Back to Journals » Infection and Drug Resistance » Volume 16
Molecular Epidemiology of Group B Streptococcus Isolates from Pregnant Women with Premature Rupture of Membranes in Fuzhou, China
Authors Liang B , Chen H , Yu D, Zhao W, Cai X, Qiu H, Xu L
Received 20 October 2022
Accepted for publication 17 December 2022
Published 14 January 2023 Volume 2023:16 Pages 269—278
DOI https://doi.org/10.2147/IDR.S393935
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Bin Liang,1,* Huiyu Chen,2,* Donghong Yu,3,4 Wantong Zhao,1,3 Xiaoling Cai,1,3 Huahong Qiu,2 Liangpu Xu1
1Medical Genetic Diagnosis and Therapy Center, Fujian Key Laboratory for Prenatal Diagnosis and Birth Defect, Fujian Maternity and Child Health Hospital College of Clinical Medicine for Obstetrics & Gynecology and Pediatrics, Fujian Medical University, Fuzhou, People’s Republic of China; 2Laboratory Department, Fujian Maternity and Child Health Hospital College of Clinical Medicine for Obstetrics & Gynecology and Pediatrics, Fujian Medical University, Fuzhou, People’s Republic of China; 3Fujian Obstetrics and Gynecology Hospital, Fuzhou, Fujian, People’s Republic of China; 4Medical Research Center, Fujian Maternity and Child Health Hospital College of Clinical Medicine for Obstetrics & Gynecology and Pediatrics, Fujian Medical University, Fuzhou, Fujian, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liangpu Xu; Huahong Qiu, Fujian Maternity and Child Health Hospital, Fuzhou, 350001, People’s Republic of China, Tel +86-0591-87554929 ; +86-0591-87604121, Email [email protected]; [email protected]
Objective: This study investigated the molecular epidemiology of Group B Streptococcus (GBS) in pregnant women with premature rupture of membranes (PROM) in Fuzhou region of China as a source of clinical reference.
Methods: GBS isolates were obtained from pregnant women with PROM. All isolates were genotyped, serotyped, and tested for drug-resistance and virulence genes using PCR and DNA sequencing. Antibiotic susceptibility testing was performed using the Vitek® 2 automated system.
Results: Among the 140 GBS isolates, seventeen sequence types (STs) were identified, of which ST19 (20.0%) was the most prevalent, followed by ST862, ST10, and ST12. Three clonal complexes (CC19, CC10 and CC1) were identified. The predominant serotype was III (45.7%), followed by V (23.6%), Ib (18.6%), Ia (7.1%), and II (3.6%). The prevalence of multidrug resistance was 72.8% (102/140). All isolates were susceptible to penicillin G, ampicillin, quinupristin, linezolid, vancomycin, and tigecycline. The majority of isolates were resistant to erythromycin (70.0%), clindamycin (72.1%), and tetracycline (81.4%), and 28.6% of isolates were resistant to levofloxacin and moxifloxacin. Of the 98 erythromycin-resistant strains, mreA, ermB, mefA, mefE, ermA, and ermTR were detected in 100%, 70.4%, 49.0%, 22.4%, 13.3%, and 9.2%, respectively. No linB was detected among 101 clindamycin-resistant strains. Of the 114 tetracycline-resistant strains, tetM, tetK, tetL and tetO were detected in 52.6%, 61.4%, 7.9%, and 23.7%, respectively. Regarding virulence genes, all strains carried rib and hylB, followed by scpB (98.6%), and bca (80.7%), whereas only one strain carried bac.
Conclusion: ST19/III and ST862/III were the most prevalent GBS subtypes. Penicillin G remains a first-line antibiotic for intrapartum antibiotic prophylaxis and treatment of GBS infections. The prevalence of resistance to clindamycin, erythromycin, and tetracycline is high among GBS isolates in the Fuzhou region. ST862 and ST651 are emerging animal origin STs in human infections, and may become potential zoonotic threats.
