Back to Journals » Infection and Drug Resistance » Volume 13
Molecular Characterization of Carbapenem/Colistin-Resistant Acinetobacter baumannii Clinical Isolates from Egypt by Whole-Genome Sequencing
Authors Fam NS, Gamal D , Mohamed SH , Wasfy RM, Soliman MS , El-Kholy AA , Higgins PG
Received 27 October 2020
Accepted for publication 3 December 2020
Published 16 December 2020 Volume 2020:13 Pages 4487—4493
DOI https://doi.org/10.2147/IDR.S288865
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Nevine S Fam,1 Doaa Gamal,1 Sara H Mohamed,2 Reham M Wasfy,1 May S Soliman,3 Amani A El-Kholy,3 Paul G Higgins4
1Department of Microbiology, Theodor Bilharz Research Institute, Giza, Egypt; 2Department of Microbiology, National Organization for Drug Control and Research, Giza, Egypt; 3Clinical Pathology Department, Faculty of Medicine, Cairo University, Cairo, Egypt; 4Institute for Medical Microbiology, Immunology and Hygiene, University of Cologne, Cologne 50935, Germany
Correspondence: May S Soliman
Clinical and Chemical Pathology Department, Faculty of Medicine, Cairo University, El Saray Street Manial, El Manial, 11956-Manial, Cairo, Egypt
Tel +20 21068800242
Email [email protected]
Purpose: The rise of carbapenem-resistant A. baumannii (CRAB) is considered a public health problem limiting the treatment options. Our current work studied the emergence and mechanisms of colistin-resistance among CRAB isolates in Egypt.
Materials and Methods: Seventeen clinically recovered A. baumannii were identified and screened for their antimicrobial susceptibilities using VITEK-2 system. Colistin susceptibility was evaluated using broth microdilution, and characterization of carbapenem/colistin resistance determinants was performed using whole-genome sequencing (Illumina MiSeq).
Results: About 52.9% (9/17) were colistin-resistant. PCR results revealed that all isolates carried blaOXA-51-like genes, blaOXA-23-like was detected in 82.3% (14/17) and blaNDM in 23.5% (4/17). Two isolates harboured blaGES-35 and blaOXA-23. Furthermore, genome analysis of seven isolates revealed six belonged to international clone 2 (IC2) while the remaining isolate was a singleton (ST158), representing a clone circulating in Mediterranean/Middle Eastern countries.
Conclusion: The emergence and high incidence of colistin-resistance among CRAB clinical isolates in Egypt are alarming because it further limits therapy options and requires prudent antimicrobial stewardship and stringent infection control measures. Whole-genome sequence analyses suggest that the resistance to colistin was associated with multiple mutations in the pmrCAB genes. The high incidence of the high-risk lineage IC2 harbouring blaOXA-23-like as well as blaNDM is also of concern.
Keywords: colistin resistance, pmrCAB, cgMLST, ST158, WGS
Introduction
Recently carbapenem-resistant Acinetobacter baumannii (CRAB) was placed on top of the list of priority pathogens for research and development of novel antibiotics.1 Multidrug-resistant (MDR) A. baumannii infections are a burden on healthcare facilities owing to limited antibiotic treatment options.2,3 Carbapenem hydrolysing class D beta-lactamases, also known as oxacillinases (OXA), are the main mechanism of resistance to carbapenems in CRAB. The OXA carbapenemases found in A. baumannii are divided into six phylogenetic subgroups: the intrinsic OXA-51-like and the acquired OXA-23-like, OXA-24/40-like, OXA-58-like which are the most common, and the less common OXA-143-like and OXA-235-like.4,5
To treat CRAB, colistin is prescribed as the last resort. However, resistance towards colistin among A. baumannii has become a serious problem as there are no alternative available therapeutic options.6 There are now reports from different regions of the world of pan-drug-resistant (PDR) A. baumannii.7
The primary resistance mechanism is mediated through the modification of the lipid A moiety of lipopolysaccharide (LPS) through the addition of phosphoethanolamine (PEtN) through overexpression and/or mutations in the two-component regulatory system PmrA and PmrB. Other mechanisms have been described and include the loss of LPS caused by either insertional inactivation of lipid A biosynthesis genes or mutations, but this appears to be limited to in-vitro studies.8 A. baumannii requires discrete genetic events to develop an adequate colistin resistance level, which are up-regulation of pmrAB, point mutation in pmrB, as well as expression of pmrC, leading to addition of PEtN to lipid A.9 Also vacJ, pldA, ttg2C, pheS mutations and a conserved hypothetical protein were reported to be involved in colistin resistance.10 Four putative colistin-resistant genes: A1S_1983, hepA, A1S_3026, and rsfS were also identified A. baumannii ATCC 17978.11 Recently, plasmid-encoded resistance to polymyxin, mediated through the mcr genes (mcr-1 to mcr-8) were described mainly in Enterobacteriaceae,12,13 and several mcr variants were recently found in Acinetobacter spp., mcr-114,15 and mcr-4.3.15,16
The occurrence of carbapenem and colistin resistance was previously reported in Egypt among MDR A. baumannii.17,18 But currently, there is little or no data regarding colistin-resistant A. baumannii in Egypt. Therefore, the current work conducted to study colistin resistance emergence and mechanisms among CRAB clinical isolates in Egypt using genetic methods including whole-genome sequencing (WGS).
