Back to Journals » Infection and Drug Resistance » Volume 12
Molecular epidemiology of ESBL-producing E. coli and K. pneumoniae: establishing virulence clusters
Authors Surgers L, Boersma P, Girard PM, Homor A, Geneste D, Arlet G, Decré D, Boyd A
Received 4 July 2018
Accepted for publication 13 November 2018
Published 31 December 2018 Volume 2019:12 Pages 119—127
DOI https://doi.org/10.2147/IDR.S179134
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Laure Surgers,1,2 Peter Boersma,3 Pierre-Marie Girard,1,4 Audrey Homor,5 Delphine Geneste,2 Guillaume Arlet,2,5 Dominique Decré,2,5 Anders Boyd4
1Infectious Diseases Department, Saint-Antoine Hospital, APHP, Paris, France; 2Sorbonne University, INSERM, U1135, Centre d’Immunologie et des Maladies Infectieuses, CIMI Team 13, Paris, France; 3Department of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN, USA; 4INSERM, Sorbonne Université, Institut Pierre Louis d’Épidémiologie et de Santé Publique, Paris, France; 5Bacteriology Department, Saint-Antoine Hospital, APHP, Paris, France
Objective: To genetically characterize clusters of virulence factors (VFs) among extended spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae and assess whether these clusters are associated with genetic determinants or clinical outcomes.
Methods: One hundred forty-eight E. coli and 82 K. pneumoniae clinical isolates were obtained from 213 patients in Paris, France. Isolates underwent ESBL characterization, MultiLocus Sequence Typing (MLST) typing and phylogenetic group identification. Detection of ten E. coli and seven K. pneumoniae VF-encoding genes were assessed, from which a k-medians partition algorithm with Jaccard similarity measure was used to construct clusters.
Results: CTX-M was the predominant ESBL and susceptibility to trimethoprim–sulfamethoxazole (32%), ciprofloxacin (22%) and aminoglycosides (32%) was low. In E. coli, there were five identified clusters, with significantly different distributions of ESBL-sequence type (P<0.001), ST131 (P<0.001) and phylogenetic group (P=0.001) between clusters. “Siderophore exclusive”, “siderophore exclusive with iroN ” and “adhesin sfa/papGIII-rich” clusters had higher 12-month mortality rates compared to others (49% vs 22%, respectively, P=0.02). In K. pneumoniae, three different clusters, with significantly different distributions of aminoglycoside-sensitivity (P<0.004), MLST-type (P<0.001) and relaxase plasmids (P=0.001) were described.
Conclusion: Distinct clusters of E. coli and K. pneumoniae VFs are observed within ESBL-producing isolates and are strongly associated with several genetic determinants. Their association with overall morbidity and mortality requires further evidence.
Keywords: ESBL, virulence, mortality, E. coli, K. pneumoniae
Introduction
Extended spectrum β-lactamase (ESBL)-producing Enterobacteriaceae first emerged during the 1980s and with their extensive transmission have now become pandemic.1 Coupled with their limited treatment options and difficulty in patient management, these infections have led to increasing morbidity and mortality and represent a major public health concern.2–4 Since 2000, Enterobacteriaceae producing CTX-M-ESBL have been predominant, yet the reasons for such wide dissemination, including in the community, remain speculative.1
Genetic characteristics of strains and plasmids carrying ESBL genes could contribute to dissemination of ESBL-producing Enterobacteriaceae. First, plasmids do not generally contain genes necessary for bacterial vitality. Nonetheless, they sometimes contain genes conferring selective advantages able to resist host responses (ie, detoxification, antibiotic resistance, virulence) and could account for some of the circulating strains observed to date.5 Second, the intrinsic virulence of ESBL-producing strains, much like any bacteria, is principally determined by the presence of virulence genes encoding for virulence factors (VFs).6 The relationship between VF and ESBL-producing Enterobacteriaceae remain unclear. Finally, certain virulent phylogenetic groups of Escherichia coli are described, such as B2 and D. Their capacity to spread has already been described, as observed with B2 O25b:H4-ST131 CTX-M-15 producing E. coli.7
Only few studies to date have attempted to understand the type of VFs factors shared within strains and whether clusters of VF are associated with other genetic or even clinical characteristics. We aimed to genetically characterize two Enterobacteriaceae most frequently established with carrying ESBL genes, E. coli and K. pneumoniae.8 We then used a statistical technique to establish linkage between VF within the two species. Finally, we assessed the association of cluster groups with genetic determinants and clinical outcomes.