Keywords: Group B Streptococcus, molecular epidemiology, premature rupture of the membranes, antibiotic resistance, China
Background
Streptococcus agalactiae, also known as Group B Streptococcus (GBS), commonly colonizes the lower gastrointestinal and genitourinary tracts of pregnant women with a prevalence of 10–35%.1 Maternal GBS colonization may contribute to preterm delivery, maternal fever, and premature rupture of membranes (PROM), which may lead to ascending infection and early-onset disease (EOD) in the neonate, including neonatal pneumonia, sepsis, and meningitis.2 Universal GBS screening at 35–37 weeks gestation, and intrapartum antibiotic prophylaxis for GBS-positive women effectively prevents neonatal GBS-EOD.3 Clinically, penicillin is a first-line antibiotic for IAP and treatment of GBS infections; however, GBS strains with reduced susceptibility to penicillin have been reported in Hong Kong,4 Japan,5,6 USA,7 and Korea.8 In addition, the prevalence of resistance to second-line antibiotics is high among GBS isolates in China. A systematic review and meta-meta-analysis of the incidence of GBS disease in infants, and the prevalence of antimicrobial resistance (AMR) in China revealed that the highest prevalence of resistance was tetracycline (median 98.0%, interquartile range [IQR] 80.0–100%), followed by clindamycin (73.3%, IQR 62.6–78.7%), erythromycin (64.4%, IQR 56.6–75%), and ciprofloxacin (25.0%, IQR 9.1–35.2%).9 GBS has also been reported to have a high prevalence of AMR in some other countries in recent years.10–13 The high level of AMR in China may be attributable to the widespread use of antibiotics in past decades and the development of resistance mutations under the selection pressure of antibiotic treatment for GBS disease. Thus, it is critical to determine the antibiotic susceptibility profile of GBS isolates in clinical practice and to select antibiotics accordingly.
One strategy for preventing invasive GBS disease is to develop specific vaccines.14 Capsular polysaccharide (CPS) is the main virulence factor of GBS strains, and is recognized as a candidate antigenic epitope for vaccines.15 To date, 10 known serotypes (Ia, Ib, and II–IX) have been categorized based on their CPS composition, among which serotype III is the most common among GBS strains inducing invasive disease.16–18 According to the PubMLST website (https://pubmlst.org/organisms/streptococcus-agalactiae), a total of 1954 sequence types (STs) have been identified (up to June 15, 2022) using multilocus sequence typing (MSLT). Previous studies have found regional and ethnic variations in the incidence of GBS infection and serotype and genotype distribution in pregnant women.1,19,20 In pregnant women, different GBS serotypes and genotypes have different levels of pathogenicity and are associated with different levels of risk of neonatal GBS-EOD.21 Therefore, knowledge of the serotypes, genotypes, virulence factors, and drug-resistance profiles of GBS strains in a region could help clinicians to identify high-risk pregnant women and infants for monitoring and treatment, and benefit the development of specific GBS vaccines. This study aimed to describe the molecular characteristics and drug-resistance profile of GBS colonization among pregnant women admitted to a large maternity hospital with premature rupture of membranes, to provide data to support the prevention and treatment of GBS colonization in the region.
Materials and Methods
Study Setting and Sample Collection and Identification
The study was conducted at Fujian Provincial Maternity and Child Health Hospital in Fuzhou, Fujian, China, from June 2020 to April 2021. The hospital delivers almost 20,000 infants annually. As universal screening of pregnant women for GBS at 35–37 gestational weeks has not been implemented in the hospital, the GBS isolates were obtained from pregnant women hospitalized with PROM.
The GBS detection was performed as follows: vaginal or cervical swabs were inoculated onto Columbia blood agar (Bioivt, Zhengzhou, China) and incubated at 37°C in 5% CO2 for 18–24 h. Colonies that were rounded, gray, translucent, with a smooth surface, with or without beta-hemolytic activity, on blood agar were considered suspected GBS and were selected to undergo confirmatory testing using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (BD MALDI Bruker Biotyper, Bremen, Germany). GBS identification was performed by staff in the bacteriological laboratory of Fujian Maternity and Child Health Hospital.
Genomic DNA Extraction
For subsequent molecular assays, a single GBS colony was selected from the blood agar culture, and mixed with 1.0 mL of sterile normal saline in a 1.5 mL centrifuge tube to prepare the bacterial suspension. Genomic DNA was extracted by heat lysis. Briefly, 0.5 mL of GBS bacterial suspension was centrifuged at 13,000 × g for 10 min and then the precipitate was suspended in 100 μL DNA extracting solution (10 mM Tris-HCl, pH 8.5, 1 mM EDTA, and 1% Triton X-100). The suspension was heated at 99°C for 10 min, followed by centrifugation at 13,000 × g for 10 min. The supernatant was stored at −20°C until use.