Materials and Methods
Bacterial Strains
Seventeen CRAB clinical strains (non-duplicate) collected from Theodor Bilharz Research Institute (TBRI) hospital during the period from 2015 to 2016 were re-used in the current work. Previously, CRAB were identified by API 20E and VITEK-2 compact system and susceptibility to different antimicrobials was performed using VITEK-2 compact system card N 222 (bioMérieux, France).19,20 Isolates were confirmed as A. baumannii by gyrB multiplex PCR and presence of blaOXA-51-like as previously described.21
Colistin Susceptibility Testing
Colistin susceptibility was previously evaluated by the recommended broth microdilution (BMD) method which was performed according to the CLSI procedures22 using colistin sulphate antibiotic powder (LKT Laboratories, USA).17 Isolates were considered susceptible (S) if MIC is ≤2 μg/mL, and resistant (R) if ≥4 μg mL-1 according to CLSI breakpoints.23
PCR Amplification for Carbapenem Resistance Genes
Crude cell lysates prepared from boiling overnight cultures were used for PCR template. Detection of carbapenemase-encoding genes (blaOXA-51-like, blaOXA-58-like, blaNDM-1-like, blaGES-like, blaOXA-23-like, blaOXA-40-like,) was previously performed by PCR.18,24
Whole-Genome Sequencing (WGS)
Seven colistin-resistant isolates were available for WGS. Using the QIAamp DNA Mini Kit (Qiagen, Germany), extraction of bacterial DNA from freshly sub-cultured colonies was done according to the manufacturer’s instructions, and the DNA concentration was measured using a Denovix Fluorometer (Denovix, USA). The genomic DNA was stored at −20°C. A volume of 1ng total of bacterial DNA was used in the library preparation. Sequencing libraries were prepared using the Nextera XT library prep kit (Illumina, San Diego, CA, USA) for a 300bp paired-end sequencing run on an Illumina MiSeq sequencer.
The program Velvet as part of the SeqSphere v6.0.2 software (Ridom GmbH, Münster, Germany) was used for de novo assembly, core genome MLST (cgMLST), and to determine the clonal lineage as described previously.25 To determine differences in specific ORFs in each draft genome sequence, the genome assemblies were aligned to the A. baumannii reference strain ACICU, which is colistin-susceptible and belongs to the most widespread clonal lineage IC2 involved in outbreaks worldwide. Resistome analysis including detection of colistin resistance determinants (eg, mcr-4.3) was carried out using ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/), as well as by performing a BLAST search using the Reference Gene Catalog listings of mcr genes as queries (NCBI BioProject PRJNA313047). Beta-lactamases were identified using the online beta-lactamase database (http://www.bldb.eu/). Raw reads generated were submitted to the Sequencing Read Archive (https://www.ncbi.nlm.nih.gov/sra/) of the National Center for Biotechnology Information (NCBI) under the SRA accession number SRP131448 and BioProject PRJNA431710.
Results
Bacterial Profile and Colistin Susceptibility Testing
The study investigated 17 CRAB strains that were isolated from respiratory samples (n=7; 41.18%), pus (n=5, 29.42%) and urine (n=4, 23.53%), and central venous catheter (n=1; 5.88%). The ICU was the main source of isolates with an isolation rate of 6/17 (35.29%). The distribution of A. baumannii isolates according to gender showed that 9 (52.94%) of the strains were from males and 8 (47%) were from females with no significant difference (p >0.05). According to CLSI breakpoints, nine of 17 isolates (52.9%) were defined as colistin resistant. By PCR, in addition to the intrinsic blaOXA-51-like that was present in all isolates, blaOXA-23-like was the most common carbapenem resistance determinant detected (14/17; 82.3%), followed by blaNDM-like that was found in four isolates (4/17; 23.5%); one isolate harboured both blaOXA-23-like and blaNDM-like. Two isolates harboured blaGES-like and blaOXA-23-like. Neither blaOXA-58-like, nor blaOXA-40-like were identified in any isolates.
