Back to Archived Journals » Research and Reports in Medicinal Chemistry » Volume 6
Molecular docking, synthesis, and antimycobacterial activities of pyrrolyl hydrazones and their copper complexes
Authors Joshi S, Dixit S, Gadag S, Kulkarni V, Aminabhavi T
Received 3 July 2015
Accepted for publication 23 October 2015
Published 30 December 2015 Volume 2016:6 Pages 1—14
DOI https://doi.org/10.2147/RRMC.S90564
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Xing-Cong Li
Shrinivas D Joshi, Sheshagiri R Dixit, Shivprasad Gadag, Venkatrao H Kulkarni, Tejraj M Aminabhavi
Novel Drug Design and Discovery Laboratory, Department of Pharmaceutical Chemistry, SET’s College of Pharmacy, Dharwad, Karnataka, India
Abstract: A novel series of pyrrole derivatives were designed and synthesized with an aim to overcome the growing antitubercular resistance and develop more potent antimicrobial agents. In this pursuit, a novel series of 4-(1H-pyrrol-1-yl)benzoic acid hydrazide Schiff bases were synthesized and reacted with copper acetate to form the respective copper complexes. The reaction of ethyl 4-(2,5-dimethyl-1H-pyrrol-1-yl)benzoate/ethyl-4-(1H-pyrrol-1-yl)benzoate with hydrazine hydrate produced ethyl 4-(2,5-dimethyl-1H-pyrrol-1-yl)benzohydrazide/ethyl 4-(1H-pyrrol-1-yl)benzohydrazide. The reaction of these hydrazides with different aldehydes yielded N’-(arylidene)-4-(2,5-dimethyl-1H-pyrrol-1-yl)benzohydrazides/N’-(arylidene)-4-(1H-pyrrol-1-yl)benzohydrazides. Furthermore, these Schiff bases were reacted with copper acetate to produce respective copper complexes. All the synthesized compounds were screened for antitubercular activity using microplate alamar blue assay method that showed reasonably good minimum inhibitory concentration (MIC) values ranging from 3.12 to 50 µg/mL compared to the standard drugs like pyrazinamide (MIC =3.125 µg/mL) and streptomycin (MIC =6.25 µg/mL). The selected compounds were evaluated for antibacterial activity against Staphylococcus aureus, Bacillus subtilis, Klebsiella pneumoniae, and Escherichia coli using the broth microdilution assay method. It was found that these compounds exhibited good antibacterial activities in the MIC range of 3.125 to >100 µg/mL when compared against standard drugs like ciprofloxacin and norfloxacin.
Keywords: synthesis, Surflex-Dock, copper complexes, antimicrobial activities
Introduction
Tuberculosis (TB) is a deadly disease caused by mycobacterium of the “tuberculosis complex”, including primarily Mycobacterium tuberculosis as well as Mycobacterium bovis and Mycobacterium africanum.1,2 The 19th World Health Organization TB report indicates that TB is one of the world’s deadliest transmissible diseases with 9 million new patients in 2013 and 1.5 million deaths, as well as 400,000 deaths allied due to co-infection with human immunodeficiency virus (HIV). The highest rates per capita occurred in African region (25%), while South-East Asia, and Western Pacific, regions account for approximately 81% of all the total cases.3 The disease is aggravated by the worldwide continuous emergence of multidrug-resistant strains of M. tuberculosis (MDR-TB), extensively drug-resistant tuberculosis and totally drug-resistant tuberculosis,4,5 and also occurrence of HIV, resulting in acquired immune deficiency syndrome has prompted the prevalence of TB, causing 50% of deaths among the HIV-infected patients due to coinfection with M. tuberculosis, thereby accelerating the collapse of immune system. The importance and extent of drug-resistant strains have increased the concern that TB may once again become a deadly disease.6,7 Moreover, the increasing incidence of TB in immunocompromised patients along with longer durations of therapy emphasizes the need for new drugs to extend the range of effective TB treatment options.8–10 Due to such increased microbial resistance, new classes of antimicrobial agents with novel mechanisms are needed to fight against MDR infections.
M. tuberculosis contains mycolic acids that are unusually long chain α-alkyl β-hydroxy fatty acids of 60–90 carbons.11 The antitubercular (anti-TB) drugs such as isoniazid12 and ethionamide13 have shown to target the synthesis of these mycolic acids, which are the central constituents of mycobacterial cell wall. Among the enzymes involved in fatty acid synthesis II pathway, enoyl acyl carrier protein reductase (ENR) is one of the key catalysts, which catalyzes NADH-dependent reduction of 2-trans-enoyl-ACP (acyl carrier protein) to yield NAD+ and reduced enoyl thioester-ACP substrate, which inhibits mycolic acid synthesis. Studies in the past have established that InhA gene is the primary molecular target for anti-TB drugs.14,15
Coordination or metal complexes that consist of a central metal ion coordinates with the surrounding array of bound molecules, called ligands or complexing agents. Compounds containing transition metals form coordination complexes.16–18 Metal complexes of Schiff base ligands have been investigated as a model for the enzyme active sites,19,20 including DNA cleavage systems,21,22 and as antibacterial23–25 or anticancer26 drugs. Metal complexes of O-, S-, and N-chelating ligands have gained a significant attention because of their remarkable physicochemical properties, prominent biological activities, and due to their metalloenzyme active sites;27,28 they are also used as anticancer, anti-TB, antibacterial, antifungal, and antioxidant agents.29–31
Recently, we have reported on the synthesis as well as molecular modeling (two- and three-dimensional Quantitative Structure–Activity Relationship) studies on pyrrolyl Schiff bases as potent enoyl ACP reductase inhibitors.32–35 Molecules containing imine linkage and pyrrole as structural fragments have been widely explored in drug design studies.
This work aims to find new chemical entities having pyrrole as the core structure that can inhibit enoyl ACP reductase enzyme along with their in vitro anti-TB activity. Figure 1 represents some of the marketed drugs, which bear imine linkage (−N=CH−) and pyrrole ring. Figure 2 indicates a multistep outline by combining classification techniques and molecular docking to understand the structural features that affect binding of pyrrolyl hydrazones and their complexes with enoyl ACP reductase receptor. All the synthesized compounds were docked into the enzyme active site. This computational workflow gives imminent structural characteristics that affect the binding and inhibitory activity of these analogs on enoyl ACP reductase.