Multilocus Sequence Typing
The MSLT analysis was carried out in accordance with the Streptococcus agalactiae MLST protocol. Seven housekeeping genes (adhP, pheS, atr, glnA, sdhA, glcK, and tkt) were amplified by PCR using oligonucleotide primers, as described previously.22 The resulting PCR products were sequenced by Sangon Biotech Co. Ltd. (Shanghai, China). The data were then submitted to the GBS MLST database (https://pubmlst.org/organisms/streptococcus-agalactiae) to assign alleles at the seven loci, and each isolate was classified according to the ST. Strains were assigned to a clonal complex (CC) according to an MLST profile downloaded from the PubMLST website (see the Supplemental Material).
Serotyping
Each isolate was serotyped based on the capsule type using a real-time PCR assay, as described previously.23 The isolate was defined as non-typeable (NT) if the real-time PCR assay failed to identify any serotype.
Antimicrobial Resistance Testing
Antimicrobial resistance testing was performed using the Vitek® 2 automated system (bioMérieux, Marcy L’Étoile, France). The samples were tested for susceptibility to penicillin G, ampicillin, levofloxacin, moxifloxacin, erythromycin, clindamycin, quinuptine, linezolid, tetracycline, vancomycin, and tigecycline. All susceptibility tests and the interpretation of the results were performed according to the guidelines and criteria established by the Clinical and Laboratory Standard Institute (CLSI), 2017 standard. We used Staphylococcus aureus (ATCC 29213) as a quality-control strain in each set of tests to ensure the accuracy of the results.
Detection of Drug-Resistance Genes
PCR testing using multipair primers (Table S1) for seven genes (mreA, ermA, ermB, ermC, mefA, mefE and ermTR), one gene (linB), and four genes (tetM, tetK, tetL and tetO) was used to detect drug-resistance genes for erythromycin, clindamycin, and tetracycline, respectively. The PCR products were analyzed by gel electrophoresis and sequenced by the Sangon Biotech Co., Ltd. (Shanghai, China). The positive products were confirmed using the Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Detection of Virulence Genes
Five virulence genes, scpB, hylB, rib, bca and bac were detected by PCR using the primer pairs (Table S1). The PCR products were analyzed using gel electrophoresis and sequenced by the Sangon Biotech Co., Ltd. The positive products were confirmed using BLAST.
Statistical Analysis
The statistical analysis was performed using SPSS 24.0 (IBM Corp., Armonk, NY, USA). The Chi-square test or Fisher’s exact test was used testing categorical data for statistical significance. P-values <0.05 were considered to be statistically significant.
Results
A total of 140 non-duplicate GBS isolates were isolated from vaginal/cervical swabs of pregnant women with PROM. Seventeen ST types were identified by MLST analysis. Specifically, the predominant ST type was ST19 (20.0%, 28/140), followed by ST862 (14.3%, 20/140), ST10 (12.1%, 17/140), ST12 (11.4%, 16/140), ST17 (7.1%, 10/140), and ST27 (7.1%, 10/140). Three CCs were identified: CC19 (27.1%, 38/140), CC10 (25.0%, 35/140) and CC1 (6.4%, 9/140) (Figure 1).
 |
Figure 1 Distribution diagram of MLST for all the GBS strains. Abbreviations: MLST, multilocus sequence typing; ST, sequence type; CC, clonal complex. |
Five serotypes were identified among 140 strains. The most prevalent serotype was III, accounting for 45.7% (64/140) of isolates, followed by V (23.6%, 33/140), Ib (18.6%, 26/140), Ia (7.1%, 10/140), II (3.6%, 5/140) and non-typeable (NT) (1.4%, 2/140) (Table 1).
 |
Table 1 Genotype Diversity of Five Serotype Distribution Among 140 Strains |
The minimum spanning tree showed good concordance between certain STs and serotypes. The most prevalent ST-serotype combinations were ST19/III (13.6%, 19/140) and ST862/III (13.6%, 19/140). The majority of ST862 (95%, 19/20), ST17 (90%, 9/10), ST651 (100%, 3/3), and ST19 (68%, 19/28) isolates were serotype III, whereas the majority of ST529 and ST24 isolates were serotype V. ST23 and ST103 accounted for 80% of serotype Ia isolates, and ST12 and ST10 accounted for 88.5% of serotype Ib isolates (Figure 2 and Table 1).
