Back to Journals » Infection and Drug Resistance » Volume 15
Molecular Characterization of Klebsiella pneumoniae Isolated from Sputum in a Tertiary Hospital in Xinxiang, China
Authors Hao Y, Jiang Y, Ishaq HM, Liu W, Zhao H, Wang M, Yang F
Received 12 April 2022
Accepted for publication 15 June 2022
Published 18 July 2022 Volume 2022:15 Pages 3829—3839
DOI https://doi.org/10.2147/IDR.S370006
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yuqi Hao,1 Yong’ang Jiang,1 Hafiz Muhammad Ishaq,2 Wenke Liu,1 Huajie Zhao,1 Mingyong Wang,3 Fan Yang1
1Xinxiang Key Laboratory of Pathogenic Biology, Department of Pathogenic Biology, School of Basic Medical Sciences, Xinxiang Medical University, Xinxiang, People’s Republic of China; 2Faculty of Veterinary and Animal Sciences, Muhammad Nawaz Shareef University of Agriculture, Multan, Pakistan; 3Xinxiang Key Laboratory of Immunoregulation and Molecular Diagnostics, School of Laboratory Medicine, Xinxiang Medical University,, Xinxiang, People’s Republic of China
Correspondence: Fan Yang; Mingyong Wang, Email [email protected]; [email protected]
Background: In clinical practice, Klebsiella pneumoniae (K. pneumoniae) is a common opportunistic pathogen responsible for nosocomial infection. This study aimed to analyze the trend of antimicrobial susceptibility and virulent characteristics of K. pneumoniae isolated from sputum. In clinics, data of the current study will help in the clinical treatment of K. pneumoniae infection.
Results: The current research showed the resistance rates of the 20 K. pneumoniae isolates against 13 antibiotics ranged from 15.0% to 80.0%. The detection rate of extended spectrum β-lactamases (ESBLs) was up to 55%, while blaSHV was the most prevalent ESBLs genes. Four strains (25.0%) of K. pneumoniae presented hypermucoviscous phenotype (HMV). Moreover, 18 strains (90.0%) showed the stronger biofilm-forming ability. wzi, wabG, fimH, mrkD were the most prevalent virulence genes in current research. Ten strains were found capsule typing and the higher genetic diversity of colonizing K. pneumoniae in this region. K19 exhibited a strong positive correlation with imipenem resistance, while K1 showed strong correlations with magA. Furthermore, HMV phenotype showed significantly negative correlations with multidrug-resistant.
Conclusion: In the hospital, the antibiotic resistance of K. pneumoniae (isolated from sputum samples) has a serious concern. Additionally, strains of K. pneumoniae show the higher genetic diversity.
Keywords: Klebsiella pneumoniae, antimicrobial resistance, resistant genes, virulent genes, biofilm-forming
Introduction
K. pneumoniae (Klebsiella pneumoniae) is a common clinical conditional pathogen. It is an important etiological agent of nosocomial infections. It may cause many infectious diseases, such as pneumonia, bloodstream infections, urinary tract infections, and osteomyelitis.1 It is second to Escherichia coli in terms of detection rate among gram-negative bacteria.2 With the widespread use and sometimes abuse of antibiotics in the clinic, resistant strains increase each year. In particular, the emergence of multidrug-resistant (MDR) strains led to the failure of clinical antibacterial treatment and prolong the course of the disease.3 This situation may increase the medical costs of patients and the mortality of inpatients, which has emerged as an urgent threat to public health. Various virulence factors are utilized in the survival and immune escape of K. pneumoniae infection,4 such as capsular polysaccharide (CPS), lipopolysaccharide (LPS), fimbriae, iron acquisition and biofilm, etc. K. pneumoniae carrying different virulence factors that show the different pathogenic and clinical characteristics.5 The number of studies of clinically isolated K. pneumoniae has been increased dramatically in recent years.6–8 The emergence of MDR K. pneumoniae especially ESBLs-producing strains and carbapenem-resistant strains (CRKP) has brought more difficult for the treatment of K. pneumoniae infections in clinics.9,10 Moreover, K. pneumoniae is thought as an important vehicle for reservoir and transmission resistant and virulent genes,11 Thus, a better understanding and monitoring of these isolates could help limit the spread of antimicrobial resistance. However, the study on antibiotic resistance and molecular characteristics of K. pneumoniae isolated from Xinxiang city, was scarce. Here, we identify the antibiotic resistance and molecular characteristics of K. pneumoniae isolated from clinical sputum samples. This study aimed to better comprehend the molecular epidemiological characteristics of K. pneumoniae in this region, which has great clinical significance for preventing and controlling the K. pneumoniae infection and transmission.
