Back to Journals » Infection and Drug Resistance » Volume 14
Molecular Characterization of Carbapenemase-Producing Klebsiella pneumoniae Isolated from Egyptian Pediatric Cancer Patients Including a Strain with a Rare Gene-Combination of β-Lactamases
Authors Osama D , El-Mahallawy H, Mansour MT, Hashem A, Attia AS
Received 1 October 2020
Accepted for publication 5 January 2021
Published 29 January 2021 Volume 2021:14 Pages 335—348
DOI https://doi.org/10.2147/IDR.S284455
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Dina Osama,1 Hadir El-Mahallawy,2 Mohamed Tarek Mansour,3 Abdelgawad Hashem,4,5 Ahmed S Attia4
1Department of Microbiology and Immunology, Faculty of Pharmacy, October University for Modern Sciences and Arts (MSA), Cairo, Egypt; 2Department of Clinical Pathology, National Cancer Institute, Cairo University, Cairo, Egypt; 3Department of Virology and Immunology, National Cancer Institute, Cairo University, Cairo, Egypt; 4Department of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Cairo, Egypt; 5Department of Microbiology and Immunology, Faculty of Pharmacy, The British University in Egypt, Shorouk City, Egypt
Correspondence: Ahmed S Attia
Department of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Room #D404, Kasr El-Ainy Street, Cairo 11562, Egypt
Tel +20-10-65344060
Fax +20-2-23628246
Email [email protected]
Purpose: Healthcare-associated infections caused by multi-drug-resistant (MDR) pathogens are a global threat. We aim to assess the clonal relatedness among carbapenemase-producing Klebsiella pneumoniae (CPKP) strains infecting Egyptian pediatric cancer patients.
Materials and Methods: Identification and antimicrobial susceptibility testing of 149 Gram-negative isolates obtained from pediatric cancer patients were performed by VITEK 2. Genes encoding carbapenemases and extended-spectrum β-lactamases were detected by PCR and verified by DNA sequencing of representative samples. The transferability of the plasmids harboring blaOXA-48, from representative clinical samples, was evaluated by performing a conjugation experiment followed by PCR and MIC shift determination. Clonal relationships among the blaOXA-48-harboring K. pneumoniae isolates were determined by enterobacterial repetitive intergenic consensus (ERIC)-PCR and pulsed-field gel electrophoresis (PFGE).
Results: Carbapenem resistance was observed in 59% of the isolates. The most prevalent species was K. pneumoniae (45.6%) and 57% of them were isolated from ICU. Fifty-nine % of the K. pneumoniae isolates were carbapenemase-producers and blaOXA-48 was detected in (58%) of them. One isolate co-harbored blaOXA-48, blaNDM-1, and blaIMP-1 genes for the first time in Egypt. PCR and meropenem MIC shift confirmed the success of the transferability of representative plasmids to E. coli K12. ERIC and PFGE identified 93% and 100% of the K. pneumoniae with a similarity coefficient ≥ 85%, respectively, including strains with indistinguishable patterns, suggesting possible clonal dissemination.
Conclusion: Our findings underline the dissemination of diverse clones of MDR CPKP among Egyptian pediatric cancer patients. Hence, routine molecular characterizations followed by strict implementation of infection control measures are crucial to tackling this threat.
Keywords: ERIC-PCR, PFGE, blaOXA-48, clonal dissemination, Egypt, carbapenem-resistant
Introduction
Bacterial infections have been implicated as one of the most frequent complications that threaten immunosuppressed cancer patients. As those patients are frequently hospitalized for treatment purposes.1,2 Gram-negative bacteria such as Enterobacteriaceae, mainly K. pneumoniae and E. coli followed by Acinetobacter and Pseudomonas, are commonly reported as infecting pathogens in cancer patients.3,4 In 2019, the CDC listed carbapenem-resistant Enterobacteriaceae (CRE) as an urgent threat.5 Of special concern, the emergence of carbapenem-producing K. pneumoniae (CPKP) which is responsible for hospital-acquired infection (HAI) outbreaks in cancer wards that became the main reason for the increased rates of mortality among the hospital settings.6
The spread of Enterobacteriaceae is attributed to their ability in carrying resistance plasmids coding for enzymes that interfere with important antibiotics used as empiric therapy in febrile neutropenic patients; cephalosporins and carbapenems.7 Particularly, those that produce carbapenemase enzymes including, K. pneumoniae carbapenemases (KPCs), Metallo-β–lactamases (MBL), and oxacillinase (OXA-48). Carbapenemases are commonly categorized using the Ambler classification scheme. KPCs are the most common carbapenemases identified, and they are members of class A. They are commonly found in Enterobacteriaceae and can be occasionally identified in P. aeruginosa or A. baumannii. Class B Metallo β-lactamases (MBLs), commonly include (NDM, VIM, and IMP) enzymes. In addition, class D carbapenemases include OXA-48-like enzymes, which are generally produced by Enterobacteriaceae, while other variants are frequently produced by A. baumannii. Other mechanisms that may contribute to carbapenem resistance in Enterobacteriaceae include; reduced uptake of β-lactam drugs by selectively altering the cell membrane porin channels, decreased membrane permeability, efflux pumps, and the presence of various broad-spectrum β-lactamases (eg, ESBL and/or AmpC cephalosporinase).8–10