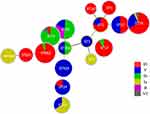 |
Figure 2 Minimum spanning tree of the relationship between STs and serotypes. Abbreviations: ST, sequence type. NT, non-typeable. |
All isolates were susceptible to penicillin G, ampicillin, quinupristin, linezolid, vancomycin, and tigecycline, whereas most of the isolates were resistant to erythromycin (70.0%), clindamycin (72.1%), tetracycline (81.4%), levofloxacin (28.6%) and moxifloxacin (28.6%) (Table 2). Overall, 102 (72.8%) isolates were multidrug resistant (MDR); for example, all the erythromycin-resistant strains were also resistant to clindamycin. Of the clindamycin-resistant strains, 96% (70/73) were resistant to erythromycin, and 4% (3/73) had an intermediate level of resistance to erythromycin. Moreover, 28.6% of strains were co-resistant to levofloxacin and moxifloxacin. Notably, the minimum inhibitory concentration (MIC) of tetracycline-resistant strains was relatively high, with an MIC required to inhibit 50% of organisms (MIC50) ≥16 μg/mL.
 |
Table 2 Antimicrobial Susceptibility and MIC Distributions of GBS Isolates |
The antibiotic-resistance profile varied by ST (Table S2), and all ST17, ST19 and ST529 isolates were MDR strains. Almost all (>90%) of ST12, ST17, ST19, ST27, and ST529 isolates were resistant to tetracycline. The predominant STs of levofloxacin-resistant and moxifloxacin-resistant were ST19 (96%) and ST10 (65%). Similar comparisons by serotype showed that the majority (>80%) of serotype Ib and V isolates were resistant to erythromycin and clindamycin (Table S3).
Among the GBS isolates, 70.0% (98/140), 72.1% (101/140), and 81.4% (114/140) strains were erythromycin-, clindamycin-, and tetracycline-resistant, respectively (Table 3). Of the 98 erythromycin-resistant strains, mreA was detected in 100% (98/98), followed by ermB (70.4%, 69/98), mefA (49.0%, 48/98), mefE (22.4%, 22/98), ermA (13.3%, 13/98) and ermTR (9.2%, 9/98), whereas no ermC-carrying strains were detected. Of the 101 clindamycin-resistant strains, no linB-carrying strains were detected. Of the 114 tetracycline-resistant strains, tetM, tetK, tetL and tetO was detected in 52.6% (60/114), 61.4% (70/114), 7.9% (9/114), and 23.7% (27/114), respectively. Of the erythromycin-resistant strains mreA-ermB, mreA-ermB-mefA and mreA-mefA-mefE, was detected in 41.8% (41/98), 18.4% (18/98), and 14.3% (14/98), respectively (Table 4). Of the isolates carrying mreA-ermB, the MIC was 0.5–≥8 μg/mL, with an MIC50 ≥4 μg/mL. Of the isolates carrying mreA-ermB-mefA, the MIC was 0.5–≥8 μg/mL, with a MIC50 ≥8 μg/mL. Of the isolates carrying mreA-mefA-mefE, the MIC was 2–≥8 μg/mL, and 71% had a MIC ≥8 μg/mL.
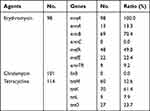 |
Table 3 The Drug-Associated Gene Detection for Erythromycin-, Clindamycin-, and Tetracycline-Resisistant Strains |
 |
Table 4 Resistance Genes Profile and Minimum Inhibitory Concentration in 98 Erythromycin-Resistant Strains |
Five virulence genes were detected in the 140 strains. All strains carried rib and hylB, followed by scpB (98.6%, 138/140), and bca (80.7%, 113/140), whereas only one strain carried bac. The prevalence of bca varied from 45–100% among the seven main STs (Table 5). However, there were no statistically significant differences in the prevalence of the five virulence genes among the different serotypes (data not shown).
 |
Table 5 Virulence Gene Detection in 140 Isolates |
Discussion
GBS colonization in late pregnancy is associated with PROM, perinatal infection, and neonatal GBS-EOD. The GBS colonization rate varies according to ethnicity, region, and individual factors. A previous study reported a GBS colonization rate of 5.3% (29/537) among women in late pregnancy in Fuzhou, China,24 but the molecular characteristics such as the serotype distribution, MLST analysis, drug-resistance genes, and virulence genes were not reported. In this study, we describe the molecular characteristics and drug susceptibility profile of GBS colonized in pregnant women with PROM.