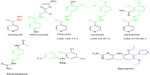  | Figure 1 Reported anti-TB drugs obtained from PubMed database. |
  | Figure 2 Designed anti-TB drugs (pyrrolyl Schiff bases and proposed structures of their complexes). |
Experimental section
General information
The melting points of synthesized compounds were determined by the capillary method in a paraffin bath/digital melting point apparatus (Shital Scientific, Mumbai, India). Fourier transform infrared (FTIR) spectra were recorded using a Bruker spectrophotometer (Bruker, Billerica, MA, USA) using KBr pellets, and the values are expressed in cm−1. The 1H and 13C NMR spectra were recorded using a Bruker AVANCE II 400/100 MHz instrument (Bruker) in deuterated chloroform (CDCl3) and dimethylsulfoxide (DMSO-d6) solvents using trimethylsilane as an internal standard. Chemical shifts are expressed as δ values (ppm), and splitting of the NMR spectra is termed as singlet (s), doublet (d), triplet (t), quartet (q), and multiplet (m). Mass spectra (MS) were recorded using Waters-Q-Tof Premier-HAB213 (Waters Ltd, Manchester, UK), and all the compounds have shown the MS data that correspond with the assumed structures. Analytical thin-layer chromatography (TLC) was performed on precoated TLC sheets of silica gel 60F254 (Merck, Darmstadt, Germany) and visualized by the long- and short-wavelength UV lamps.
Molecular docking studies
Molecular docking studies were carried out36 using the Sybyl-X, version 2.0 (Tripos International, St Louis, MO, USA), run on a Intel® Core™ i3-2130 CPU @ 3.40 GHz processor (Intel, Santa Clara, CA, USA) on a Windows-7 professional workstation. When docking was performed with default settings, it revealed a number of possible conformations and orientations for the inhibitors at the binding site. Understanding of the binding site conformations helped us to understand the important interactions that could stabilize ligand–receptor complex. Surflex-Dock (Tripos International) adopted the empirical scoring function using the patented searching engine37,38 for molecular docking. The crystal structure of M. tuberculosis InhA inhibited by PT70 (5-hexyl-2-[2-methylphenoxy]phenol) was selected from the Protein Data Bank ([PDB entry code 2X22] extracted from the Brookhaven Protein Database http://www.rcsb.org/pdb) and these were used in the initial docking. Co-crystallized ligand and water molecules were removed from the structure, while the essential hydrogen atoms were added and side chains were fixed during the protein preparation. The 3D structures of pyrrole derivatives were constructed using the standard geometric parameters of Sybyl-X 2.0 software (Tripos International), and the structure was subjected to energy minimization. The MMFF94 (Merck Molecular Force Field) charges were calculated for the ligand, while Amber7 FF02 (University of California) was used for the protein. Then, it was subjected to energy minimization following the gradient termination of Powell method for 3,000 iterations using the Tripos force field39 with nonbonding cut-off value set at 8.0 and dielectric constant set at 1.0. Then, ligand-based docking was introduced to generate the “protomol” (it is an object-oriented, component-based framework for molecular dynamics simulations), and all the inhibitors were docked within the prepared protein.
In order to identify the ligand–protein interactions, the top pose and protein were loaded into the work area and MOLCAD (Molecular Computer Aided Design) program was employed to visualize the binding mode between the protein and the ligand. MOLCAD calculates and exhibits the surfaces of channels and cavities as well as separating surface between the protein subunits.40,41 MOLCAD program aims to create molecular surface by using the fast Connolly’s method, a matching cube algorithm to engender the surface.
Synthesis
General procedure for the synthesis of ethyl 4- (2,5-dimethyl/1H-pyrrol-1-yl)benzoates (2a, 2b)
To a solution of ethyl 4-aminobenzoate 1 (0.1 mol) in glacial acetic acid (15 mL), acetonyl acetone/2,5-dimethoxytetrahydrofuran (0.1 mol) was added and the reaction mixture was heated under reflux for 45 minutes between 150°C and 160°C. After cooling, the reaction mixture was poured onto crushed ice and neutralized with a saturated solution of sodium bicarbonate. The residue obtained was filtered, washed with water, and finally recrystallized with ethanol.42
General procedure for the synthesis of 4-(2,5dimethyl/1H-pyrrol-1yl)benzohydrazides (3a, 3b)
A mixture of ester (2a, b) (0.015 mol) and 99% hydrazine hydrate (10 mL) was refluxed with ethanol as a solvent for 3 hours. The crude product was obtained upon cooling, filtered, and recrystallized from ethanol to get benzohydrazides (3a, b).42
General procedure for the synthesis of N′-(arylidene)-4-(2,5-dimethyl-1H-pyrrol-1-yl)benzohydrazides (4a–4k)
Equimolar quantities of 4-(2,5-dimethyl-1H-pyrrol-1-yl)benzohydrazide (2a) (0.1 mol) and substituted aldehydes (0.1 mol) with a catalytic amount of glacial acetic acid were refluxed for 4–8 hours using ethanol solvent. The precipitate formed after cooling was collected by filtration, washed with hot ethanol, dried, and recrystallized using ethanol solvent to obtain the compounds (4a–4k) in good yields.
4-(2,5-Dimethyl-1H-pyrrol-1-yl)-N′-(3-methoxybenzylidene)benzohydrazide (4a)
Yield, 79%; mp 177°C–179°C; IR (KBr): 3183 (NH), 1649 (C=O), 1608 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 2.00 (s, 6H, −2CH3), 3.77 (s, 3H, −OCH3), 5.91 (s, 2H, pyrrole-C3, C4−H), 6.91–8.03 (m, 8H, phenyl-C2, C3, C5, C6 and methoxyphenyl-C2, C4, C5, C6−H), 8.37 (s, 1H, −N=CH−), 10.27 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 13.09, 55.23, 106.47, 110.89, 117.46, 121.14, 128.20, 128.56, 128.81, 129.61, 132.07, 135.07, 142.30, 150.00, 159.81, 164.45; MS (EI): m/z = found 348.17 [M+ +1]; calcd. 347.16. Anal. C21H21N3O2.
N′-(2,4-dimethoxybenzylidene)-4-(2,5-dimethyl- 1H-pyrrol-1-yl)benzohydrazide (4b)
Yield, 80%; mp 188°C–190°C; IR (KBr): 3200 (NH), 1640 (C=O), 1606 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 2.04 (s, 6H, −2CH3), 3.73–3.90 (m, 6H, −OCH3), 5.92 (s, 2H, pyrrole-C3, C4−H), 6.43–6.69 (m, 3H, methoxyphenyl-C3, C5, C6−H), 7.26–8.15 (m, 4H, phenyl-C2, C3, C5, C6−H), 8.54 (s, 1H, −N=CH−), 9.28 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 17.91, 60.44, 102.69, 110.57, 111.23, 120.37, 132.50, 133.67, 137.67, 146.37, 149.65, 164.23, 167.60, 168.05; MS (EI): m/z = found 378.18 [M+ +1]; calcd. 377.17. Anal. C22H23N3O3.
4-(2,5-Dimethyl-1H-pyrrol-1-yl)-N′-(3,4,5-trimethoxybenzylidene)benzohydrazide (4c)
Yield, 72%; mp 225°C–227°C; IR (KBr): 3184 (NH), 1641 (C=O), 1607 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 2.04 (s, 6H, −2CH3), 3.84–3.93 (m, 9H, −3OCH3), 5.93 (s, 2H, pyrrole-C3, C4−H), 7.09 (s, 2H, methoxyphenyl-C2, C6−H), 7.32 (t, 2H, phenyl-C2, C6−H), 7.99 (d, 2H, phenyl-C3, C5−H), 8.29 (s, 1H, −N=CH−), 9.69 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 13.10, 56.09, 60.89, 104.66, 106.61, 128.20, 128.73, 129.24, 132.05, 140.21, 142.39, 149.78, 153.46, 164.61; MS (EI): m/z = found 408.19 [M+ +1]; calcd. 407.18. Anal. C23H25N3O4.