Whole-Genome Sequencing
Out of seven colistin-resistant isolates that were sequenced, five were ST2 using the Pasteur MLST scheme and the remaining two isolates, R14 and R41, were ST158 and ST570, respectively. cgMLST analysis showed that two of the ST2 isolates were identical, with the remaining ST2 isolates differing by 64–296 alleles (Figure 1) suggesting that colistin resistance is not wholly the result of transmission events. The ST570 strain, a single locus variant (SLV) of ST2, had almost 700 alleles different from its nearest ST2 strains, but is still considered to be an IC2 isolate (Figure 1). The ST158 isolate differed in >2666 alleles from the IC2 isolates in this study. cgMLST analysis against our in-house database of A. baumannii revealed that R14 had 72 and 86 allelic differences from two isolates, collected from Egypt and Republic of Ireland, respectively, and is considered to be a singleton (data not shown).
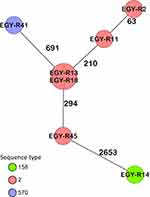 |
Figure 1 Minimum spanning tree generated by cgMLST coloured by ST (Pasteur) based on 3487 targets. Numbers between the nodes indicate the number of alleles that are different. |
Resistance genes analysis revealed that all the tested strains harbored multiple antibiotic resistance genes against most of the antibiotic groups (Table 1), particularly aminoglycoside modifying enzymes. Of note, all the IC2 isolates had blaOXA-66, the IC2 variant of the intrinsic blaOXA-51-like gene, while R14, possessed blaOXA-65. Furthermore, 5 variants of the intrinsic cephalosporinase (ADC) were found (Table 1). Other beta-lactamases found in the isolates included blaTEM-1, blaNDM-1, and blaGES-35.
 |
Table 1 Resistomes in Seven Colistin-Resistant A. Baumannii Isolates by WGS |
Published data suggest that multiple mutations in pmrCAB genes cause reduction of colistin susceptibility. We therefore investigated the pmrCAB operon by comparing the tested strains with the reference one (A. baumannii ACICU). Several mutations leading to amino acid substitutions were detected in pmrCAB. All IC2 isolates had the same pmrA gene sequence as ACICU, while R14 had an Ile18-Thr substitution. In pmrB, four isolates were identical to ACICU, while isolates R11 and R2 show an Ala-138-Thr substitution, and R14 has a Val444-Ala substitution (amino acid adjacent to the stop codon). The six IC2 isolates had an identical pmrC, however, this differed from the ACICU pmrC with an Ala354-Ser substitution. Isolate R14 had several substitutions when compared to ACICU, Ile42-Val, Ile115-Val, Leu150-Phe, and Thr515-Lys (Table 2). Interestingly, compared to the seven colistin-resistant isolates we have investigated, ACICU has a 25 amino acid deletion, YQIPENLKKKWCKDGECYDDILIDS, at position 324 (Figure 2).
 |
Table 2 Amino Acids Substitution in PmrCAB System in Seven Sequenced Isolates in Comparison to ACICU Strain |
 |
Figure 2 Alignment of the PmrC amino acid sequences based on whole-genome sequencing. ST2 isolates are represented by isolate R11. |
Other possible resistance mechanisms associated with colistin resistance include mutations in IpxA, IpxC, IpxD, vacJ, pheS, zndP, PldA, Ttg2C, and H-NS. However, in all cases, these genes were unchanged when compared to the colistin susceptible ACICU, with the exception of lpxC where although we did observe several mutations, they were all silent and did not change the amino acid sequences, except in strain 14 which had an Ile61-Val substitution in LpxA.
Discussion
In this study, we have investigated colistin resistance in CRAB isolates from Egypt. As in previous studies OXA-23 is by far the most widespread carbapenem resistance determinant and was found in 82% of CRAB isolates, followed by NDM-1 (23.5%).26,27 Analysis of the sequenced genomes revealed that six of the isolates were IC2, the most common lineage found worldwide. One isolate was ST158, an ST which has three entries in the PubMLST database, submitted from Turkey, Iraq and Egypt, but does not currently represent any of the international clones, but it appears to represent a clone circulating in Mediterranean/Middle Eastern countries.