Methods
Bacterial Isolates
Twenty strains of K. pneumoniae (19 from sputum and 1 from alveolar lavage fluid) were collected between July and November 2020 from the Affiliated People’s Hospital of Xinxiang Medical University, Henan Province, China. The patients had been suffering from common underlying diseases, including chronic obstruction, pneumonia, septic chest, lung abscess, craniocerebral injury, coronary artery disease, gastrointestinal bleeding, cerebral infarction, and cirrhosis. The strains were part of the routine laboratory procedures in the hospital. All isolates were identified by staining, biochemical tests and the VITEK-2 compact system (bioMerieux, Craponne, France). K. pneumoniae ATCC 700603 was used as a standard control strain for species identification. The isolates were marked numerically as KP1-KP20.
Antibiotics and Reagents
Following antibiotics were used in this study: Ceftazidime (CAZ, 30μg), Ceftazidime-clavulanic acid (CAC, 30μg/10μg), Cefotaxime (CTX, 30μg), Ceftazidime-clavulanic acid (CTC, 30μg/10μg), Cefoxitin (CFX, 30μg), Aztreonam (AZT, 30μg), Piperacillin-tazobactam (PIT, 100μg/10μg), Piperacillin (PIP, 100μg), Imipenem (IPN, 10μg), Doxycycline (DOX, 30μg), Chloramphenicol (CHL, 30μg), Ciprofioxacin (CIP, 5μg), Levofloxacin (LVF, 5μg), Gatifloxacin (GAT, 5μg), Kanamycin (KAN, 30μg). Luria-Bertani (LB) agar plates and Columbia blood agar plates were purchased from Guangzhou Huankai Microbiology Technology Limited Company. The reagents of polymerase chain reaction (PCR) were purchased from Kangwei Century Co., Ltd. The primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. The primer sequences are shown in Tables 1 and 2.
 |
Table 1 PCR Primer Sequence Information of Antimicrobial Resistance Genes |
 |
Table 2 PCR Primers Sequence Information of Virulence Genes |
ESBLs Phenotype Confirmation Tests
ESBLs phenotypes of 20 Strains of K. pneumoniae were detected by the combined disc test (CDT) recommended by the Clinical & Laboratory Standards Institute (CLSI) 2020.12 Briefly, the bacteria with turbidity equivalent to 0.5 McFarland standards were swabbed onto Mueller-Hinton (MH) agar plates containing 30 μg of CTX and CAZ, with and without 10 μg of clavulanic acid, and were placed independently, 25 mm apart (center to center) on a lawn culture. However, K. pneumoniae ATCC 700603 was used as a standard control strain. The plates were incubated at 37°C for 16–18 h. Isolates were considered ESBL positive if the inhibition zone measured around one of the combination discs was at least 5 mm larger than the corresponding cephalosporin disc.
Antimicrobial Susceptibility Testing
The sensitivity of K. pneumoniae isolates against 13 kinds of antibiotics was determined by the disc diffusion method recommended by CLSI.12 Antibiotic susceptibility results were determined according to the CLSI 2020 standard. Moreover, Escherichia coli ATCC 35218 was used as a standard control strain. We considered strains resistant to at least three antimicrobial classes as MDR strains.13
Detection of Antimicrobial Resistance Genes
Bacterial DNA template was extracted by using the water boiling method. Briefly, bacteria were inoculated, and cultured overnight in LB liquid medium at 37°C. There was 1 mL bacterial culture added into the 1.5 mL EP tube, centrifuged at 12,000 RPM for 2 min, and discarded the supernatant. Then, the sediments were resuspended in 500 μL water, centrifuged again under the aforementioned conditions, and discarded the supernatant. After Adding 200 μL water, boiled at 100°C, 20–30 min to lysate thallus. Freezing for 10–15 min and centrifuged at 12,000 rpm at 4°C for 15 min. The supernatant (genomic DNA) was extracted into the new EP tube and stored at −20°C. Sterile water was used as blank control, and the following resistant genes of K. pneumoniae were amplified by PCR, including encode ESBLs (blaCTX-M, blaTEM, blaSHV), carbapenemase genes (blaKPC, blaNDM, blaOXA-48, blaIMP), quinolones resistance genes (qnrA, qnrB, oqxA, oqxB), and aminoglycoside resistance gene (aac). The corresponding gene sequences were taken from NCBI and completed primers designed as primer 5.0. The primer sequences of the resistant genes are shown in Table 1.