A total of 17 STs were identified among the 140 GBS strains, with ST19, ST862, ST10, ST12, ST17, and ST27 being higher prevalent. These six predominant STs are similar to those of women from Xiamen (Fujian, China) in late pregnancy.21 Notably, ST862 was the second most prevalent type in Fuzhou and Xiamen, but it has not been reported in studies conducted in other parts of China.25–27 Five serotypes (Ia, Ib, II, III, and V) were identified in 98.6% of the GBS strains isolated. A recent review reported that 85% serotypes of GBS isolates from pregnant women in worldwide were Ia, Ib, II, III, and V,28 and that these serotypes accounted for 96% and 93% of GBS strains in the United States and Europe, respectively. These results suggested that the development of a broad-coverage GBS vaccine (Ia, Ib, II, III, and V) would provide protection against infection by the majority of strains worldwide.
ST19/III and ST862/III were the most prevalent in this region, each accounting for 13.6%. ST862, ST17, ST651, and ST190 were mainly associated with serotype III; ST529 and ST24 were mainly associated with serotype V; ST23 and ST103 were mainly associated with serotype Ia; and ST12 and ST10 were mainly associated with serotype Ib. As the sample sizes and types of patients recruited were varied in previous studies,21,29–31 it is not possible to compare the results of this study with those of previous studies.
Clinically, penicillin is still recognized as a first-line antibiotic for IAP and treatment of GBS infections. In this study, all the isolates were susceptible to penicillin G, ampicillin, quinupristin, linezolid, vancomycin, and tigecycline. Considering the 0.7–10% prevalence of penicillin allergies,32 clindamycin, erythromycin, and levofloxacin are important alternatives. However, this study showed a high prevalence of resistance to clindamycin (72.1%), erythromycin (70.0%) and tetracycline (81.4%), suggesting that these agents should not be used as alternatives in patients with penicillin allergies in the absence of drug susceptibility testing. The levofloxacin-resistance rate in this study (28.6%) is similar to that reported in Shanghai (35.5%),33 but higher than that reported in other countries.34,35 Differences in antibiotic use could be a major contributor to geographic differences in the prevalence of antimicrobial resistance. From a public health perspective, having additional drugs with which to treat GBS would be useful. This requires taking actions to mitigate AMR in areas with a high prevalence of AMR, such as drug susceptibility testing to select appropriate agents for IAP and treatment, instead of empirical drug use.
All erythromycin-resistant strains carried mreA, followed by ermB, mefA, mefE, ermA, and ermTR, whereas no ermC-carrying strains were identified. Furthermore, most erythromycin-resistant strains carried mreA-ermB, mreA-ermB-mefA or mreA-mefA-mefE, which tended to have higher MIC50 levels. These results suggest that the “efflux pump”, mediated by mreA and mefA/E, is an important resistance mechanism. The high prevalence of ermB suggests that GBS commonly uses target methylation as the mechanism of macrolide resistance. No linB was detected among the 101 clindamycin-resistant strains, suggesting that clindamycin resistance is related to other resistance mechanisms. Of the 114 tetracycline-resistant strains, tetM, tetK, tetL and tetO were detected. This suggests that tetracycline resistance is mediated by ribosomal protection proteins encoded mainly by the tetM or tetO genes and “efflux pumps” encoded by the tetK or tetL genes.
GBS virulence is also important for the development of vaccines and for understanding its pathogenicity. All strains carried rib and hylB, and the majority of strains carried scpB, and bca, whereas only one strain carried bac. Moreover, the prevalence of bca varied among the seven main STs, and was lowest for ST862 but highest for ST12. These results suggest that GBS bacterial surface proteins encoded by rib, hylB, and scpB may be candidates for antigenic epitopes of vaccines, whereas bac with a very low detection rate (<1%) is not a suitable candidate.