4-(2,5-Dimethyl-1H-pyrrol-1-yl)-N′- (2-nitrobenzylidene)benzohydrazide (4d)
Yield, 72%; mp 161°C–163°C; IR (KBr): 3212 (NH), 1649 (C=O), 1607 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 2.04 (s, 6H, −2CH3), 5.92 (s, 2H, pyrrole-C3, C4−H), 7.26–8.89 (m, 9H, nitrophenyl-C3, C4, C5, C6 and phenyl-C2, C3, C5, C6 and −N=CH−), 10.29 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 13.04, 106.51, 124.63, 128.25, 128.53, 129.58, 130.49, 131.56, 133.63, 142.53, 144.63, 147.95, 164.35; MS (EI): m/z = found 363.14 [M+ +1]; calcd. 362.14. Anal. C20H18N4O3.
4-(2,5-Dimethyl-1H-pyrrol-1-yl)-N′- (3-phenoxybenzylidene)benzohydrazide (4e)
Yield, 75%; mp 215°C–218°C; IR (KBr): 3191 (NH), 1642 (C=O), 1607 (C=N).
4-(2,5-Dimethyl-1H-pyrrol-1-yl)-N′- (4-methoxybenzylidene)benzohydrazide (4f)
Yield, 75%; mp 122°C–125°C; IR (KBr): 3216 (NH), 1650 (C=O), 1607 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 2.02 (s, 6H, −2CH3), 3.82 (s, 3H, −OCH3), 5.82 (s, 2H, pyrrole-C3, C4−H), 7.01 (d, 2H, methoxyphenyl-C3, C5−H), 7.40 (d, 2H, phenyl-C2, C6−H), 7.71 (d, 2H, methoxyphenyl-C2, C6−H), 8.05 (d, 2H, phenyl-C3, C5−H), 8.41 (s, 1H, −N=CH−), 11.82 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 12.90, 55.26, 106.46, 114.32, 126.79, 127.53, 128.61, 128.73, 141.05, 147.81, 162.36, 171.24; MS (EI): m/z = found 348.17 [M+ +1]; calcd. 347.16. Anal. C21H21N3O2.
4-(2,5-Dimethyl-1H-pyrrol-1-yl)-N′- (4-methylbenzylidene)benzohydrazide (4g)
Yield, 73%; mp 186°C–188°C; IR (KBr): 3222 (NH), 1666 (C=O), 1607 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 2.02 (s, 6H, −2CH3), 2.36 (s, 3H, −CH3), 5.83 (s, 2H, pyrrole-C3, C4−H), 7.27 (d, 2H, methylphenyl-C3, C5−H), 7.41 (d, 2H, methylphenyl-C2, C6−H), 7.65 (d, 2H, phenyl-C2, C6−H), 8.06 (d, 2H, phenyl-C3, C5−H), 8.43 (s, 1H, −N=CH−), 11.90 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 12.90, 21.01, 106.47, 127.10, 127.52, 127.92, 129.43, 131.54, 132.52, 139.95, 141.11, 147.97, 162.49; MS (EI): m/z = found 332.17 [M+ +1]; calcd. 331.17. Anal. C21H21N3O.
N′-(2-Chlorobenzylidene)-4-(2,5-dimethyl-1H-pyrrol-1-yl)benzohydrazide (4h)
Yield, 85%; mp 150°C–153°C; IR (KBr): 3184 (NH), 1656 (C=O), 1607 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 2.00 (s, 6H, −2CH3), 5.80 (s, 2H, pyrrole-C3, C4−H), 7.36 (d, 4H, chlorophenyl-C3, C4, C5, C6−H), 7.45 (d, 2H, phenyl-C2, C6−H), 8.06 (d, 2H, phenyl-C3, C5−H), 8.67 (s, 1H, −N=CH−), 12.18 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 12.84, 106.40, 126.88, 127.20, 127.47, 127.80, 129.68, 131.17, 131.52, 132.10, 133.37, 141.37, 143.99, 162.57; MS (EI): m/z = found 352.12 [M+ +1]; calcd. (351.11). Anal. C20H18ClN3O.
4-(2,5-Dimethyl-1H-pyrrol-1-yl)-N′-(4-isopropylbenzylidene)benzohydrazide (4i)
Yield, 83%; mp 220°C–222°C; IR (KBr): 3178 (NH), 1637 (C=O), 1607 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 1.26–1.31 (m, 6H, isopropyl −2CH3), 2.04 (d, 6H, −2CH3), 2.90–2.97 (m, 1H, isopropyl-CH), 5.94 (s, 2H, pyrrole-C3, C4−H), 7.24–7.33 (m, 4H, isopropylphenyl-C2, C3, C5, C6−H), 7.68 (d, 2H, phenyl-C2, C6−H), 8.01 (d, 2H, phenyl-C3, C5−H), 8.36 (s, 1H, −N=CH−), 9.78 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 18.48, 24.49, 28.79, 101.19, 122.92, 123.29, 126.18, 127.01, 136.97, 145.05, 146.44, 159.15; MS (EI): m/z = found 360.20 [M+ +1]; calcd. 359.20. Anal. C23H25N3O.
N′-((1H-Indol-3-yl)methylene)-4-(2,5-dimethyl- 1H-pyrrol-1-yl)benzohydrazide (4j)
Yield, 65%; mp 165°C–167°C; IR (KBr): 3102 (NH), 1789 (C=O), 1607 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 2.02 (s, 6H, −2CH3), 5.83 (d, 2H, pyrrole-C3, C4−H), 6.62 (d, 1H, indole-C2−H), 7.14–7.81 (m, 4H, indole-C5, C6, C7, C8−H), 8.06 (d, 2H, phenyl-C2, C6−H), 8.27–8.34 (m, 2H, phenyl-C3, C5−H), 8.64 (s, 1H, −N=CH−), 11.58 (s, 1H, NH), 11.65 (s, 1H, indole-NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 12.92, 99.49, 106.42, 111.81, 120.17, 120.40, 120.80, 122.10, 127.55, 127.87, 128.52, 129.04, 129.54, 130.44, 133.12, 137.03, 140.79, 145.10, 161.94; MS (EI): m/z = found 357.17 [M+ +1]; calcd. 356.16. Anal. C22H20N4O.
N′-Butylidene-4-(2,5-dimethyl-1H-pyrrol-1-yl)benzohydrazide (4k)
Yield, 70%; mp 133°C–135°C; IR (KBr): 3275 (NH), 1656 (C=O), 1608 (C=N).