Materials and methods
Study design
Data were obtained from a cross-sectional study conducted at Saint-Antoine Hospital, Assistance Publique-Hôpitaux de Paris, Paris, France from April 2012 to April 2013.8 Biological samples testing positive for an ESBL-producing E. coli or K. pneumoniae, with the exception of rectal swabs, from patients attending in- or outpatient clinics processed at the Department of Microbiology were included. Information was given to each patient for the use of stored samples and personal data related to non-interventional research. All data have been treated anonymously. In accordance with French Public Health law, ethical approval was not required for this study.
Clinical data
A patient was defined as “infected” if his attending physician decided to treat with antibiotics or as “colonization” otherwise. Portal of entry was classified as lung, urinary tract (UTI), digestive tract or unknown according to the attending physician. Acquisition of ESBL-producing Enterobacteriaceae was characterized as follows: hospital-acquired, if the first positive sample was detected >48 hours after admission; health care-associated, if the first positive sample was detected ≤48 hours after admission and the patient underwent hospitalization within 3 months prior or community-acquired, if the first positive sample was detected ≤48 hours of admission without any recent hospitalization. Data on transfer to the intensive care unit (ICU) and all-cause deaths within 12 months after entry into care were obtained from electronic patient medical records. Only infected patients had available data on ICU transfer.
Bacterial and genetic characterization of isolated strains
Species identification
After routine culture at 37°C in Trypticase soy and Drigalski agar, species were identified using the API20E system (BioMérieux, Marcy l’Etoile, France) or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
Antibiotic resistance
Antibiotic susceptibility patterns were determined by standard diffusion on Mueller–Hinton agar (Bio-Rad Laboratories Inc., Hercules, CA, USA), and results were interpreted according to the Comité de l’Antibiogramme de la Société Française de Microbiologie (CA-SFM)/EUCAST.31
ESBL detection and sequencing
ESBL production was detected using the double-disk test for synergy between clavulanic acid and extended-spectrum cephalosporins (ceftazidime and cefotaxime) on Muller–Hinton agar. For strains with a positive result, ESBL characterization was performed on stored samples using specific PCR amplification and sequencing after bacterial DNA extraction using an InstaGeneMatrix® kit (Bio-Rad).9,10 For E. coli strains, phylogenetic group was characterized using a quadruplex PCR method.11 A pabB and trpA allele-specific PCR was used to detect, among B2 strains, those belonging to the pandemic O25b:H4-ST131 clone.12
VFs
PCR was used on stored samples to detect the presence of genes encoding VF in the core genome or in plasmids, including adhesins (papC, papG, including papG alleles, sfa/foc and ibeA), toxins (hlyC and cnf1) and iron capture systems (fyuA, iroN and iucC).13 Sequences were analyzed using the program SeqScape® (Thermo Fisher Scientific, Waltham, MA, USA).15 Seven VF genes, including the regulator of mucoid phenotype A (rmpA), fimbrial adhesion (mrkD), iron capture systems (entB, kfu, ybtS and iutA) and factor associated with allantoin metabolism (allS) as well as capsular serotypes K1 and K2 were determined.16
MultiLocus Sequence Typing (MLST) typing
MLST typing was performed on stored samples using the international K. pneumoniae MLST typing scheme.14
Relaxases
Plasmid characterization was performed on stored samples by transferring ESBL genes to azide or rifampin-resistant E. coli J53 as a recipient and selecting transconjugants on Drigalski agar supplemented with sodium azide (100 mg/L) or rifampin (256 mg/L) and cefotaxime (2 mg/L) for CTX-M-producing strains or ticarcillin (125 mg/mL) for TEM- and SHV-producing strains. If resistance genes were unable to be transferred by conjugation, plasmids carrying the ESBL gene were extracted with DNA Plasmid Miniprep Kit ® (Qiagen NV, Venlo, the Netherlands) and transferred by electroporation in E. coli DH10B cells. Plasmids were characterized using PCR-based replicon typing, and multireplicons were studied using the Kado method.17,18
Statistical analysis
Unless otherwise stated, all comparisons were performed using a Pearson’s chi-squared test or Fisher’s exact test for categorical variables and Kruskal–Wallis rank test for continuous variables. In comparing bacterial characteristics, individual isolates were considered without accounting for patient correlation for those with more than one isolate. In comparing patient-level characteristics, only the last patient visit was considered for those with multiple isolates.