Of note, ST862 and ST651 are emerging zoonotic STs which are the main cause of bovine mastitis.36 ST862 has become the second most prevalent type in Fujian province (including Fuzhou and Xiamen). As mentioned above, 95% of ST862 and all the ST651 strains were identified as serotype III. For ST862 strains, the resistance rate to erythromycin, clindamycin, and tetracycline were 60%, 65% and 65%, respectively, and a low carry rate (45%) of bca virulence gene was observed. As bovine S. agalactiae may be affected by the selection pressure of antibiotic treatment for mastitis, further information is needed on the antibiotic resistance profile of S. agalactiae STs of animal origin in the Fuzhou region.
This study had some limitations. First, the D-test was not performed on GBS strains showing resistance to erythromycin and clindamycin to test for inducible macrolide resistance. Second, no cases of neonatal GBS-EOD were diagnosed during the study period, because all pregnant women with PROM received IAP, and no cases of neonatal GBS infection occurred.
Conclusions
We determined the molecular epidemiology of GBS in pregnant women with PROM in Fuzhou City, China. The predominant STs were ST19, ST862, ST10, and ST12, and the three CCs were CC19, CC10, and CC1. The most prevalent serotypes were III, V, and Ib. All isolates were susceptible to penicillin G, ampicillin, quinupristin, linezolid, vancomycin, and tigecycline, and highly resistant to erythromycin, clindamycin, tetracycline, levofloxacin, and moxifloxacin. The common erythromycin-resistance genes were mreA, ermB, mefA and mefE, and the common tetracycline-resistance genes were tetK and tetM. The predominant virulence genes were rib, hylB, scpB, and bca. It should draw the attention that ST862 and ST651 as emerging zoonotic STs that may become a potential threat. This study provides a comprehensive description of the molecular epidemiology of GBS in the Fuzhou region, and the results can be used to inform the prevention and treatment of perinatal GBS disease.
Abbreviations
CPS, capsular polysaccharide; EOD, early-onset disease; GBS, Group B Streptococcus; AMR, antimicrobial resistance; MLST, multilocus sequence typing; PROM, premature rupture of the membranes; ST, sequence type; CC, clonal complex.
Data Sharing Statement
The data that support the findings of this study are openly available in figshare.2014. (https://doi.org/10.6084/m9.figshare.20584314.v2).
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of the Fujian Maternity and Child Care Hospital (No.2019–066). Informed consent was obtained from all the participants prior to enrollment. Our study was carried out followed the principles outlined in the Declaration of Helsinki.
Acknowledgments
We are grateful to the patients for their collaboration.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, drafting and revising the article, gave final approval of the version to be published, have agreed on the journal to which the article will be submitted and agreed to be accountable for all aspects of the work. The final manuscript was reviewed and approved by all authors.
Funding
This work was supported by grants from Fujian Provincial Natural Science Foundation (2020J05275); Fujian Provincial Maternity and Child Health Hospital [grant no. YCXM 19-15]; and Fujian Provincial Maternity and Child Health Hospital [grant no. YCXM 20-30].