General procedure for the synthesis of N′-(arylidene)-4-(1H-pyrrol-1-yl)benzohydrazides (5a–5f)
Equimolar quantities of 4-(1H-pyrrol-1-yl)benzohydrazide (2b) (0.1 mol) and substituted aldehydes (0.1 mol) with a catalytic amount of glacial acetic acid were refluxed for 4–8 hours using ethanol solvent. The precipitate formed after cooling was collected by filtration, washed with hot ethanol, dried, and recrystallized using ethanol as a solvent to obtain compounds (5a–5f) in good yields.
N′-(3-Methoxybenzylidene)-4-(1H-pyrrol-1-yl)benzohydrazide (5a)
Yield, 65%; mp 165°C–167°C; IR (KBr): 3182 (−NH), 1650 (C=O), 1609 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 3.83 (s, 3H, −OCH3), 6.31 (t, 2H, pyrrole-C3, C4−H), 6.99 (d, 1H, methoxyphenyl-C4−H), 7.27–7.41 (m, 5H, methoxyphenyl-C2, C5, C6 and pyrrole-C2, C5−H), 7.69 (d, 2H, phenyl-C2, C6−H), 8.05 (d, 2H, phenyl-C3, C5−H), 8.45 (s, 1H, −N=CH−), 11.87 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 54.97, 111.06, 116.06, 118.75, 120.11, 129.30, 129.58, 135.63, 142.29, 147.66, 159.44, 162.42; MS (EI): m/z = found 320.13 [M+ +1]; calcd. 319.19. Anal. C19H17N3O2.
N′-(2,4-Dimethoxybenzylidene)-4-(1H-pyrrol-1-yl)benzohydrazide (5b)
Yield, 80%; mp 215°C–220°C; IR (KBr): 3195 (NH), 1644 (C=O), 1607 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 3.84–3.92 (q, 6H, −OCH3), 6.31 (t, 2H, pyrrole-C3, C4−H), 6.58 (t, 2H, methoxyphenyl-C3, C5−H), 7.36 (t, 2H, pyrrole-C2, C5−H), 7.63 (d, 2H, phenyl-C2, C6−H), 7.91 (d, 1H, methoxyphenyl-C6−H), 8.07 (d, 2H, phenyl-C3, C5−H), 8.75 (s, 1H, −N=CH−), 11.70 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 55.64, 97.96, 105.59, 111.18, 118.89, 129.35, 129.91, 130.82, 142.91, 145.27, 159.46, 162.84, 163.58; MS (EI): m/z = found 350.15 [M+ +1]; calcd. 349.14. Anal. C20H19N3O3.
4-(1H-Pyrrol-1-yl)-N′-(3,4,5-trimethoxybenzylidene)benzohydrazide (5c)
Yield, 72%; mp 225°C–230°C; IR (KBr): 3197 (NH), 1645 (C=O), 1608 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 3.88 (s, 9H, −OCH3), 6.40 (t, 2H, pyrrole-C3, C4−H), 6.98–7.15 (m, 4H, methoxyphenyl-C2, C6 and pyrrole-C2, C5−H), 7.48 (t, 2H, phenyl-C2, C6−H), 7.95 (s, 2H, phenyl-C3, C5−H), 8.28 (s, 1H, −N=CH−), 9.43 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 56.22, 60.89, 104.67, 111.58, 118.86, 129.56, 140.13, 143.32, 149.77, 153.49, 164.42; MS (EI): m/z = found 380.16 [M+ +1]; calcd. 379.15. Anal. C21H21N3O4.
N′-(2-Nitrobenzylidene)-4-(1H-pyrrol-1-yl) benzohydrazide (5d)
Yield, 72%; mp 195°C–200°C; IR (KBr): 3222 (NH), 1650 (C=O), 1607 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 6.32 (t, 2H, pyrrole-C3, C4−H), 7.41 (t, 2H, pyrrole-C2, C5−H), 7.63–7.80 (m, 4H, nitrophenyl-C3, C4, C5, C6−H), 8.04–8.23 (m, 4H, phenyl-C2, C3, C5, C6−H), 8.95 (s, 1H, −N=CH−), 12.22 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 111.07, 118.67, 124.35, 128.85, 129.43, 130.18, 133.26, 142.47, 142.69, 147.99, 162.55; MS (EI): m/z =335.11 [M+ +1]; calcd. 334.11. Anal. C18H14N4O3.
N′-(3-Phenoxybenzylidene)-4-(1H-pyrrol-1-yl)benzohydrazide (5e)
Yield, 75%; mp 220°C–223°C; IR (KBr): 3188 (NH), 1671 (C=O), 1607 (C=N).
N′-Butylidene-4-(1H-pyrrol-1-yl)benzohydrazide (5f)
Yield, 70%; mp 210°C–215°C; IR (KBr): 3236 (NH), 1651 (C=O), 1608 (C=N); 1H NMR (400 MHz, CDCl3) δ (ppm): 0.95–0.99 (m, 3H, CH2−CH2−CH3), 1.53–1.59 (m, 2H, CH2−CH2−CH3), 2.27–2.32 (m, 2H, CH2−CH2−CH3), 6.30 (t, 2H, pyrrole-C3, C4−H), 7.36 (t, 2H, pyrrole-C2, C5−H), 7.64 (d, J =8.64 Hz, 2H, phenyl-C2, C6−H), 7.75–7.78 (t, 1H, −N=CH−), 7.99 (d, J =8.64 Hz, 2H, phenyl-C3, C5−H), 11.44 (s, 1H, NH); 13C NMR (100 MHz, CDCl3) δ (ppm): 13.88, 19.97, 29.69, 111.34, 119.62, 128.61, 129.68, 143.03, 153.47, 163.76; MS (EI): m/z = found 256.14 [M+ +1]; calcd. 225.14. Anal. C15H17N3O.
General procedure for the synthesis of copper complexes of N′-(arylidene)-4-(2,5-dimethyl- 1H-pyrrol-1-yl)benzohydrazides (6a–6k)
Compounds (6a–6k) were synthesized by refluxing a mixture of N′-(arylidene)-4-(2,5-dimethyl-1H-pyrrol-1-yl)benzohydrazides (4a–k) (0.1 mol) and copper acetate (0.2 mol) in ethanol for 8–10 hours, and completion of the reaction was monitored by TLC. The reaction mixture was cooled, the separated solid was collected by filtration and washed with cold ethanol. The product was dried and purified by column chromatography using chloroform:petroleum ether in the ratio of 8:2 as the mobile phase.
Bis[4-(2,5-dimethyl-1H-pyrrol-1-yl)-N′-(3-methoxybenzylidene)benzohydrazide] copper (II) anhydride (6a)
Yield, 56%; mp >300°C; IR (KBr): 3109, 2924 (Ar−CH) 1632 (C=N). MS (EI): m/z = found 757.37 [M+ +1]; calcd. 756.35. Anal. (C21H21N3O2)2Cu; UV (λmax): 290.