Determination of Mucinous Phenotype
The mucinous phenotype of K. pneumoniae was analyzed by “String-forming test” according to the previous method.14 Briefly, K. pneumoniae was transferred to Columbia blood plate for overnight culture and incubated at 37°C. Dip the colony was with the inoculum ring, then lift the inoculum ring. If the adhesive wire formed larger than 0.5 cm, it was considered positive; otherwise, it was negative. The strain with high mucilage phenotype was considered positive in the “String-forming test”.
Analysis of Biofilm Formation Ability
Biofilm-forming ability was measured by determining adhesion to flat-bottomed microtiter plates (96-well).15 Briefly, each well of the 96-well microtitration plates was filled with 200 μL sterile broth liquid medium. During biofilm formation (18 h incubation), bacterial cultures were added to each well (1:100 in liquid medium, 200 μL liquid medium). Sterile LB liquid medium was used as a standard blank control. After incubation at 37°C for 48 h, total cell mass was measured as absorbance at 570 nm (OD1) while the blank was OD10. Each well was washed 3 times with phosphate buffer saline (PBS), dried for 1 h at 60°C and stained for 20 min with 200 μL of 1% crystal violet. After removing the crystal violet solution, each well was washed with PBS 4 times to remove the remaining stain. Air-dried was done in the aseptic processing table for 30 mins, and each well was added with 200 μL of 95% ethanol. After vibrating for 30 min, the absorbance was measured at 570 nm (OD2), while the blank was OD20. The biofilm formation capacity was calculated by using B=(OD2-OD20)/(OD1-OD10). All the strains were classified based on the adherence capabilities into the following categories: nonbiofilm producers (B < 0.1), weak biofilm producers (B ≥ 0.1), moderate biofilm producers (0.1 < B ≤ 1.0), strong biofilm producers (B > 1.0).
Analysis of Capsular Serotypes and Virulence Genes
Amplifying wzi genes for detecting capsular polysaccharide (antigen K) serotype is a new method for capsular serotyping, which has been used in the laboratory. In this study, wzi primers were provided for expanded PCR amplification according to the previous study,16 and the products were sent to Bioengineering (Shanghai) incorporated company for sequencing. Sequencing results were submitted Institut Pasteur website17 for comparative analysis, obtained strain wzi parting and part of capsular serotyping. Virulence genes were detected by PCR, including encoding capsular polysaccharide (wzi), adhesin (fimH, markD), lipopolysaccharide (wabG), mucous phenotypic related genes (rmpA, magA), and ferritin genes (iucA, iutA). PCR primer sequence of virulence genes has been shown in Table 2.
Statistical Analysis
Clustal W 2.1 was used to complete the alignment of wzi gene sequence. Initial phylogenetic trees were constructed using MEGA 7 based on the neighbor-joining method (500 bootstrap replicates) and Jukes-Cantor distance. Pearson’s correlation coefficient was calculated by applying bivariate correlation analysis with SPSS software version 19.0. The correlation in the heat map of the generated data was computed by using Hiplot.
Results
Antimicrobial Susceptibility Testing
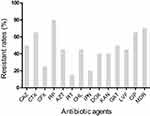
The resistance of 20 strains of K. pneumoniae to 13 commonly used antibiotics has been shown in Figure 1. Overall, K. pneumoniae isolated from various underlying diseases showed high antibiotic resistance rates ranging from 15.0% to 80.0%. However, multi-drug resistance rates were upto 70.0%. The strains showed the highest resistance to PIP (80.0%), CTX (65.0%) and CIP (65.0%) respectively. The lowest resistance was to PIT (15.0%), followed by IPN (20.0%) and CFX (25.0%). The resistance rate of other antibiotics was about 50.0%.