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Russell NJ, Seale AC, O’Driscoll M, et al. Maternal colonization with Group B Streptococcus and serotype distribution worldwide: systematic review and meta-analyses. Clin Infect Dis. 2017;65(suppl_2):S100–S111. doi:10.1093/cid/cix658
2. Edmond KM, Kortsalioudaki C, Scott S, et al. Group B streptococcal disease in infants aged younger than 3 months: systematic review and meta-analysis. Lancet. 2012;379(9815):547–556. doi:10.1016/S0140-6736(11)61651-6
3. Naat G, Encourage PC. Prevention of Group B Streptococcal early-onset disease in newborns: ACOG committee opinion, number 797. Obstet Gynecol. 2020;135(2):e51–e72. doi:10.1097/AOG.0000000000003668
4. Chu YW, Tse C, Tsang GK, So DK, Fung JT, Lo JY. Invasive group B Streptococcus isolates showing reduced susceptibility to penicillin in Hong Kong. J Antimicrob Chemother. 2007;60(6):1407–1409. doi:10.1093/jac/dkm390
5. Kimura K, Suzuki S, Wachino J, et al. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Antimicrob Agents Chemother. 2008;52(8):2890–2897. doi:10.1128/AAC.00185-08
6. Nagano N, Nagano Y, Toyama M, et al. Nosocomial spread of multidrug-resistant group B streptococci with reduced penicillin susceptibility belonging to clonal complex 1. J Antimicrob Chemother. 2012;67(4):849–856. doi:10.1093/jac/dkr546
7. Dahesh S, Hensler ME, Van Sorge NM, et al. Point mutation in the group B streptococcal pbp2x gene conferring decreased susceptibility to beta-lactam antibiotics. Antimicrob Agents Chemother. 2008;52(8):2915–2918. doi:10.1128/AAC.00461-08
8. Yi A, Kim CK, Kimura K, et al. First Case in Korea of Group B Streptococcus with reduced penicillin susceptibility harboring amino acid substitutions in penicillin-binding protein 2X. Ann Lab Med. 2019;39(4):414–416. doi:10.3343/alm.2019.39.4.414
9. Ding Y, Wang Y, Hsia Y, Russell N, Heath PT. Systematic review and meta-analyses of incidence for Group B Streptococcus disease in infants and antimicrobial resistance, China. Emerg Infect Dis. 2020;26(11):2651–2659. doi:10.3201/eid2611.181414
10. Hayes K, O’Halloran F, Cotter L. A review of antibiotic resistance in Group B Streptococcus: the story so far. Crit Rev Microbiol. 2020;46(3):253–269. doi:10.1080/1040841X.2020.1758626
11. Francois Watkins LK, McGee L, Schrag SJ, et al. Epidemiology of Invasive Group B Streptococcal infections among nonpregnant adults in the United States, 2008–2016. JAMA Intern Med. 2019;179(4):479–488. doi:10.1001/jamainternmed.2018.7269
12. Maeda T, Takayama Y, Fujita T, et al. Comparison between invasive and non-invasive streptococcus agalactiae isolates from human adults, based on virulence gene profiles, capsular genotypes, sequence types, and antimicrobial resistance patterns. Jpn J Infect Dis. 2021;74(4):316–324. doi:10.7883/yoken.JJID.2020.761
13. Van Du V, Dung PT, Toan NL, et al. Antimicrobial resistance in colonizing group B Streptococcus among pregnant women from a hospital in Vietnam. Sci Rep. 2021;11(1):20845. doi:10.1038/s41598-021-00468-3
14. Wang CH, Kung WJ, Lee CH, et al. High rates of colonization and antimicrobial resistance of group B streptococcus highlight the need for vaccination even after implementation of guidelines for intrapartum antibiotic prophylaxis. Vaccine. 2022;40(2):282–287. doi:10.1016/j.vaccine.2021.11.069
15. Heath PT. Status of vaccine research and development of vaccines for GBS. Vaccine. 2016;34(26):2876–2879. doi:10.1016/j.vaccine.2015.12.072
16. Zhu Y, Wu J, Zheng X, et al. Etiological serotype and genotype distributions and clinical characteristics of group B streptococcus-inducing invasive disease among infants in South China. BMC Pediatr. 2020;20(1):146. doi:10.1186/s12887-020-02048-2
17. Nanduri SA, Petit S, Smelser C, et al. Epidemiology of invasive early-onset and late-onset Group B Streptococcal disease in the United States, 2006 to 2015: multistate laboratory and population-based surveillance. JAMA Pediatr. 2019;173(3):224–233. doi:10.1001/jamapediatrics.2018.4826
18. Furfaro LL, Chang BJ, Payne MS. Perinatal Streptococcus agalactiae Epidemiology and Surveillance Targets. Clin Microbiol Rev. 2018;31(4):e00049–18. doi:10.1128/CMR.00049-18
19. Shabayek S, Spellerberg B. Group B Streptococcal colonization, molecular characteristics, and epidemiology. Front Microbiol. 2018;9:437. doi:10.3389/fmicb.2018.00437
20. Shabayek S, Ferrieri P, Spellerberg B. Group B Streptococcal Colonization in African countries: prevalence, capsular serotypes, and molecular sequence types. Pathogens. 2021;10:12. doi:10.3390/pathogens10121606
21. Yao Z, Jiayin W, Xinyi Z, et al. Identification of Group B Streptococcus serotypes and genotypes in late pregnant women and neonates that are associated with neonatal early-onset infection in a South China Population. Front Pediatr. 2020;8:265. doi:10.3389/fped.2020.00265
22. Jones N, Bohnsack JF, Takahashi S, et al. Multilocus sequence typing system for group B streptococcus. J Clin Microbiol. 2003;41(6):2530–2536. doi:10.1128/JCM.41.6.2530-2536.2003
23. Alhhazmi A, Pandey A, Tyrrell GJ. Identification of Group B Streptococcus Capsule type by use of a dual phenotypic/genotypic assay. J Clin Microbiol. 2017;55(9):2637–2650. doi:10.1128/JCM.00300-17
24. Huang LR, Wang Y, Wang WQ, Ying W, Wang LB, Pan HY. Drug resistance analysis of Streptococcus agalactiae in urogenital tract of women in late pregnancy in Fuzhou area. Chin J Health Lab Tec. 2020;30(20):2470–2472.