Bis[N′-(2,4-dimethoxybenzylidene)-4-(2,5-dimethyl-1H-pyrrol-1-yl)benzohydrazide] copper (II) anhydride (6b)
Yield, 52%; mp >300°C; IR (KBr): 2924, 2854 (Ar−CH) 1604 (C=N). MS (EI): m/z = found 817.42 [M+ +1]; calcd. 816.40. Anal. (C22H23N3O3)2Cu; UV (λmax): 300.
Bis[4-(2,5-dimethyl-1H-pyrrol-1-yl)-N′-(3,4,5-trimethoxybenzylidene)benzohydrazide] copper (II) anhydride (6c)
Yield, 54%; mp >300°C; IR (KBr): 3067, 2967 (Ar−CH) 1603 (C=N). MS (EI): m/z = found 877.47 [M+ +1]; calcd. 876.45. Anal. (C23H25N3O4)2Cu; UV (λmax): 294.
Bis[4-(2,5-dimethyl-1H-pyrrol-1-yl)-N′-(2-nitrobenzylidene)benzohydrazide] copper (II) anhydride (6d)
Yield, 64%; mp >300°C; IR (KBr): 2923, 2853 (Ar−CH) 1607 (C=N). MS (EI): m/z = found 787.31 [M+ +1]; calcd. 786.29. Anal. (C20H18N4O3)2Cu; UV (λmax): 300.
Bis[4-(2,5-dimethyl-1H-pyrrol-1-yl)-N′-(3-phenoxybenzylidene)benzohydrazide] copper (II) anhydride (6e)
Yield, 67%; mp >300°C; IR (KBr): 3064, 2918 (Ar−CH) 1603 (C=N). MS (EI): m/z = found 881.51 [M+ +1]; calcd. 880.49. Anal. (C26H23N3O2)2Cu; UV (λmax): 299.
Bis[4-(2,5-dimethyl-1H-pyrrol-1-yl)-N′- (4-methoxybenzylidene)benzohydrazide] copper (II) anhydride (6f)
Yield, 60%; mp >300°C; IR (KBr): 2993, 2920 (Ar−CH) 1604 (C=N); MS (EI): m/z = found 757.37 [M+ +1]; calcd. 756.35. Anal. (C22H22N3O2)2Cu; UV (λmax): 290.
Bis[4-(2,5-dimethyl-1H-pyrrol-1-yl)-N′-(4-methylbenzylidene)benzohydrazide] copper (II) anhydride (6g)
Yield, 62%; mp >300°C; IR (KBr): 3096, 2919 (Ar−CH) 1604 (C=N); MS (EI): m/z = found 725.25 [M+ +1]; calcd. 724.35. Anal. (C21H21N3O)2Cu; UV (λmax): 300.
Bis[N′-(2-chlorobenzylidene)-4-(2,5-dimethyl- 1H-pyrrol-1-yl)benzohydrazide] copper (II) anhydride (6h)
Yield, 65%; mp >300°C; IR (KBr): 3063, 2920 (Ar−CH) 1606 (C=N). MS (EI): m/z = found 766.20 [M+ +1]; calcd. 765.19. Anal. (C22H22N3O2)2Cu; UV (λmax): 280.
Bis[4-(2,5-dimethyl-1H-pyrrol-1-yl)-N′- (4-isopropylbenzylidene)benzohydrazide] copper (II) anhydride (6i)
Yield, 58%; mp >300°C; IR (KBr): 2919, 2850 (Ar−CH) 1608 (C=N); MS (EI): m/z = found 781.31 [M+ +1]; calcd. 780.46. Anal. (C23H25N3O)2Cu; UV (λmax): 300.
Bis[N′-((1H-indol-3-yl)methylene)-4-(2,5-dimethyl-1H-pyrrol-1-yl)benzohydrazide] copper (II) anhydride (6j)
Yield, 55%; mp >300°C; IR (KBr): 3063, 2922 (Ar−CH) 1616 (C=N). MS (EI): m/z = found 775.39 [M+ +1]; calcd. 774.37. Anal. (C22H20N4O)2Cu; UV (λmax): 310.
Bis[N′-butylidene-4-(2,5-dimethyl-1H-pyrrol-1-yl)benzohydrazide] copper (II) anhydride (6k)
Yield, 70%; mp >300°C; IR (KBr): 3038, 2924 (Ar−CH) 1592 (C=N). MS (EI): m/z = found 629.34 [M+ +1]; calcd. 628.27. Anal. (C18H23N3O2)2Cu; UV (λmax): 285.
General procedure for the synthesis of copper complexes of N′-(arylidene)-4-(1H-pyrrol-1-yl)benzohydrazides (7aM–7f)
Compounds (7a–7f) were synthesized by refluxing a mixture of N′-(arylidene)-4-(1H-pyrrol-1-yl)benzohydrazide (5a–5f) (0.1 mol) and copper acetate (0.2 mol) using ethanol as a solvent for 8–10 hours. Completion of the reaction was monitored by TLC. The reaction mixture was cooled, and the separated solid was collected by filtration and washed with cold ethanol. The final product was dried and purified by column chromatography using chloroform:petroleum ether in the ratio of 8:2 as a mobile phase.
Bis[N′-(3-methoxybenzylidene)-4-(1H-pyrrol-1-yl)benzohydrazide] copper (II) anhydride (7a)
Yield, 57%; mp >300°C; IR (KBr): 3066, 2922 (Ar−CH) 1590 (C=N); MS (EI): m/z = found 701.26 [M+ +1]; calcd. 700.24. Anal. (C19H17N3O2)2Cu; UV (λmax): 290.
Bis[N′-(2,4-dimethoxybenzylidene)-4-(1H-pyrrol-1-yl)benzohydrazide] copper (II) anhydride (7b)
Yield, 55%; mp >300°C; IR (KBr): 2960, 2924 (Ar−CH) 1604 (C=N); MS (EI): m/z = found 761.31 [M+ +1]; calcd. 760.30. Anal. (C18H23N3O2)2Cu; UV (λmax): 310.
Bis[4-(1H-pyrrol-1-yl)-N′-(3,4,5-trimethoxybenzylidene)benzohydrazide] copper (II) anhydride (7c)
Yield, 52%; mp >300°C; IR (KBr): 3025, 2920 (Ar−CH) 1605 (C=N); MS (EI): m/z = found 821.36 [M+ +1]; calcd. 820.35. Anal. (C21H21N3O4)2Cu; UV (λmax): 300.
Bis[N′-(2-nitrobenzylidene)-4-(1H-pyrrol-1-yl)benzohydrazide] copper (II)anhydride (7d)
Yield, 59%; mp >300°C; IR (KBr): 3033, 2956 (Ar−CH) 1591 (C=N); MS (EI): m/z = found 731.20 [M+ +1]; calcd. 730.19. Anal. (C18H24N4O3)2Cu; UV (λmax): 310.
Bis[N′-(3-phenoxybenzylidene)-4-(1H-pyrrol-1-yl)benzohydrazide] copper (II) anhydride (7e)
Yield, 60%; mp >300°C; IR (KBr): 3067, 2927 (Ar−CH) 1603 (C=N); MS (EI): m/z = found 825.40 [M+ +1]; calcd. 824.38. Anal. (C24H19N3O2)2Cu; UV (λmax): 290.