In order to define clusters of VF, we employed a k-medians clustering algorithm with the Jaccard similarity measure (specified for binomially distributed data).19 Since VFs are different between bacterial species, the algorithm was run separately for E. coli and K. pneumoniae. The optimal number of clusters was tested using a range of k ={2, 3, …, 10}, while the final choice of k was based on maximizing the Caliński and Harabasz pseudo-F index and evaluating meaningful distributions of VFs (ie, avoiding excess clusters with sparse prevalence of specific VFs). After obtaining the optimal k, cluster groups were assigned to each observation based on the similarity measure above.
A P-value of <0.05 was considered significant. All analysis was conducted using STATA statistical package (v13.0, College Station, TX, USA).
Results
Description of bacterial genetic characteristics
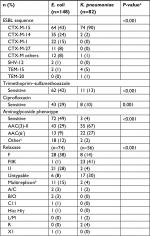
In total, 148 E. coli and 82 K. pneumoniae ESBL-producing strains with available genetic data were included. Isolates were obtained in 198 patients: 5 were infected with the same species (2 with 2 E. coli and 3 with 2 K. pneumoniae) and 11 were infected with E. coli and K. pneumoniae; all of which were collected during different episodes. Genetic characteristics are detailed in Table 1. Isolates were sensitive to trimethoprim–sulfamethoxazole, ciprofloxacin and aminoglycosides in 32%, 22% and 32%, respectively. Among all strains, CTX-M-15 (60%) was the most common ESBL enzyme and was highly prevalent in K. pneumoniae (90%). Of the 148 E. coli strains, 51 (34%) were ST131. Of the 82 K. pneumoniae strains, MLST classes were distributed as follows: ST29 (n=19), ST323 (n=16), ST147 (n=11), ST405 (n=5) and other (n=31). One strain of K. pneumoniae had a K2 capsular serotype harboring rmpA, defined as “hypervirulent”, and was multiresistant (ST86 CTX-M-3 producing K2).20
One hundred thirty plasmids were characterized in electroporants or transconjugants from E. coli (n=74) and K. pneumoniae (n=56), with the most common being IncF plasmids (28%). Of note, 12/21 E. coli carrying IncI1 plasmids were CTX-M-1 producers. Eleven E. coli had multireplicons (n=10 Inc F/I1 and n=1 Inc I1/A/C).
Identification of VFs’ clusters within bacterial species
The distribution of E. coli and K. pneumoniae VF is reported in Table 2. For E. coli VFs, five clusters were identified (pseudo-F index =45.7). The first cluster without any VF genes (defined as “no VF”) was observed in 19 strains (13%). The second cluster included strains with almost only siderophores fyuA and iucC (defined as “siderophore exclusive”) (n=62, 42%), while the third one included these VF genes with iroN siderophore gene (n=29, 20%) (defined as “siderophore exclusive with iroN”). Finally, two other clusters were identified with abundant adhesin VF genes: one cluster with a higher proportion of sfa/foc and papGIII VF genes (n=14, 3%) (defined as “adhesin sfa/papGIII-rich”) and the other with all papC and a higher proportion of papGII VF genes (n=24, 16%) (defined as “adhesin papC/papGII-rich”). Between these clusters, the “adhesin sfa/papGIII-rich” cluster was also found to have a higher prevalence of the toxin VF genes cnf1 and hly, whereas the “adhesin papC/papII-rich” cluster had a higher proportion of iucC VF genes (Table 2).
Three distinct clusters were identified for K. pneumoniae (pseudo-F index =154.9) (Table 2). The first cluster (n=40, 49%) was defined by the absence of VF except for mrkD and entB (defined as “KP common VF”). Two other clusters contained strains that also included mostly ybtS (n=27, 33%) (defined as “KP common VF with ybtS”) or exclusively kfu VF genes (n=15, 18%) (defined as “KP common with kfu”).