ESBL Phenotype and Resistance Genes of K. pneumoniae
Table 3 describes the distribution of antibiotic resistance genes in clinical isolated K. pneumoniae. Strains producing ESBLs were common in the region with a detection rate of 55.0%, while blaSHV was the most widely distributed gene, but the other two genes were less frequently detected. The blaKPC type carbapenemase gene was detected in only three strains. Furthermore, the other types of carbapenemase genes were not detected in the samples. Among plasmid-mediated quinolone resistance (PMQR) genes, qnrA was not detected, while oqxAB was present in almost all strains. Genes aac and qnrB were scattered, with detection rates of 40.0% and 25.0%, respectively, shown in Table S1. blaCTX-M +blaTEM+aac+oqxAB gene strains were widely distributed about (30.0%), which possesses multidrug-resistant strains. The 2/3 strains showed co-production of blaKPC+blaSHV+oqxAB among blaKPC-producing strains. Overall, the detection multidrug-resistant rate was higher in strains isolated from pulmonary infectious diseases, which account for 87.5% (7/8).
 |
Table 3 Distribution of Antibiotic Resistance Genes and ESBL Phenotype in 20 Strains of K. pneumoniae |
Virulence Phenotypes and Genes of K. pneumoniae
The results of the crystal violet staining showed that all 20 K. pneumoniae could form biofilms, while 18 strains (90.0%) showed strong biofilms. String test showed that 5 strains (25.0%) presented the HMV phenotype. Table 4 characterizes the distribution of the virulence genes of K. pneumoniae strains isolated from various underlying diseases. The wabG gene encoding with LPS and wzi encoding with CPS were found in all strains. The genes fimH and mrkD encoding with pili were detected at 90% and 85%, respectively. The most prevalent virulence genes iucA and iutA (encoding and regulating with aerobactin)18 were detected at 35.0% and 30.0%, respectively. Overall, the strains extracted from patients with craniosynostosis were more virulent and had a higher level of all virulence gene detection rate, accounting for 50% (3/6).
 |
Table 4 Distribution of Virulence Genes of K. pneumoniae Isolated from Sputum |
Capsular Serotypes and wzi Phylogenetic Tree
Among 20 K. pneumoniae isolates, 10 strains were identified as K1 (2 strains), K19 (2 strains), K17, K23, K24, K46.61, K14.64, and K43 (1 strain). In contrast, the other 10 strains were unknown capsular serotyping, which belonged to wzi 168, wzi84, wzi679, wzi206, wzi187, wzi209, wzi150, wzi516, wzi275, and wzi401. Each sequence corresponds indicated a distinct wzi allele. The corresponding capsular types followed the allele number. The phylogenetic tree showed a high genetic diversity with more branches of the sample wzi gene and significant differences were observed between strains isolated from the samples and the same families (Figure 2).
Correlation Analysis
Pearson’s correlation analysis was performed to assess the existence of the potential correlation between antibiotic-resistant phenotypes and molecular characteristics of K. pneumoniae strains. Moreover, Figure 3 suggests that the MDR is positively significant correlated with blaTEM (r = 0.535, p < 0.05), blaCTX-M (r = 0.480, p < 0.05) and aac (r = 0.535, p < 0.05), while negative correlations with HMV (r = −0.630, p < 0.01) and rmpA (r = −0.630, p < 0.01). HMV exhibited positive correlations with rmpA (r = 1.000, p < 0.01), iucA (r = 0.545, p < 0.05), iutA (r = 0.630, p < 0.01) and magA (r = 0.577, p < 0.01). SBF was positively correlated with mrkD (r = 0.327). Some molecular types displayed positive or negative correlations with several resistance and virulence genes. K19 and CR (r = 0.667, p < 0.01), blaKPC (r = 0.793, p < 0.01) displayed a strong positive correlation with SBF (r = −1.000, P < 0.01), and a negative correlation with mrkD (r = −0.327). K1 was completely correlated with magA (r = 1.000, p < 0.01). According to Figure 3, virulence genes rmpA and iutA (r = 0.630, p < 0.01) shows significantly positive correlation, and all antibiotic resistance phenotypes have a huge correlation with their genes.