Bis[N′-butylidene-4-(1H-pyrrol-1-yl)benzohydrazide]copper (II) anhydride (7f)
Yield, 70%; mp >300°C; IR (KBr): 2959, 2929 (Ar−CH) 1607 (C=N); MS (EI): m/z = found 572.19 [M+]; calcd. 572.16. Anal. (C15H17N3O)2Cu; UV (λmax): 280.
Biological evaluation
In vitro antitubercular activity
Antitubercular activity of the synthesized compounds was evaluated43 using the MABA and was performed in black, clear-bottomed, 96-well microplates in order to minimize background effects. Outer perimeter wells were filled with sterile water to prevent the dehydration in the experimental wells. These plates were then filled with 100 μL of Middle brook 7H9 broth (Sigma-Aldrich, St Louis, MO, USA) and serial dilutions of the compounds were made directly on the plate from 100 to 0.2 μg/mL. Then, these plates were covered and sealed with parafilm and incubated at 37°C for 5 days. After this, 25 μL of the freshly prepared 1:1 mixture of alamar blue reagent and 10% Tween-80 was added to the plate and incubated for 24 hours. A blue color in the well was interpreted as no bacterial growth, but pink color was scored as the growth of the bacteria.
In vitro antibacterial activity
The in vitro antibacterial activity of compounds was assessed44,45 against Gram-positive (S. aureus [ATCC-12598], Bacillus subtilis [ATCC-6633]) and Gram-negative bacteria (Klebsiella pneumoniae [ATCC-29665], Escherichia coli [ATCC-25922]) using the broth microdilution method.
Preparation of media
Sterilization of media and glassware
The media used was Mueller-Hinton agar (Thermo Fisher Scientific, Waltham, MA, USA) and the nutrient agar was sterilized in conical flasks of a suitable capacity by autoclaving at 15 lb pressure for approximately 20 minutes. The test tubes and pipettes were sterilized in hot air oven at 160°C for 1 hour.
Preparation of test compounds
Serial dilutions of the compounds and reference drugs were prepared in Mueller-Hinton agar (Thermo Fisher Scientific). Drugs (1 mg) were dissolved in dimethylsulfoxide/CDCl3 (1 mL). Further, progressive dilutions with the melted Mueller-Hinton agar (Thermo Fisher Scientific) were performed to obtain the required concentrations of 0.2, 0.4, 0.8, 1.6, 3.125, 6.25, 12.5, 25, 50, and 100 μg/mL of test compounds.
Preparation of inoculums
Test organisms were subcultured onto nutrient agar and incubated overnight at 35°C. The tubes that contained 2 mL of Muller-Hinton agar were inoculated with five or more colonies from the agar plate and turbidity was adjusted to match a 1 McFarland standard (105 cfu/mL) and incubated at 37°C for 18 hours. The MIC was determined as the lowest concentration of the compound yielding no visible growth on the plate. To ensure that solvent had no effect on the bacterial growth, a control experiment was performed with the test medium supplemented with DMSO/CDCl3 at the same dilutions as used in the experiments. The DMSO/CDCl3 did not show any effect on the microorganisms in the concentration ranges studied.
Results and discussion
The synthetic route for the synthesis of new targeted derivatives is depicted in Figures 3 and 4. In a condensation process, pyrrole ring (2a–b) was constructed via the reaction of ethyl 4-aminobenzoate 1 with acetonyl acetone/2,5-dimethoxytetrahydrofuran by refluxing with acetic acid. These esters were converted to hydrazides (3a–b) via hydrazinolysis of (2a–b) with hydrazine hydrate. Preparation of Schiff bases (4a–k and 5a–f) in good yields was achieved via condensation reactions (3a–b) with different aldehydes in a refluxing ethanol (Figure 3).
  | Figure 3 Synthetic route for the synthesis of pyrrolyl hydrazones. |
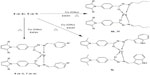  | Figure 4 Synthetic route for the synthesis of metal complexes. |
FTIR spectra of Schiff bases (4a–k) and (5a–f), absorption bands of carbonyl groups showed in the region 1,637–1,789 cm−1 in addition to peaks in the region 1,606–1,609 cm−1 that are attributed to the presence of C=N group. The 1H nuclear magnetic resonance (NMR) spectra of (4a–k) and (5a–f) showed signals of methine proton in the region δ 8.29–8.67 ppm, while protons of −NH were observed in the region δ 9.28–12.18 ppm; a singlet signal approximately δ 2.00–2.04 ppm is attributed to protons of methyl groups at C2 and C5 positions of pyrrole ring. Furthermore, 13C NMR spectra of (4a–k) and (5a–f) showed resonating signals at δ 168 ppm due to carbonyl groups. Signals in the region of δ 141–150 ppm are attributed to methine group; the signals for methyl groups are observed at δ 12.84–18.48 ppm.
Schiff bases (4a–k) and (5a–f) were reacted with copper acetate in a refluxing ethanol medium to furnish the targeted copper complexes (6a–k) and (7a–f), respectively.
FTIR spectra
FTIR spectra of metal complexes are compared with those of ligands to determine coordination sites involved in the bond formation. The presence of some guide peaks in the spectrum of ligand enables to understand this aspect. These peaks change either in their positions and/or their intensities upon chelation, but some peaks disappear after chelation.
The carbonyl stretching bands are found at approximately 1,637–1,789 cm−1 (4a–k and 5a–f), which have disappeared, indicating the involvement of carbonyl group in the bond formation with the metal, while two −C=N peaks are observed in the FTIR spectra of the metal complex. From these observations, it can be concluded that two carbonyl groups of ligand have undergone mesomerism, since the mesomeric form containing −C=N−N−O− arrangement might be involved in the bond formation with the central metal. Further, this type of mesomerism was proved by the appearance of two −C=N− stretching bands in the region of 1,630–1,604 cm−1 (medium band). From the FTIR spectra, it was found that hydrazide nitrogen is involved in the coordinate bond formation with the metal (6a–k and 7a–f).
UV–visible spectroscopy
UV (ultraviolet)–visible spectroscopy was used to analyze the geometry of the metal complexes. To identify metal complexes, generally UV frequency is considered at >300 nm. By this study, we confirm that the synthesized metal complexes have tetrahedral geometry. The stability of copper complexes was performed in different buffer solutions, and complexes were found to be stable.
Mass spectrometry
Mass spectroscopy was used to determine the ratio of ligands involved in the complex formation with the central metal atom as well as to estimate the mole reactants used in the synthesis. It also gives appropriate information about the involvement of water or chlorine molecule during complexation process. In this study, neither water nor chlorine was involved in the formation of the coordination sphere.