Carbapenem resistance is considered a great threat as they are usually combined with other resistance mechanisms for a wide range of antimicrobial agents; thus, infections with carbapenemase-producing Enterobacteriaceae are associated with high mortality rates. Hospital-acquired infection outbreaks are also associated with Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) that have been identified as a major cause of the MDR phenotype, because of their wide diversity of enzymes, including TEM, SHV, and CTX-M.3,7,11,12 Investigation of infection outbreaks, the transmission of microorganisms between the environment and the patients, and epidemiological surveillance have become an important tool in monitoring local dissemination of resistant bacterial strains within the healthcare facilities.1,13,14 Accordingly, phenotypic methods have been replaced by molecular typing methods that examine the relatedness of microorganisms at the genomic level.15 To date, few studies have focused on carbapenem resistance in pediatric patients, especially in critically ill children.16 Accordingly, in this study, the clonality of CPKP strains causing infections in pediatric cancer patients was investigated using two different molecular typing methods; Enterobacterial Repetitive Intergenic Consensus (ERIC) PCR and pulsed-field gel electrophoresis (PFGE).
Methods
Bacterial Strains and Microbiological Methods
The epidemiology of carbapenem-resistant Enterobacteriaceae was studied in a 320-bed tertiary care hospital (Children’s Cancer Hospital) for pediatric cancer patients between November 2014 and August 2016, in Cairo, Egypt. A total of 149 non-duplicated Gram-negative isolates were collected. All isolates were identified by both biochemical testing and VITEK 2 automated system using the ID-GNB card for identification of Gram-negative bacilli according to the manufacturer’s instructions (bioMérieux, France). The study approval was obtained from the Research Ethics Committee of the Faculty of Pharmacy, Cairo University (approval no. MI-1203). No patients’ consent form was required as all the isolates were obtained from the hospital microbiology lab after being grown on MacConkey and Trypticase Soy Agar, for routine hospital work, then stored at −80°C in LB medium supplemented with 15% v/v glycerol.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing to a panel of antibiotics according to the hospital routine was performed using the Kirby Bauer disc diffusion method.17 Minimum inhibitory concentration (MIC) for amikacin, cefepime, cefoxitin, cefotaxime, ceftriaxone, gentamicin, ciprofloxacin, tobramycin, meropenem, and piperacillin-tazobactam were determined by the VITEK 2 system using the AST-GN card (bioMérieux) and confirmed for meropenem, imipenem, and colistin using the E-test strips (bioMérieux). The interpretation was according to the CLSI breakpoint values.18 E. coli ATCC 25,922 and ATCC 35,218 were used as negative and positive quality control strains for susceptibility testing, respectively.19
Phenotypic Detection of Carbapenemases
According to the CLSI recommendations, Modified Hodge Test (MHT), Carba NP (RAPIDEC, bioMérieux), and the modified Carbapenem Inactivation Method (mCIM) tests were used to assess the ability of the carbapenem-resistant isolates to produce carbapenemases.18,20,21 For the MHT
K. pneumoniae ATCC BAA-1705 and ATCC BAA-1706 were used as a positive and negative control, respectively. While, K. pneumoniae ATCC 700,603 was used as a positive control for ESBL.19
Molecular Analysis of Antibiotic Resistance Determinants
Total DNA was extracted using the boiling method.22 An in-house PCR test for carbapenemase-encoding genes was performed using specific primers for (blaKPC, blaVIM, blaNDM, blaOXA-48, and blaIMP), as well as the ESBLs (blaCTX-M, blaTEM, and blaSHV). Briefly, PCR reactions were carried out in a total volume of 25 μL reaction mixture containing 1 µL (~50 ng/mL) bacterial DNA template, 12.5 µL DreamTaq Green PCR Master Mix (2X) (Thermo Fisher Scientific), and 10 µM (1 µL) from each primer, and nuclease-free water to complete the reaction to 25 µL. PCR cycling conditions have been optimized at different annealing temperatures (Ta) for different primer sets (Table 1). PCR reactions were carried out using a gradient PCR thermocycler (Analytik, Jena, Germany). A reaction with no DNA-template was used as a negative control, while K. pneumoniae ATCC 700,603 was used as ESBL-positive control.19 In order to verify that the obtained PCR products are indeed the intended gene fragments, representative samples of the PCR products were subjected to DNA sequencing analyses. The FinchTV software was used to analyze the sequencing data. The DNA sequence of each gene fragment was compared to the sequences in the GenBank nucleotide database using the BLAST tool (http://www.ncbi.nlm.nih.gov/blast/).23
 |
Table 1 List of Primers Used in the Current Study |
Molecular Typing
ERIC Typing
Selected K. pneumoniae isolates that harbor the blaOXA-48 gene were analyzed for typing purposes using the ERIC-PCR. Genomic DNA was extracted by the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The PCR reaction contained 12.5 µL of DreamTaq Green PCR Master Mix (2X), 50 ng of bacterial DNA template, 2 µL from the ERIC primers (Table 1), and nuclease-free water to complete the total volume to 25 µL. According to the protocol described by Bilung and colleagues24 with slight modifications, the conditions used were as follows: initial denaturation for 15 min at 95°C followed by 35 cycles of denaturation for 1 min at 95°C, annealing for 1 min at 45°C, the extension for 8 min at 72°C, and a final extension for 15 min at 72°C.