Association of bacterial genetic characteristics with VF clusters
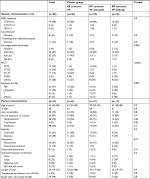
Clusters of E. coli VF were strongly associated with other genetic determinants and antimicrobial resistance (Table 3). Significant differences in phylogenetic groups were noted (P=0.001); however, there was considerable diversity between clusters for all other phylogenetic groups. Distribution of ESBL types was also significantly different between clusters (P<0.001), with the largest differences in CTX-M-15 prevalence. ST131 was significantly more prevalent in the “siderophore exclusive” and “adhesin papC/papGII-rich” clusters compared to all others (P<0.001). More specifically, ST131 compared to non-ST131 isolates had a higher prevalence of the siderophore VFs fyuA (100% vs 57%, respectively, P<0.001) and iucC (94% vs 56%, respectively, P<0.001), reflecting the “siderophore exclusive” cluster, as well as the adhesin VF papGII (25% vs 7%, respectively, P=0.002), reflecting the “adhesin papC/papGII-rich” cluster. There were significant differences between clusters in the proportion of strains with ciprofloxacin (P=0.001) and trimethoprim–sulfamethoxazole-resistance (P=0.03), while no difference was observed with aminoglycoside resistance phenotype.
Clusters of K. pneumoniae VF also appeared strongly associated with bacterial characteristics (Table 4). There were no significant differences in ESBL types between clusters (P=0.9). Nevertheless, significant differences in the distribution of MLST types were observed (P<0.001). ST147 and ST323 were more frequently observed in the “KP common VF” cluster, ST405 in the “KP common VF with kfu” cluster (with proportion harboring kfu VF at 100% compared to 10% for all other MLST types, P<0.001) and ST29 in the “KP common VF with ybtS” cluster (with proportion harboring ybts VF at 95% vs 22% for all other MLST types, P<0.001). Distribution of relaxase-encoding plasmids was also significantly different between clusters (P=0.001). No significant differences were observed in the proportion of isolates resistant to ciprofloxacin (P=0.9) and trimethoprim/sulfamethoxazole (P=0.5), but isolates from the “KP common VF” cluster appeared to have a higher proportion of the AAC(6′) aminoglycoside-resistant phenotype compared to others (P=0.008).
Patient characteristics and clinical outcomes between VF clusters
A comparison of the five E. coli clusters found no significant differences in patient level characteristics or measured clinical outcomes (Table 3). Nevertheless, a significant difference in the distribution of suspected source of infection at admission was noted between clusters (P=0.01), with more frequent urine and digestive sources for clusters with more prevalent and varied VF (“siderophore exclusive with iroN” and “adhesin papC/papGII-rich” clusters). In addition, “siderophore exclusive”, “siderophore exclusive with iroN” and “adhesin sfa/papGIII-rich” clusters had the highest 12-month mortality rate compared to others (P=0.02), while only the “siderophore exclusive with iroN” cluster having a high proportion with bacteremia (21%).
A comparison of the three K. pneumoniae clusters identified significant differences in proportion with recent travel (P=0.02) (Table 4). Furthermore, the “KP common VF with ybtS” had the largest proportion of patients recently hospitalized (91%), whereas the “KP common VF” and “KP common VF with kfu” had the lowest (56%) (P=0.02). No other significant differences in other patient characteristics or clinical outcomes were found (Table 4).
Discussion
With the widespread dissemination of ESBL-producing Enterobacteriaceae, data on the genetic characteristics of strains are needed. In this large collection, we confirm the preponderance of CTX-M-producing microorganisms, of which CTX-M15 was the most common. CTX-M enzymes were particularly frequent in K. pneumoniae, as described by others.1 We confirmed the high proportion of ST131 E. coli stains, first described in 2008 and now widespread.7
A wide body of literature has indicated that many VFs are observed in complex combinations among both E. coli and K. pneumoniae species, while the significance of these groupings vis-à-vis genetic characteristics or their clinical ramifications remains fairly understudied.6,21 Here, we used a simple form of network analysis in which clusters showing the closest similarity between pairwise combinations of VFs are identified. Other studies, limited to animal populations, have used similar clustering techniques to describe groups of VF.22,23
E. coli clusters ranged from absence of all screened VF to increasing complexity of concomitant VF. The strains with the least number of VF originated from the “no VF”, “siderophore exclusive” and “siderophore exclusive with iroN” clusters. These strains were mostly implicated in UTI, particularly the “siderophore exclusive” cluster. This is surprising given that bacteria from these three clusters harbored few adhesins, which are generally involved in UTIs. Given that our study population had a high risk of mortality, the strong presence of E. coli as a UTI might be more linked with opportunistic conditions rather than the VFs themselves.24
In terms of clusters with increasing VF complexity, the “adhesin papC/papGII-rich” and “siderophore exclusive” clusters appears to bring together strains that are from the B2 phylogenetic group and ST131. “Siderophore exclusive” cluster frequently included CTX-M-15 producers or harboring fluoroquinolone resistance, and whose ESBL-producing genes are carried by the IncF plasmid. This cluster closely corresponds to the successful O25b:H4-ST131 CTX-M-15 clone.7 The “adhesin sfa/papGIII-rich” cluster contained strains with Incl1 plasmids carrying CTX-M-1-producing genes. These strains have been observed in both humans and poultry, but in contrary to the O25b:H4-ST131 CTX-M-15 clone, were sensitive to fluoroquinolones.25–27
Overall mortality appeared to be higher in certain VF clusters, namely “siderophore exclusive”, “siderophore exclusive with iroN” and “adhesin sfa/papGIII-rich”, but no relationship was observed in ICU transfer rates. It should be mentioned, however, that patients in our study population had multiple co-morbidities and were for the most part immunosuppressed, which likely contributed to a higher risk of overall mortality in general. Other research has in fact shown that host factors and portal of entry likely outweigh bacterial determinants in predicting the severity of E. coli bacteremia.8,24 In another study of E. coli bacteremia among pregnant women, lower virulence scores were actually associated with more severe episodes.28 The link between these groups of VF and mortality will need to be addressed in larger studies.