Molecular docking
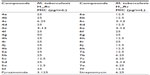
All the conformations were minimized using Tripose force field. The atomic charges were calculated using MMFF94 (Merck Molecular Force Field) method, while Amber7 FF02 (University of California, San Francisco, CA, USA) was used for the protein. Among the synthesized compounds, compounds 4c, 4e, 6c, 7b, and 7d have shown the better scoring function which indicates the binding affinity of these compounds at the active site of the receptor. As depicted in Figure 5, the hydrogen of the amide linkage of compound 4e makes three H-bonding interactions with the cofactor NAD+1270 (NH…O-NAD+1270). Compound 7d makes one H-bonding interaction with NAD+1270, ie, oxygen of −NO2 by interacting with hydrogen of NAD+1270 (Figure 6). The results of molecular docking are listed in Table 1.
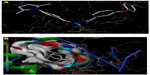  | Figure 5 (A) and (B) Docked view of ligand (compound 4e) in B chain of PDB 2X22. |
  | Figure 6 Docked view of ligand (compound 7d) in B chain of PDB 2X22. |
Biological evaluation
In vitro antitubercular activity
The compounds were evaluated in vitro for antitubercular activity against M. tuberculosis H37Rv using microplate alamar blue assay (MABA) method, with pyrazinamide and streptomycin as the reference standards. The results expressed in minimum inhibitory concentration (MIC) are listed in Table 2. Among all the tested compounds, compounds 4e, 6c, 6e, and 7d were highly active with the MIC of 3.12 μg/mL, while for those of 4c, 7b, 7c, and 7e, the MIC values were 6.25 μg/mL, and the remaining compounds showed moderate activities.
In vitro antibacterial activity
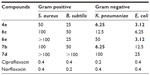
Target compounds that showed better anti-TB activities were further evaluated for in vitro antibacterial activity against both Gram-positive (S. aureus, B. subtilis) and Gram-negative (K. pneumoniae, E. coli) bacteria using broth microdilution method. Ciprofloxacin and norfloxacin were used as reference drugs. These data results are summarized in Table 3.
Based on the previously mentioned study, it was found that the presence of −NH of hydrazide and −NO2 group at ortho position of aromatic ring are essential for the binding at the active site. In summary, our results on the molecular docking and biological screening of compounds offered an excellent framework that may lead to the discovery of potent antimycobacterial agents.
Conclusion
In this work, we have reported the synthesis of new series of Schiff base and their copper complexes along with their spectral data as well as antibacterial and antitubercular activities. Compounds 6c, 6e, and 7d were found to be more active than Schiff bases against M. tuberculosis H37Rv strain, and selected compounds were screened for antibacterial activities that have shown good activity against Gram-negative bacterium, especially against E. coli, than the other tested bacteria with MIC value of 3.12 μg/mL. A marked increase was observed in antibacterial and antitubercular activities when hydrozones were converted to copper complexes, indicating the relevance of copper atom to exhibit biological activity. Molecular docking of all compounds into enoyl ACP-reductase enzyme suggested that compounds 4c, 4e, 6c, 7b, and 7d bind to cofactor NAD+. The result of this study can be utilized to further optimize and improve the potency and selectivity toward ENR enzyme by varying the basic skeleton.
Additional enzyme inhibition studies and molecular docking study involving other enzymes, viz, topoisomerase II, transamidase, dihydrofolate reductase, will offer better insights to understand the detailed binding interaction and the mechanism, this work will be undertaken in the near future.
Acknowledgments
The authors gratefully acknowledge the financial support from the Council of Scientific and Industrial Research, New Delhi, India (Letter Number 02(0139)/13/EMR-II dated 12/04/2013) and also acknowledge the partial financial support from the Vision Group on Science and Technology, Department of Information Technology, Biotechnology and Science and Technology, Bangalore (File VGST/P-3/SMYSR/GRD-277/2013-14/, dated January 28, 2014). We thank Mr HV Dambal, President, SET’s College of Pharmacy, Dharwad, India, for providing facilities. We also thank Dr KG Bhat, Maratha Mandal’s Dental College, Hospital and Research Centre, Belgaum, India, for antitubercular and antibacterial tests. The Director of SAIF Punjab University, Chandigarh, Punjab, India, and the Director of Indian Institute of Technology, Kanpur, India, have provided NMR and mass spectral data. The authors are grateful to Mr Ravi N Nadigir for his technical assistance. One of the authors (Sheshagiri R Dixit) thanks the authorities of Rajiv Gandhi University of Health Sciences, Bangalore, India.
Disclosure
The authors report no conflicts of interest in this work.
References
Wolinsky E. Tuberculosis. In: Wyngaarden JB, Smith LH Jr, Bennett JC, editors. Cecil Textbook of Medicine. Vol 2. 19th ed. Philadelphia, PA: WB Saunders Company; 1992:1733–1742. | |
Sensi P, Grass IGG. Antimycobacterial agents. In: Burger A, Wolff ME, editors. Burger’s Medicinal Chemistry and Drug Discovery. Vol 2. 5th ed. New York, NY: John Wiley and Sons; 1996:575–635. | |
World Health Organization. Global Tuberculosis Report. Geneva, Switzerland: WHO; 2013. | |
Zumla A, Nahid P, Cole ST. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov. 2013;12:388–404. | |
Goldman RC, Plumley KV, Laughon BE. The evolution of extensively drug resistant tuberculosis (XDR-TB): history, status and issues for global control. Infect Disord Drug Targets. 2007;7:73–91. | |
Benatar SR. Extensively drug resistant tuberculosis – problem will get worse in South Africa unless poverty is alleviated. Br Med J. 2006; 333(7570):705. | |
Lawn SD, Wilkinson R. Extensively drug resistant tuberculosis – a serious wake-up call for global health. Br Med J. 2006;333:559–560. | |
Dahle UR. Extensively drug resistant tuberculosis – beware patients lost to follow-up. Br Med J. 2006;333(7570):705. | |
Gandhi NR, Moll A, Sturm AW, et al. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. | |
Manissero D, Fernandez K. Extensive drug-resistant TB: a threat for Europe? Euro Surveill. 2006;11:E060928.2. | |
Takayama K, Wang C, Besra GS. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev. 2005;18:81–101. | |
Takayama K, Wang L, David HL. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1972;2(1):29–35. | |