Discussion
It has been reported that the antibiotic resistance of K. pneumoniae isolated from sputum has increased year by year.6–8 But molecular characteristics studies are still very rare. In the current study, 20 strains of K. pneumoniae (19 from sputum and 1 from alveolar lavage fluid) were collected from the affiliated people’s hospital of Xinxiang Medical University, Henan Province, China. The antibiotic susceptibility results showed that the antibiotic resistance of sputum isolates of K. pneumoniae was severe, with the highest resistance rate of 80.0% for PIP. While lower resistance rates for IPN and PIT 20.0% and 15.0%, respectively. Moreover, ESBLs-producing positive strains were found up to 55.0%. So, these results are consistent with the existing literature.6,19,20 The aforementioned literature also reported that resistance rates of cephalosporins and quinolone antibiotics rates were below 40.0% a while carbapenems were below 10.0%. In the current research, K. pneumoniae strains exhibited a higher resistance prevalence than the literature cited above. Current study results suggest that this difference may be related to the spread of the region’s antimicrobial resistance genes due to frequent antibiotic use.21 Therefore, further experimental work is still to be needed to explore the antimicrobial resistance genes and the local antibiotic use.
Many antibiotic-resistant genes are involved in the drug resistance of K. pneumonia. The current study elaborates the mechanism of antibiotic resistance which is being used in clinical practice. β-lactam antibiotics are the most commonly used antibiotics in the clinic, among which ESBLs production is the primary mechanism of antibiotic resistance. In this study, all ESBLs-producing strains were MDR strains that are used in the current study. These findings support the conclusion that ESBLs-producing bacteria’s resistance was enhanced, agreeing with the reported studies.22,23 In the current research, blaSHV is more widely distributed among genes encoding resistance to ESBLs, which may be the main gene type in the prevalence of ESBLs in our region. The underlying mechanism of antibiotic resistance is the production of carbapenemases which can hydrolyze penicillins, cephalosporins, monocyclic β lactamides, aminoglycosides, quinolones, and other antibiotics.24 The modified Hodge test and PCR were used to confirm the phenotype and genes of carbapenemase. The results showed that only 3 K. pneumoniae of producing carbapenemases were detected with KPC gene in this study, and these 3 strains were resistant to all antibiotics except Kanamycin, chloramphenicol and doxycycline. The details of antibiotics have been described in Table S2. Fluoroquinolones are a class of powerful broad-spectrum antibiotics that have been used to treat severe or antibiotic-resistant infections since the end of the 20th century.25 However, in recent years, with frequent use and abuse of antibiotics, the resistance rate of the pathogen to fluoroquinolones has been increasing and is currently up to 50%. Reported studies by Liam S. Redgrave et al showed that the prevalence of PMQR genes among the strains was not affected by quinolone antibiotics selection,25 which may be the crucial factor in the rapid spread of resistance. In current experimental work, qnrA was not detected, qnrB and aac were scattered, while the detection rate of oqxAB was higher, which is parallel with some reported literature.26,27
The emergence of MDR strains would be hindered the clinical treatment of infection caused by K. pneumoniae. The MDR study mechanism has great importance in reducing the fatality rate, improving the spread of MDR strains, and delaying the occurrence of pan-resistant strains. The results show that the rate of K. pneumoniae in MDR is higher, about 70.0%. However, the antibiotic resistance genes blaCTX-M, blaTEM, and aac are closely associated with MDR strains. Also, all three genes are plasmid-mediated antibiotic resistance genes.28 So, the strains that produced all three genes together accounted for about 30.0%. It has been reported that PMQR genes (aac, qnrA, qnrB, oqxA and oqxB) are usually carried on plasmids with other resistance genes, especially ESBLs-producing plasmids.26 The results of the current study showed that ESBLs genes (blaCTX-M, blaTEM) and PMQR genes (aac) are more strongly correlated. Therefore, with combined literature26 analysis, the above mentioned genes may be located in the same plasmid for transmission. Therefore, these genes may be critical for transmitting multi-drug resistant strains in this region.
The main virulence factors of K. pneumoniae included capsular (CPS), lipopolysaccharide (LPS), pili, and iron carriers, which are also the main factors leading to the characteristics of hypervirulent K. pneumoniae (hvKP).29 Many genes encoding these virulence factors have been reported, and some representative genes were selected for amplification in the current study. The results show that HMV phenotype is strongly correlated with rmpA, mapA, iucA and iutA, which is aligned with previous studies. rmpA and magA regulate the expression of CPS, closely related to the high HMV phenotype of K. pneumonia.29,30 While iucA codes aerobactin and iutA codes aerobactin transporter18 shows higher level virulence.30 wzi and wabG have been detected in 20 strains of K. pneumoniae, wzi encodes outer membrane protein Wzi and is involved in the attachment of the CPS to the outer membrane.16 wabG gene encodes WabG protein and is involved in the synthesis of LPS.31 The detection rates of fimH and mrkD genes encoding type I and III pili are 90% and 85%, respectively. The detection rate of the above four genes is higher, which is consistent with the reported literature.6,26,32 All K. pneumoniae in this study can form the biofilms, which may be related to the source of samples. Type I and III pili are crucial factors for K. pneumoniae colonization, and fimH encodes FimH adhesion molecule at the tip of type I pili,32 which is closely related to urinary tract infection.33 mrkD encodes adhesive subunit located at the tip of type III pili,30 and mediates the binding of K. pneumoniae with organ cells and lung tissues,34 which is related to lung infection. The current study results elaborate that SBF and mrkD showed a positive correlation, which supports our study.