ERIC-PCR fingerprint was analyzed using BioNumerics 7.6 software (Applied Maths, Belgium), based on the Dice similarity coefficient and using the unweighted pair group method with arithmetic mean (UPGMA).25 Band position tolerance was 1.5%, and the optimization was 1%. Isolates with a similarity coefficient ≥85% were considered to belong to the same cluster.
PFGE Typing
Pulsed-field gel electrophoresis (PFGE) was performed using 50 U XbaI restriction enzyme (NEB, USA). Macro-restriction of genomic DNA was carried out to establish clonal relationships between K. pneumoniae resistant isolates, according to the PulseNet PFGE protocol from the Centers for Disease Control and Prevention (CDC) with minor modifications.26 Electrophoresis was performed on a 1% low-melting agarose gel (Sigma-Aldrich, USA) using a CHEF DR II apparatus (Bio-Rad, USA). PFGE conditions were as follows: pulse times ranged from 5 to 40s over 18 h at 6.0 V/cm and at 14°C. DNA fragment patterns interpretation was carried out according to the Tenover criteria.27 DNA fragment patterns were normalized using the bacteriophage Lambda PFG ladder (NEB). A dendrogram was generated based on the dice coefficient determination, using the UPGMA with 1.5% band matching tolerance and optimization in BioNumerics software version 7.6.
Conjugation Experiments
To determine the transferability of the blaOXA-48 gene, broth mating assay was implemented using the Luria-Bertani (LB) broth medium, E. coli K12 served as the recipient strain and two clinical isolates harboring the blaOXA-48 gene served as the donor cells as previously described.28–30 Transconjugants were selected on MacConkey agar plates containing ampicillin (100 µg/mL) and meropenem (50 µg/mL). The success of acquiring the resistance plasmid was tested by performing a PCR on the E. coli K12 before and after conjugation. In addition, the MIC of meropenem for the transconjugants was compared to the donors and E. coli K12 to further confirm the transferability of the resistance gene. The experiment was performed using two K. pneumoniae strains as donors as they exhibited the highest MIC value towards meropenem (isolates 116 and 128) in the collected isolates.
Statistical Analyses
Statistical analyses were carried out using the software package SPSS® v23 (SPSS Inc., USA). Categorical variables were described using the Chi-square test. Interquartile range and mean were used for continuous variables, according to data distribution. A P-value of <0.05 was considered significant. The genetic diversity and discriminatory index (DI) for the typing techniques for both ERIC and PFGE were calculated using Hunter and Gaston’s formula.31 DI values range from 0 to 1; the least and the most possible biodiversity in the population.
Results
Clinical and Epidemiological Characteristics
During the study period, a total of 149 non-duplicated Enterobacteriaceae isolates were obtained from various clinical specimens. The strains were isolated from 140 pediatric cancer patients, with an age range (2 weeks–17 years, median 6 months). The majority of the patients were males 81 (58%). K. pneumoniae n=68 (45.6%), was the most prevalent species. Accordingly, these isolates would be the focus of the current study. The distribution of the K. pneumoniae isolates among gender, different clinical specimens, and wards is shown in (Table 2).