There has been an increase in the use of polymyxins (especially colistin) over the last decade because they are generally active against multidrug-resistant A. baumannii and thus are used to treat infections as a last resort antibiotic.28 However, the use of colistin has resulted in colistin resistance becoming an emerging global phenomenon.29 Detection of colistin resistance in A. baumannii by MIC using BMD is the only method approved by both the CLSI and by the European Committee on Antimicrobial Susceptibility Testing (EUCAST).23,30 In our study, nine of 17 isolates (52.9%) were colistin resistant by BMD, this resistance percentage was higher than that was recently reported in Egypt (5%),31 India (7.2%)32 and (9%),33 Pakistan (9.6%),15 Taiwan (10.1%),34 Turkey (28%),35 and Sweden (36.36%).36 While lower than the percentage of 56% that reported in Greece.37 In Enterobacteriaceae, colistin resistance is often associated with acquisition of resistance determinants like mcr-1.12,13,38 However, this is not the case with A. baumannii. Although there are a few reports of mcr-genes in A. baumannii, they are still rare and we did not detect any MCR-variant in the sequenced isolates.14,15,39,40 Instead, in nearly all the reported cases, colistin resistance in A. baumannii appears to be mediated through chromosomal changes. Mutations in the lipid A encoding genes including lpxA, lpxC, and lpxD, causing marked loss of LPS but this is likely to be confined to the laboratory. In the present study, we found the lpx genes to be intact and identical between colistin susceptible and resistant IC2 isolates. The sporadic ST158 isolate did carry an Ile61-Val amino acid substitution in LpxA, but this could be a lineage-related polymorphism, which has been noted previously.41,42
The two-component system PmrAB considered an important mechanism which is a response regulator and sensor kinase which regulates pmrC, the gene product of which adds PEtN to lipid A. Point mutations in pmrA and pmrB can lead to upregulation of pmrC, resulting in the addition of PEtN to lipid A, leading to reduced colistin susceptibility.28 In the present study, we compared the pmrCAB operon of our colistin-resistant isolates to the colistin susceptible ACICU strain to identify unique sequences only found in resistant isolates.41 Using this approach we found some amino acid substitutions in pmrB in three isolates, although strain-14 is the sporadic isolate and the different pmrB could be a lineage-specific trait and needs further investigation.
A recent study from Egypt reported mutations in pmrCAB genes were detected in colistin-resistant strains when compared with those in A. baumannii ATCC 17978.31 In the current study, we chose to compare our strains with the colistin susceptible standard one (A. baumannii ACICU) as the comparator because all but one of our isolates were ST2, the same as ACICU. However, we had similar results; the ATCC 17978 PmrAB were identical to that from ACICU, and ATCC 17978 did not have the 25 amino acid deletion in PmrC.
Conclusion
Colistin resistance is an emerging problem in CRAB isolates in our region. Molecular analysis by WGS showed that our strains were mainly IC2 except one strain that was singleton with an ST found circulating in the Middle East and showed different resistant profiles than the other strains. The high incidence of the high-risk lineage IC2 harboring blaOXA-23 as well as blaNDM is also of concern. Examining pmrCAB operon for colistin resistance, while we do find several amino acid changes in the operon, it remains to be proven that these are the cause of resistance and not an association. Therefore, we conclude that more investigations are in needed to understand of colistin resistance mechanism in A. baumannii.
Data availability
Repositories:
Raw reads generated were submitted to the Sequencing Read Archive (https://www.ncbi.nlm.nih.gov/sra/) of the National Center for Biotechnology Information (NCBI) under the SRA accession number SRP131448 and BioProject PRJNA431710.
Acknowledgments
We acknowledge the Science and Technology Development Fund (STDF) Project ID 22838 granted to Clinical Pathology Department, Kasr El-Aini School of Medicine, Cairo University and Project 96 D of Microbiology Department, Theodor Bilharz Research Institute (TBRI) for funding this work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. WHO. Global priority list of antibioticresistant bacteria to guide research, discovery, and development of new antibiotics. 2018.