To analyze K. pneumoniae, basic information about the isolated strains was taken from Xinxiang City, Henan Province, between July and November 2020. Overall, strains isolated from pulmonary infectious diseases tend to exhibit greater drug resistance, which is relatively more virulent strains isolated from craniosynostosis (more common and critical). The high rate of resistance in pulmonary infectious diseases may be related to the use of β-lactamase inhibitor antibiotics prior to sampling.21 β-lactamase inhibitor antibiotics are more commonly available in the community in Henan Province. So, this phenomena may lead to the occurrence of drug-resistant bacteria in the community. Strains of current research collected from the People’s Hospital of Xinxiang City, Henan Province (not the community) provide a possible history of antibiotic usage in the patients. Therefore, it is necessary to analyze the local dissemination of drug-resistant genes by incorporating antibiotic abuse in the local community. Patients with craniosynostosis are often associated with severe clinical symptoms, which may be related to the high virulence of the strains.
wzi, is a conserved open reading frame. So the first of four conserved gene blocks (wzi-wza-wzb-wzc) were found in all group 1 K-antigen serotypes.35 Sequencing of the wzi genotype allows rapid prediction of K-serotypes, and this method can be used to determine K-types.36–38 In this study, we found wzi typing and K-antigen serotypes and some strains by wzi sequence analysis. Moreover, only exact matches were selected for analysis to ensure accurate experiments. wzi sequencing are not sufficient for accurate serotyping. Therefore, K serotypes of some strains could not be determined. Clear serotyping by immunological methods or sequencing the entire polysaccharide capsule synthesis gene cluster (35 kbp) is necessary for further genetic characterization. In the current study, wzi phylogenetic tree shows the genetic diversity of the experimental samples and the low probability of intra-sectional strain transmission. So, it indicates a representative sample.
Correlation analysis shows a strong relationship between the distribution of iutA and rmpA, which may be related to the fact that both the genes are located in the same pLVPK plasmid.39 It has been reported that magA encodes CPS polymerase specific to K1 type,40 K1 serotype and magA also shows an absolute correlation in this study. Serotype K19 is strongly correlated with blaKPC, which might be the main epidemic serotype of carbapenase in the local area. The antibiotic resistance of K. pneumoniae generally negatively correlates with the distribution of virulence characteristics. Thus, the multiresistant strains exhibited negative correlations with virulence phenotypes and genes. The HMV phenotype is negatively correlated with antibiotic resistance phenotypes and genes, which is consistent with previously reported literature.41 KP13 is a multiresistant strain of capsular serotype K1, carrying three types of ESBLs antibiotic-resistant genes in the current study, which shows serious concern with public health. The prevalence mechanism of antibiotic resistance and virulence factors has crucial implications for the patients.
Conclusions
In conclusion, our study describes antibiotic resistance and virulence characteristics of 20 Strains of K. pneumoniae isolated from the sputum samples in Xinxiang, China. The results show that local K. pneumoniae has severe drug resistance and contains antibiotic resistance genes (blaCTX-M, blaTEM and aac). Virulence genes magA, rmpA, iucA and iutA are crucial influencing factors of HMV. In general, virulence exhibited significant negative correlations with antibiotic resistance factors, and some strains exist simultaneously. Therefore, it is necessary to investigate their transmission mechanism to effectively slow down the emergence of such strains, ultimately reducing the medical treatment costs.
Data Sharing Statement
The datasets supporting the conclusions of this article are included within the article (and the Supplementary Materials).
Ethics Approval and Consent to Participate
This research was carried out in accordance with Declaration of Helsinki. The informed consent have been voluntarily obtained from the participants and participants have been informed of the study including any of the benefits and risks involved. The research was approved by the Ethics Committee of Xinxiang Medical University.