25. Li J, Ji W, Gao K, et al. Molecular characteristics of group B Streptococcus isolates from infants in southern mainland China. BMC Infect Dis. 2019;19(1):812. doi:10.1186/s12879-019-4434-0
26. Wang P, Tong JJ, Ma XH, et al. Serotypes, antibiotic susceptibilities, and multi-locus sequence type profiles of Streptococcus agalactiae isolates circulating in Beijing, China. PLoS One. 2015;10(3):e0120035. doi:10.1371/journal.pone.0120035
27. Jiang H, Chen M, Li T, Liu H, Gong Y, Li M. Molecular characterization of streptococcus agalactiae causing community- and hospital-acquired infections in Shanghai, China. Front Microbiol. 2016;7:1308. doi:10.3389/fmicb.2016.01308
28. Melin P, Efstratiou A. Group B streptococcal epidemiology and vaccine needs in developed countries. Vaccine. 2013;31(Suppl 4):D31–42. doi:10.1016/j.vaccine.2013.05.012
29. Lo CW, Liu HC, Lee CC, et al. Serotype distribution and clinical correlation of Streptococcus agalactiae causing invasive disease in infants and children in Taiwan. J Microbiol Immunol Infect. 2019;52(4):578–584. doi:10.1016/j.jmii.2017.09.002
30. Kao Y, Tsai MH, Lai MY, et al. Emerging serotype III sequence type 17 group B streptococcus invasive infection in infants: the clinical characteristics and impacts on outcomes. BMC Infect Dis. 2019;19(1):538. doi:10.1186/s12879-019-4177-y
31. Lu B, Li D, Cui Y, Sui W, Huang L, Lu X. Epidemiology of Group B streptococcus isolated from pregnant women in Beijing, China. Clin Microbiol Infect. 2014;20(6):O370–3. doi:10.1111/1469-0691.12416
32. Neugut AI, Ghatak AT, Miller RL. Anaphylaxis in the United States: an investigation into its epidemiology. Arch Intern Med. 2001;161(1):15–21. doi:10.1001/archinte.161.1.15
33. Yan Y, Hu H, Lu T, et al. Investigation of serotype distribution and resistance genes profile in group B Streptococcus isolated from pregnant women: a Chinese multicenter cohort study. APMIS. 2016;124(9):794–799. doi:10.1111/apm.12570
34. Safari D, Gultom SM, Tafroji W, et al. Prevalence, serotype and antibiotic susceptibility of Group B Streptococcus isolated from pregnant women in Jakarta, Indonesia. PLoS One. 2021;16(5):e0252328. doi:10.1371/journal.pone.0252328
35. Piccinelli G, Gargiulo F, Corbellini S, et al. Emergence of the first levofloxacin-resistant strains of Streptococcus agalactiae isolated in Italy. Antimicrob Agents Chemother. 2015;59(4):2466–2469. doi:10.1128/AAC.05127-14
36. Carvalho-Castro GA, Silva JR, Paiva LV, et al. Molecular epidemiology of Streptococcus agalactiae isolated from mastitis in Brazilian dairy herds. Braz J Microbiol. 2017;48(3):551–559. doi:10.1016/j.bjm.2017.02.004
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