2. Irvem A. Colistin-daptomycin, colistin-linezolid, colistin-vancomycin combination effects on colistin in multi-resistant Acinetobacter baumannii strains. Medical Science and Discovery. 2018;5:124–129. doi:10.17546/msd.391234
3. Lucas DD, Crane B, Wright A, et al. Emergence of high-level colistin resistance in an Acinetobacter baumannii clinical isolate mediated by inactivation of the global regulator H-NS. Antimicrob Agents Chemother. 2018;62:e02442–17. doi:10.1128/AAC.02442-17
4. Evans BA, Amyes SGB. OXA β-lactamases. Clin Microbiol Rev. 2014;27:241–263. doi:10.1128/CMR.00117-13
5. Amine A, Hazzah W, Abou-bakr A. Genetic background of carbapenem resistant Acinetobacter baumannii in a health care setting in Alexandria, Egypt. Bull High Inst Public Heal. 2011;41:311–324.
6. Lee J, Chung ES, Ko KS. Transition of colistin dependence into colistin resistance in Acinetobacter baumannii. Sci Rep. 2017;7:14216. doi:10.1038/s41598-017-14609-0
7. Bahador A, Farshadzadeh Z, Raoofian R, Mokhtaran M, Pourakbari B. Association of virulence gene expression with colistin-resistance in Acinetobacter baumannii: analysis of genotype, antimicrobial susceptibility, and biofilm formation. Ann Clin Microbiol Antimicrob. 2018;17:24. doi:10.1186/s12941-018-0277-6
8. Olaitan AO, Morand S, Rolain J. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi:10.3389/fmicb.2014.00643
9. Beceiro A, Llobet E, Aranda J, et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii by the pmrAB two-component regulatory system. Antimicrob Agents Chemother. 2011;55:3370–3379. doi:10.1128/AAC.00079-11
10. Nhu NTK, Riordan DW, Nhu TDH, et al. The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci Rep. 2016;6(1):28291. doi:10.1038/srep28291
11. Mu X, Wang N, Li X, et al. The effect of colistin resistance-associated mutations on the fitness of Acinetobacter baumannii. Front Microbiol. 2016;7:1715. doi:10.3389/fmicb.2016.01715
12. Partridge SR, Di Pilato V, Doi Y, et al. Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J Antimicrob Chemother. 2018;73(10):2625–2630. doi:10.1093/jac/dky262
13. Rebelo A, Bortolaia V, Kjeldgaard J, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23(6):17–00672. doi:10.2807/1560-7917.ES.2018.23.6.17-00672
14. Rahman M, Ahmad S. 549. First report for emergence of chromosomal borne colistin resistance gene mcr-1 in a clinical acinetobacter baumannii Isolates from India. Open Forum Infect Dis. 2019;6(Supplement_2):S261–S262. doi:10.1093/ofid/ofz360.618
15. Hameed F, Khan MA, Muhammad H, Sarwar T, Bilal H, Rehman TU. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: first report from Pakistan. Rev Soc Bras Med Trop. 2019;52:e20190237. doi:10.1590/0037-8682-0237-2019
16. Martins-sorenson N, Snesrud E, Xavier D, et al. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J Antimicrob Chemother. 2020;75(1):60–64. doi:10.1093/jac/dkz413
17. Fam NS, Mohamed SH, Gamal D, Wasfy RM, Soliman M, El-Kholy AA. Reliability of phenotypic methods for detection of colistin resistance among carbapenem-resistant Acinetobacter baumannii clinical isolates from Egypt. Germs. 2020.
18. Fam N, Gamal D, Salem D, Dahroug H, Wasfy RM, Morcos MM. Clonal diversity and high prevalence of Oxa-23 among carbapenem resistant Acinetobacter baumannii isolates in Egypt. J Biosci Appl Res. 2019;5:110–124.
19. Fam N, Gamal D, Azmy M, et al. Antimicrobial efficacy of doripenem colistin combination on carbapenem - resistant Acinetobacter baumanii isolates by E-test Agar dilution and ultrastrastructural methods. Egypt J Med Microbiol. 2017;26(1):1–7. doi:10.12816/0046266
20. Mohamed SH, Salem D, Azmy M, Fam NS. Antibacterial and antibiofilm activity of cinnamaldehyde against carbapenem-resistant Acinetobacter baumannii in Egypt: in vitro study. J Appl Pharm Sci. 2018;8:151–156. doi:10.7324/JAPS.2018.81121
21. Nowak J, Zander E, Stefanik D, et al. High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother. 2017;72(12):3277–3282. doi:10.1093/jac/dkx322
22. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing.
23. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing.
24. Fallah F, Noori M, Hashemi A, et al. Prevalence of blaNDM, blaPER, blaVEB, blaIMP, and blaVIM genes among Acinetobacter baumannii isolated from two hospitals of Tehran, Iran. Scientifica (Cairo). 2014;2014:245162.