There are certain noteworthy aspects of the K. pneumoniae strains observed in our study with three specific clones. CTX-M-15-producing genes carried on IncFIIk plasmid and ST147 resistant to quinolones clones of K. pneumoniae have been previously described.29,30 These two clones have been isolated in different services at our hospital. Additionally, we isolated 19 ST29 strains, for the most part resistant to quinolones or trimethoprim/sulfamethoxazole, phenotype AAC(3)-II and with entB, mrkD and ybtS, from patients in the gastrointestinal surgery department. Plasmids from this ST belonged to the IncF group and carried CTX-M-15 genes. There are few data in the literature to date on ST29. Rep and ERIC PCR confirmed the clonality of these strains, thus suggesting cross-transmission.
Furthermore, it is worth noting that strains observed in our study population produced for the most part CTX-M ESBL. Four E coli and five K. pneumoniae isolates were non-CTX-M producing and their presence was only observed in adhesin papC/papGII-rich VF clusters of E coli and in all VF clusters of K. pneumoniae. As expected, excluding non-CTX-M-producing isolates did not change our overall analysis (data not shown). Nevertheless, we stress that these findings stem from E. coli and K. pneumoniae with predominately CTX-M ESBL production.
Some limitations of our study need to be stated. First, there are a multitude of methods able to determine clusters (ie, hierarchical clustering, principal component analysis, latent class analysis, etc). We opted for a more simplistic method based on the few parameters (ten VF at most), binomial distribution of these parameters and relatively few numbers of patients. We cannot infer how robust these clusters would be in other settings. Second, this study was conducted at a single hospital center and the isolates obtained herein might not be representative of other settings. These results should be interpreted while keeping in mind the diverse range of departments included. Third, strains from both infection and colonization were considered and since there was substantial variability between these two groups, additional heterogeneity could have affected cluster composition. Fourth, some individuals had more than one infection with the same species but at different time points. The repeated data in these patients would not alter the cluster analysis, but could bias statistical comparisons. Given the few patients in whom this was observed (n=5), any effect from this bias would likely be minimal. Finally, K. pneumoniae isolates were not overly abundant and their identified VFs were limited in spectrum. The cluster composition, as a result, frequently led to a single VF. Larger collections of isolates would be needed to obtain more meaningful results.
Conclusion
Distinct clusters of E. coli and K. pneumoniae VF were observed within ESBL-producing isolates. These clusters were linked to EBSL-producing plasmids and phylogenetic groups for E. coli, ST and relaxase types for K. pneumoniae, and resistance to specific antibiotic agents for both species.
Further evidence in healthier populations would be needed to sufficiently address whether VF are associated with clinical outcomes or if they can be used to predict severe morbidity and mortality during treatment. Analyzing genetic factors using a clustering approach could be beneficial for future studies.
Disclosure
LS received a grant from the “Fondation pour la Recherche Médicale” (DEA20140630021). AB received post-doctoral funding from SIDACTION. PB received a Martinson-Luepker Student Travel Award from the University of Minnesota for the work presented in this manuscript. The authors report no other conflicts of interest in this work.