Funding
This research was supported by Science and Technology Research Project of Henan Province (grant 182102310553, 222102520036), the Project of Basic Medical College of Xinxiang Medical university (grant JCYXYKY202117), the Program for Innovative Research Team (in Science and Technology) in University of Henan Province (20IRTSTHN030), Natural Science Foundation of Henan Province for Distinguished Young Scholars (212300410013).
Disclosure
The authors declare that they have no competing interests.
References
1. Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae.. Front Cell Infect Microbiol. 2018;8:4. doi:10.3389/fcimb.2018.00004
2. Tantry BA, Rahiman S. Antibacterial resistance and trend of urinary tract pathogens to commonly used antibiotics in Kashmir Valley. West Indian Med J. 2012;61(7):703–707.
3. Karaiskos I, Giamarellou H. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin Pharmacother. 2014;15(10):1351–1370. doi:10.1517/14656566.2014.914172
4. Li B, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9(9):1071–1081. doi:10.2217/fmb.14.48
5. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):e00001–19. doi:10.1128/CMR.00001-19
6. Wyres KL, Nguyen TNT, Lam MMC, et al. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med. 2020;12(1):11. doi:10.1186/s13073-019-0706-y
7. Imai K, Ishibashi N, Kodana M, et al. Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study, Japan, 2014–2017. BMC Infect Dis. 2019;19(1):946. doi:10.1186/s12879-019-4498-x
8. Zhang H, Zhang G, Yang Y, et al. Antimicrobial resistance comparison of Klebsiella pneumoniae pathogens isolated from intra-abdominal and urinary tract infections in different organs, hospital departments and regions of China between 2014 and 2017. J Microbiol Immunol Infect. 2021;54(4):639–648. doi:10.1016/j.jmii.2020.03.009
9. Chong Y, Shimoda S, Shimono N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol. 2018;61:185–188. doi:10.1016/j.meegid.2018.04.005
10. Hu Y, Liu C, Shen Z, et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect. 2020;9(1):1771–1779. doi:10.1080/22221751.2020.1799721
11. Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018;45:131–139. doi:10.1016/j.mib.2018.04.004
12. Clinical and Laboratory Standards Instiute. Performance standards for antimicrobial susceptibility testing.
13. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
14. Yang F, Deng B, Liao W, Wang P, Chen P, Wei J. High rate of multiresistant Klebsiella pneumoniae from human and animal origin. Infect Drug Resist. 2019;12:2729–2737. doi:10.2147/IDR.S219155
15. Rode TM, Langsrud S, Holck A, Møretrø T. Different patterns of biofilm formation in Staphylococcus aureus under food-related stress conditions. Int J Food Microbiol. 2007;116(3):372–383. doi:10.1016/j.ijfoodmicro.2007.02.017
16. Brisse S, Passet V, Haugaard AB, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51(12):4073–4078. doi:10.1128/JCM.01924-13
17. Institut Pasteur. Bacterial Isolate Genome Sequence Database (BIGSdb); Klebsiella pneumoniae. Available from: https://bigsdb.pasteur.fr/klebsiella/. Accessed June 1, 2021.
18. Russo TA, Olson R, Macdonald U, et al. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun. 2014;82(6):2356–2367. doi:10.1128/IAI.01667-13
19. Vuotto C, Longo F, Pascolini C, et al. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J Appl Microbiol. 2017;123(4):1003–1018. doi:10.1111/jam.13533
20. Soltani E, Hasani A, Rezaee MA, et al. Virulence characterization of Klebsiella pneumoniae and its relation with ESBL and AmpC beta-lactamase associated resistance. Iran J Microbiol. 2020;12(2):98–106.