25. Higgins PG, Prior K, Harmsen D, Seifert H. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One. 2017;12:e0179228.
26. El-Sayed-Ahmed MA, Amin MA, Tawakol WM, Loucif L, Bakour S, Rolaina J-M. High Prevalence of blaNDM-1 carbapenemase-encoding gene and 16S rRNA armA methyltransferase gene among Acinetobacter baumannii clinical isolates in Egypt. Antimicrob Agents Chemother. 2015;59:3602–3605. doi:10.1128/AAC.04412-14
27. Hamidian M, Nigro SJ. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genomics. 2019;5. doi:10.1099/mgen.0.000306
28. Asif M, Alvi IA, Rehman SU. Insight into Acinetobacter baumannii: pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect Drug Resist. 2018;11:1249–1260.
29. Hua X, Liu L, Fang Y, et al. Colistin resistance in Acinetobacter baumannii MDR-ZJ06 revealed by a multiomics approach. Front Cell Infect Microbiol. 2017;7:45. doi:10.3389/fcimb.2017.00045
30. EUCAST. The European committee on antimicrobial susceptibility testing. Breakpoint tables for interpretation of MICs and zone diameters. 2019. Available from: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf.
31. Abdulzahra AT, Khalil MAF, Elkhatib WF. First report of colistin resistance among carbapenem-resistant Acinetobacter baumannii isolates recovered from hospitalized patients in Egypt. New Microbes New Infect. 2018;26:53–58. doi:10.1016/j.nmni.2018.08.007
32. Singhal L, Sharma M, Verma S, et al. Comparative Evaluation of broth microdilution with polystyrene and glass-coated plates, agar dilution, E-Test, Vitek, and disk diffusion for susceptibility testing of colistin and polymyxin B on carbapenem-resistant clinical isolates of Acinetobacter baumannii. Microb Drug Resist. 2018;24:1082–1088. doi:10.1089/mdr.2017.0251
33. Bakthavatchalam YD, Veeraraghavan B, Shankar A, Thukaram B, Krishnan DN. Evaluation of colistin and polymyxin B susceptibility testing methods in Klebsiella pneumoniae and Acinetobacter baumannii. J Infect Dev Ctries. 2018;12:504–507. doi:10.3855/jidc.9660
34. Lai -C-C, Chen Y-S, Lee N-Y, et al. Susceptibility rates of clinically important bacteria collected from intensive care units against colistin, carbapenems, and other comparative agents: results from Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART). Infect Drug Resist. 2019;12:627–640.
35. Çağlan E, Nigiz S, Sancak B, Gür D. Resistance and heteroresistance to colistin among clinical isolates of Acinetobacter baumannii. Acta Microbiol Immunol Hung. 2019;9:1–5. doi:10.1556/030.66.2019.021
36. Matuschek E, Åhman J, Webster C, Kahlmeter G. Antimicrobial susceptibility testing of colistin - evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect. 2018;24:865–870. doi:10.1016/j.cmi.2017.11.020
37. Malli E, Tsilipounidaki K, Xitsas S, Pyridou P, Papagiannitsis C, Petinaki E. Implementation of the rapid polymyxin Acinetobacter test to detect colistin-resistant Acinetobacter baumannii. Microb Drug Resist. 2019.
38. Jeannot K, Bolard A, Plésiat P. Resistance to polymyxins in gram-negative organisms. Int J Antimicrob Agents. 2017;49:526–535. doi:10.1016/j.ijantimicag.2016.11.029
39. Ahmed SS, Alp E, Hopman J, Voss A. Global epidemiology on colistin resistant Acinetobacter baumannii. J Infect Dis Ther. 2016;4:1–5. doi:10.4172/2332-0877.1000287
40. Snyman Y, Whitelaw A, Reuter S, Dramowski A, Reratilwe M, Maloba B. Clonal expansion of colistin resistant Acinetobacter baumannii isolates in Cape Town, South Africa. Int J Infect Dis. 2019.
41. Gerson S, Betts JW, Lucaßen K, et al. Investigation of novel pmrB and eptA mutations in isogenic Acinetobacter baumannii isolates associated with colistin resistance and virulence in vivo. Antimicrob Agents Chemother. 2019;63:e01586–18.
42. Gerson S, Nowak J, Zander E, et al. Diversity of mutations in regulatory genes of resistance-nodulation- cell division efflux pumps in association with tigecycline resistance in Acinetobacter baumannii. J Antimicrob Chemother. 2018;73:1501–1508. doi:10.1093/jac/dky083
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