References
Cantón R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9(5):466–475. | ||
Schwaber MJ, Navon-Venezia S, Kaye KS, Ben-Ami R, Schwartz D, Carmeli Y. Clinical and economic impact of bacteremia with extended- spectrum-beta-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2006;50(4):1257–1262. | ||
Tumbarello M, Sanguinetti M, Montuori E, et al. Predictors of mortality in patients with bloodstream infections caused by extended-spectrum-beta-lactamase-producing Enterobacteriaceae: importance of inadequate initial antimicrobial treatment. Antimicrob Agents Chemother. 2007;51(6):1987–1994. | ||
Livermore DM. Has the era of untreatable infections arrived? J Antimicrob Chemother. 2009;64(Suppl 1):i29–i36. | ||
Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303(6–7):298–304. | ||
Vila J, Sáez-López E, Johnson JR, et al. Escherichia coli: an old friend with new tidings. FEMS Microbiol Rev. 2016;40(4):437–463. | ||
Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27(3):543–574. | ||
Surgers L, Boyd A, Boelle PY, et al. Clinical and microbiological determinants of severe and fatal outcomes in patients infected with Enterobacteriaceae producing extended-spectrum β-lactamase. Eur J Clin Microbiol Infect Dis. 2017;36(7):1261–1268. | ||
Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–495. | ||
Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J Antimicrob Chemother. 2006;57(1):14–23. | ||
Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5(1):58–65. | ||
Clermont O, Dhanji H, Upton M, et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother. 2009;64(2):274–277. | ||
Bonacorsi S, Houdouin V, Mariani-Kurkdjian P, Mahjoub-Messai F, Bingen E. Comparative prevalence of virulence factors in Escherichia coli causing urinary tract infection in male infants with and without bacteremia. J Clin Microbiol. 2006;44(3):1156–1158. | ||
Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. | ||
Institut Pasteur. Klebsiella Sequence Typing [Internet]; 2018. Available from: http://bigsdb.pasteur.fr/klebsiella/klebsiella.html. | ||
Compain F, Babosan A, Brisse S, et al. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol. 2014;52(12):4377–4380. | ||
Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145(3):1365–1373. | ||
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. | ||
Hastie T. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed. New York, NY: Springer; 2009:745. | ||
Surgers L, Boyd A, Girard PM, Arlet G, Decré D. ESBL-producing strain of hypervirulent Klebsiella pneumoniae K2, France. Emerg Infect Dis. 2016;22(9):1687–1688. | ||
Li B, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9(9):1071–1081. | ||
Mateus L, Henriques S, Merino C, Pomba C, Lopes da Costa L, Silva E. Virulence genotypes of Escherichia coli canine isolates from pyometra, cystitis and fecal origin. Vet Microbiol. 2013;166(3–4):590–594. | ||
Zhu Y, Dong W, Ma J, et al. Characterization and virulence clustering analysis of extraintestinal pathogenic Escherichia coli isolated from swine in China. BMC Vet Res. 2017;13(1):94. | ||
Lefort A, Panhard X, Clermont O, et al; COLIBAFI Group. Host factors and portal of entry outweigh bacterial determinants to predict the severity of Escherichia coli bacteremia. J Clin Microbiol. 2011;49(3):777–783. | ||
Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 2011;17(6):873–880. | ||
Vila J, Simon K, Ruiz J, et al. Are quinolone-resistant uropathogenic Escherichia coli less virulent? J Infect Dis. 2002;186(7):1039–1042. | ||
Johnson JR. Microbial virulence determinants and the pathogenesis of urinary tract infection. Infect Dis Clin North Am. 2003;17(2):261–278. | ||
Surgers L, Bleibtreu A, Burdet C, et al; COLIBAFI Group. Escherichia coli bacteraemia in pregnant women is life-threatening for foetuses. Clin Microbiol Infect. 2014;20(12):O1035–O1041. | ||
Damjanova I, Tóth A, Pászti J, et al. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005 – the new “MRSAs”? J Antimicrob Chemother. 2008;62(5):978–985. | ||
Dolejska M, Brhelova E, Dobiasova H, et al. Dissemination of IncFII(K)-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from children in hospital paediatric oncology wards. Int J Antimicrob Agents. 2012;40(6):510–515. | ||
Société Française de Microbiologie. CASFM/EUCAST Recommendations 2018 V.2.0. September. Available from http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFMV2_SEPTEMBRE2018.pdf. Accessed August 15, 2018. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.