 Coharboring bla (NDM-1), bla (OXA-48) and extended-spectrum β-lactamases from Pakistan. Infect Drug Resist. 2020;13:2105–2115. doi:10.2147/IDR.S251532
42. Hameed F, Khan MA, Bilal H, Muhammad H, Tayyab Ur R. Detection of MCR-1 gene in multiple drug resistant escherichia coli and klebsiella pneumoniae in human clinical samples from Peshawar, Pakistan. Comb Chem High Throughput Screen. 2020.
43. Berrazeg M, Hadjadj L, Ayad A, Drissi M, Rolain JM. First detected human case in algeria of mcr-1 plasmid-mediated colistin resistance in a 2011 Escherichia coli isolate. Antimicrob Agents Chemother. 2016;60(11):6996–6997. doi:10.1128/AAC.01117-16
44. Doumith M, Godbole G, Ashton P, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. 2016;71(8):2300–2305. doi:10.1093/jac/dkw093
45. Quan J, Li X, Chen Y, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–410. doi:10.1016/S1473-3099(16)30528-X
46. Rhouma M, Letellier A. Extended-spectrum β-lactamases, carbapenemases and the mcr-1 gene: is there a historical link? Int J Antimicrob Agents. 2017;49(3):269–271. doi:10.1016/j.ijantimicag.2016.11.026
47. Jeannot K, Bolard A, Plésiat P. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents. 2017;49(5):526–535. doi:10.1016/j.ijantimicag.2016.11.029
48. Baloch Z, Lv L, Yi L, et al. Emergence of almost identical F36: a-:B32Plasmids Carrying bla (NDM-5) and qepA in Escherichia coli from both Pakistan and Canada. Infect Drug Resist. 2019;12:3981–3985. doi:10.2147/IDR.S236766
49. Mediavilla JR, Patrawalla A, Chen L, et al. Colistin- and carbapenem-resistant Escherichia coli Harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio. 2016;7(4). doi:10.1128/mBio.01191-16.
50. Bachiri T, Lalaoui R, Bakour S, et al. First Report of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli ST405 isolated from Wildlife in Bejaia, Algeria. Microb Drug Resist. 2018;24(7):890–895. doi:10.1089/mdr.2017.0026
51. Yang RS, Feng Y, Lv XY, et al. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a Single Muscovy Duck (Cairina moschata). Antimicrob Agents Chemother. 2016;60(11):6899–6902. doi:10.1128/AAC.01365-16
52. Rossi F, Girardello R, Morais C, et al. Plasmid-mediated mcr-1 in carbapenem-susceptible Escherichia coli ST156 causing a blood infection: an unnoticeable spread of colistin resistance in Brazil? Clinics (Sao Paulo). 2017;72(10):642–644. doi:10.6061/clinics/2017(10)09
53. Azam M, Mohsin M, Johnson TJ, et al. Genomic landscape of multi-drug resistant avian pathogenic Escherichia coli recovered from broilers. Vet Microbiol. 2020;247:108766. doi:10.1016/j.vetmic.2020.108766
54. Manges AR, Johnson JR. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin Infect Dis. 2012;55(5):712–719. doi:10.1093/cid/cis502
55. Elbediwi M, Wu B, Pan H, Jiang Z, Biswas S, Li Y. Genomic characterization of mcr-1-carrying Salmonella enterica Serovar 4,[5],12:i:- ST 34 clone isolated from Pigs in China. Front Bioeng Biotechnol. 2020;8:663. doi:10.3389/fbioe.2020.00663
56. Kuo SC, Huang WC, Wang HY, Shiau YR, Cheng MF, Lauderdale TL. Colistin resistance gene mcr-1 in Escherichia coli isolates from humans and retail meats, Taiwan. J Antimicrob Chemother. 2016;71(8):2327–2329. doi:10.1093/jac/dkw122
57. Snesrud E, He S, Chandler M, et al. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother. 2016;60(11):6973–6976. doi:10.1128/AAC.01457-16
58. Mohsin M, Van Boeckel TP, Saleemi MK, et al. Excessive use of medically important antimicrobials in food animals in Pakistan: a five-year surveillance survey. Glob Health Action. 2019;12(sup1):1697541. doi:10.1080/16549716.2019.1697541
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.