Quemard A, Laneelle G, Lacave C. Mycolic acid synthesis: a target for ethionamide in mycobacteria? Antimicrob Agents Chemother. 1992;36(6):1316–1321. | |
Banerjee A, Dubnau E, Quemard A, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. | |
Joshi SD, Dixit SR, More UA, Aminabhavi TM, Kulkarni VH, Gadad AK. Enoyl ACP reductase as effective target for the synthesized novel antitubercular drugs: a-state-of-the-art. Mini Rev Med Chem. 2014;14:678–693. | |
Aminabhavi TM, Biradar NS, Patil SB, Hoffman DE. Structural and biological studies on benzimidazolyl amino acid complexes of dimethyldichlorosilane. Inorg Chim Acta. 1986;125:125–128. | |
Jaiswal V, Gupta SR, Rastogi RB, Kumar R, Singh VP. Evaluation of antiwear activity of substituted benzoylhydrazones and their copper(II) complexes in paraffin oil as efficient low SAPS additives and their interactions with the metal surface using density functional theory. J Mater Chem A. 2015;3:5092–5109. | |
Sutradhar M, Kirillova MV, Guedes da Silva MFC, Liu CM, Pombeiro AJL. Tautomeric effect of hydrazone Schiff bases in tetranuclear Cu(II) complexes: magnetism and catalytic activity towards mild hydrocarboxylation of alkanes. Dalton Trans. 2013;42:16578–16587. | |
Nitschke JR. Construction, substitution, and sorting of metallo-organic structures via subcomponent self-assembly. Acc Chem Res. 2007;40:103–107. | |
Opstal T, Verpoort F. Synthesis of highly active ruthenium indenylidene complexes for atom-transfer radical polymerization and ring-opening-metathesis polymerization. Angew Chem Int Ed Engl. 2003;42:2876–2884. | |
Chew ST, Lo KM, Lee SK, et al. Copper complexes with phosphonium containing hydrazone ligand: topoisomerase inhibition and cytotoxicity study. Eur J Med Chem. 2014;76:397–407. | |
Joseph J, Nagashri K, Janaki GB. Novel metal based anti-tuberculosis agent: synthesis, characterization, catalytic and pharmacological activities of copper complexes. Eur J Med Chem. 2012;49:151–163. | |
Patil M, Hunoor R, Gudasi K. Transition metal complexes of a new hexadentate macroacyclic N2O4-donor Schiff base: inhibitory activity against bacteria and fungi. Eur J Med Chem. 2010;45:2981–2986. | |
Liu ZC, Wang BD, Wang B, et al. Synthesis of heterobimetallic complexes: In vitro DNA binding, cleavage and antimicrobial studies. J Med Chem. 2010;45:5353–5361. | |
Kaabi K, Zeller M, Ferretti V, Silva PPS, Nasr BC. Iron(II), Nickel(II), Copper(II) and Zinc(II) Complexes of 2,4-dinitro-6(pyridine-2-ylmethylamino) methylphenolate: synthesis, characterization and Antimicrobial Activities. Inorg Chim Acta. 2012;388:52–59. | |
Singh AP, Kaushik NK, Verma AK, Hundal G, Gupta R. Synthesis, structure and biological activity of copper(II) complexes of 4-(2-pyridylmethyl)-1,7-dimethyl-1,4,7-triazonane-2,6-dione and 4-(2-pyridylethyl)-1,7-dimethyl-1,4,7-triazonane-2,6-dione. Eur J Med Chem. 2009;44:1607–1614. | |
Horsfall JG. Principles of Fungicidal Action. Waltham, MA: Chronica Botanica Co.; 1956:141. | |
Arman P, Wain RL. Studies upon the copper fungicides: X. The role of leaf exudates in the solution of copper from Bordeaux mixture. Ann Appl Biol. 1958;46:366–371. | |
Raman N, Jeyamurugan R, Senthilkumar R, Rajkapoor B, Franzblau SG. In vivo and in vitro evaluation of highly specific thiolate carrier group copper(II) and zinc(II) complexes on Ehrlich ascites carcinoma tumor model. Eur J Med Chem. 2010;45:5438–5451. | |
Li Y, Yang ZY, Wu JC. Synthesis, crystal structures, biological activities and fluorescence studies of transition metal complexes with 3-carbaldehyde chromone thiosemicarbazone. Eur J Med Chem. 2010;45:5692–5701. | |
Sayed M, Zayed MAA, Gehad MG. Tetradentate-arm Schiff base derived from the condensation reaction of 3,3′-dihydroxybenzidine, glyoxal/diacetyl and 2-aminophenol: designing, structural elucidation and properties of their binuclear metal(II) complexes. Arab J Chem. 2010;3:103–113. | |
Joshi SD, More UA, Pansuriya K, Aminabhavi TM, Gadad AK. Synthesis and molecular modeling studies of novel pyrrole analogs as antimycobacterial agents. J Saudi Chem Soc. 2013. doi:10.1016/j.jscs.2013.09.002. | |
More UA, Joshi SD, Aminabhavi TM, Gadadd AK, Nadagouda MN, Kulkarni VH. Design, synthesis, molecular docking and 3D-QSAR studies of potent inhibitors of enoylacyl carrier protein reductase as potential antimycobacterial agents. Eur J Med Chem. 2014;71:199–218. | |
Joshi SD, Dixit SR, More UA, Rai S, Kulkarni VH. Molecular modeling, synthesis, antibacterial and antitubercular activities of some novel pyrrolyl 1,2,4-triazole derivatives. Indo Am J Pharm Res. 2014;4(5):2323–2338. | |
Joshi SD, More UA, Dixit SR, Dubey D, Tripathi A, Kulkarni VH. Discovering potent inhibitors against the enoyl-acyl carrier protein reductase (InhA) of mycobacterium tuberculosis: structure based design, synthesis and antimicrobial activity of quinoline hydrazones. Indo Am J Pharm Res. 2014;4(2):864–877. | |
Tripos International. Sybyl-X 2.0. St Louis, MO: Tripos International; 2012. | |
Jain AN. Scoring noncovalent protein-ligand interactions: a continuous differentiable function tuned to compute binding affinities. J Comput Aided Mol Des. 1996;10:427–440. | |
Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003;46:499–511. | |
Clark M, Cramer RD, Opdenbosch VN. Validation of the general purpose tripos 5.2 force field. J Comput Chem. 1989;10:982–1012. | |
Ai Y, Wang ST, Sun PH, Song FJ. Molecular modeling studies of 4,5-dihydro-1H-pyrazolo[4,3-H] quinazoline derivatives as potent CDK2/cyclin a inhibitors using 3D-QSAR and docking. Int J Mol Sci. 2010;11:3705–3724. | |
Lan P, Chen WN, Chen WM. Molecular modeling studies on imidazo[4,5-b]pyridine derivatives as Aurora A kinase inhibitors using 3D-QSAR and docking approaches. Eur J Med Chem. 2011;46:77–94. | |
Joshi SD, Vagdevi HM, Vaidya VP, Gadaginamath GS. Synthesis of new 4-pyrrol-1-yl benzoic acid hydrazide analogs and some derived oxadiazole, triazole and pyrrole ring systems: a novel class of potential antibacterial and antitubercular agents. Eur J Med Chem. 2008;43:1989–1996. | |
Lourenco MCS, deSouza MVN, Pinheiro AC, et al. Evaluation of anti-tubercular activity of nicotinic and isoniazid analogues. ARKIVOC. 2007;15:181–191. | |
Goto S, Jo K, Kawakita T, Mitsuhashi S, Nishino T, Ohsawa N. Determination method of minimum inhibitory concentrations. Chemotherapy. 1981;29:76–79. | |
Winter CA, Risley EA, Nuss GW. Carrageenin-induced edemas in hind paw of the rat as an assay for antiiflammatory drugs. Proc Soc Exp Biol Med. 1962;111:544–547. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.