21. Bian W, Chen W, Gu X, et al. Analysis of related factors of carbapenem resistant Klebsiella pneumoniae infection in patients with artificial airway. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2020;32(11):1324–1330. doi:10.3760/cma.j.cn121430-20200601-00431
22. Gautam V, Thakur A, Sharma M, et al. Molecular characterization of extended-spectrum β-lactamases among clinical isolates of Escherichia coli & Klebsiella pneumoniae: a multi-centric study from tertiary care hospitals in India. Indian J Med Res. 2019;149(2):208–215. doi:10.4103/ijmr.IJMR_172_18
23. Yazdansetad S, Alkhudhairy MK, Najafpour R, et al. Preliminary survey of extended-spectrum β-lactamases (ESBLs) in nosocomial uropathogen Klebsiella pneumoniae in north-central Iran. Heliyon. 2019;5(9):e02349. doi:10.1016/j.heliyon.2019.e02349
24. Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi:10.1128/CMR.00001-07
25. Redgrave LS, Sutton SB, Webber MA, Piddock LJ. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014;22(8):438–445. doi:10.1016/j.tim.2014.04.007
26. Silva-Sánchez J, Cruz-Trujillo E, Barrios H, et al. Characterization of plasmid-mediated quinolone resistance (PMQR) genes in extended-spectrum β-lactamase-producing Enterobacteriaceae pediatric clinical isolates in Mexico. PLoS One. 2013;8(10):e77968. doi:10.1371/journal.pone.0077968
27. Yuan J, Xu X, Guo Q, et al. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J Antimicrob Chemother. 2012;67(7):1655–1659. doi:10.1093/jac/dks086
28. Schultsz C, Geerlings S. Plasmid-mediated resistance in Enterobacteriaceae: changing landscape and implications for therapy. Drugs. 2012;72(1):1–16. doi:10.2165/11597960-000000000-00000
29. Wang G, Zhao G, Chao X, Xie L, Wang H. The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. Int J Environ Res Public Health. 2020;17(17):6278. doi:10.3390/ijerph17176278
30. Clegg S, Murphy CN, Mulvey MA, Stapleton AE, Klumpp DJ. Epidemiology and Virulence of Klebsiella pneumoniae. Microbiol Spectr. 2016;4(1):
31. Izquierdo L, Coderch N, Piqué N, et al. The Klebsiella pneumoniae wabG gene: role in biosynthesis of the core lipopolysaccharide and virulence. J Bacteriol. 2003;185(24):7213–7221. doi:10.1128/JB.185.24.7213-7221.2003
32. Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18(5):224–232. doi:10.1016/j.tim.2010.03.002
33. Hornick DB, Thommandru J, Smits W, Clegg S. Adherence properties of an mrkD-negative mutant of Klebsiella pneumoniae. Infect Immun. 1995;63(5):2026–2032. doi:10.1128/iai.63.5.2026-2032.1995
34. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
35. Rahn A, Beis K, Naismith JH, Whitfield C. A novel outer membrane protein, Wzi, is involved in surface assembly of the Escherichia coli K30 group 1 capsule. J Bacteriol. 2003;185(19):5882–5890. doi:10.1128/JB.185.19.5882-5890.2003
36. Candan ED, Aksöz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62(4):867–874. doi:10.18388/abp.2015_1148
37. Kim JH, Cho YY, Choi JY, Wi YM, Ko KS. Two distinct genotypes of KPC-2-producing Klebsiella pneumoniae isolates from South Korea. Antibiotics. 2021;10(8):911. doi:10.3390/antibiotics10080911
38. Choi M, Hegerle N, Nkeze J, et al. The diversity of lipopolysaccharide (O) and capsular polysaccharide (K) antigens of invasive Klebsiella pneumoniae in a multi-country collection. Front Microbiol. 2020;11:1249. doi:10.3389/fmicb.2020.01249
39. Tang HL, Chiang MK, Liou WJ, et al. Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur J Clin Microbiol Infect Dis. 2010;29(6):689–698. doi:10.1007/s10096-010-0915-1
40. Lin TL, Yang FL, Yang AS, et al. Amino acid substitutions of MagA in Klebsiella pneumoniae affect the biosynthesis of the capsular polysaccharide. PLoS One. 2012;7(10):e46783. doi:10.1371/journal.pone.0046783
41. Lee CR, Lee JH, Park KS, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. doi:10.3389/fcimb.2017.00483
42. Monstein HJ, Ostholm-Balkhed A, Nilsson MV, Nilsson M, Dornbusch K, Nilsson LE. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS. 2007;115(12):1400–1408. doi:10.1111/j.1600-0463.2007.00722.x
43. Piekarska K, Wołkowicz T, Zacharczuk K, et al. Co-existence of plasmid-mediated quinolone resistance determinants and mutations in gyrA and parC among fluoroquinolone-resistant clinical Enterobacteriaceae isolated in a tertiary hospital in Warsaw, Poland. Int J Antimicrob Agents. 2015;45(3):238–243. doi:10.1016/j.ijantimicag.2014.09.019
44. Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705. doi:10.1084/jem.20030857
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.