 |
Table 2 Relationship Between Different Carbapenemases and β-Lactamases Among the Klebsiella pneumoniae Isolates and Patient’s Clinical Data |
Antimicrobial Susceptibility
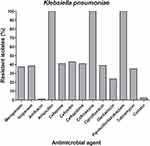
According to the MIC test results of all the collected isolated, 139 isolates were considered MDR as they were non-susceptible to three or more antimicrobial classes.32 Among those MDR isolates, 88 were classified as carbapenem-resistant Enterobacteriaceae (CRE). The majority of the CRE isolates were K. pneumoniae (59%, n=52). Overall, the collected K. pneumoniae isolates showed diminished susceptibility for the tested antibiotics; meropenem, imipenem, ampicillin, piperacillin/tazobactam, cefoxitin, ceftazidime, ceftriaxone, cefepime, ciprofloxacin, gentamicin, and tobramycin. However, marked susceptibility for amikacin, tigecycline, and colistin was observed as illustrated in (Figure 1).
Phenotypic Detection of Carbapenemases
Fifty-one (93%) and 50 (90%) K. pneumoniae isolates were labeled as CPKP, according to the results of the mCIM and Carba NP tests, respectively. However, 48 (84%) isolates were identified as CPKP with the MHT.
Molecular Analysis of Antimicrobial Resistance Genes in the K. pneumoniae Isolates
K. pneumoniae comprised the vast majority of the carbapenem-resistant isolates. Accordingly, they were subjected to further analyses to determine their genetic background. PCR screening revealed that 30, 12, 2, and 1 isolates were harboring carbapenemase genes (blaOXA-48, blaNDM, blaIMP, and blaVIM), respectively. Furthermore, β-lactamases genes (blaTEM, blaSHV, and blaCTX-M) were detected in 28, 16 and 24 K. pneumoniae isolates, respectively. Meanwhile, 63% of the K. pneumoniae isolates co-harbored more than one carbapenemase gene. It is worth mentioning that among the K. pneumoniae isolates, an isolate (no.134) co-harbored (blaOXA-48, blaNDM-1, and blaIMP-1). This specific strain was isolated from a blood specimen of a patient with hematologic malignancy admitted to the ICU. Furthermore, it was characterized by being MDR carbapenemase-producer isolate that was phenotypically positive by all the MHT, CarbaNP, and mCIM tests. It also co-harbored the ESBLs genes (blaSHV-1 and blaCTX-M15) and showed MIC value of 16 and 32 µg/mL to meropenem and imipenem, respectively. To the best of our knowledge, this is the first report of this gene combination in a strain isolated in Egypt. The distribution of the different analyzed resistance genes in all the K. pneumoniae isolates among gender, different clinical specimens, and wards is presented in Table 2. To provide a better representation of the frequencies of the carbapenemase and ESBLs genes among the carbapenem-resistant K. pneumoniae isolates the data is illustrated in Figure 2. The most common gene combinations were (blaOXA-48, blaTEM, blaSHV, and blaCTX-M) and (blaOXA-48, blaTEM). This was followed by the (blaOXA-48, blaTEM, and blaCTX-M) combination. On the other hand, the most common single gene, not present in a combination with other carbapenemase and ESBLs genes, was the blaNDM followed by blaOXA-48 and blaTEM.
Nucleotide sequences analyses of some of the obtained PCR products were compared with the non-redundant genome sequences available in the GenBank database. These analyses revealed the prevalence of blaOXA-48, blaNDM-1, blaIMP-1, blaVIM-1, blaTEM-1, blaSHV-1, and blaCTX-15 among the studied isolates. Overall, analysis of the distribution of the genetic resistance determinants indicated that the blaOXA-48 is widely spread among the included carbapenem-resistant K. pneumoniae isolates (30/52; 58%). Therefore, these strains were further investigated molecularly for their clonal relatedness.
ERIC-PCR Analysis
Dendrogram analysis of the ERIC-PCR genotyping revealed three clusters of the blaOXA-48-harbouring K. pneumoniae, one major cluster (I) included 27 isolates and the other 3 isolates demonstrated unique patterns distributed among the other 2 clusters (II and III) (Figure 3). Sixty percent of cluster I isolates exhibited similarity ≥95%; however, the 3 other isolates were ≤85% similar (Figure 3). The discriminatory power for the ERIC analysis was 0.2.
PFGE Analysis
The PFGE analysis of the 30 blaOXA-48-harbouring K. pneumoniae isolates revealed 28 distinct PFGE profiles (pulsotypes) as illustrated in the dendrogram (Figure 4). The P17 was the most common pulsotype (3/30, 10% of isolates), ie, isolates 86, 429, and 430 showed an indistinguishable pattern. Whereas, another 2 strains exhibited 99% similarity coefficient (3 and 134) that were suggested to be clonally related to the former 3 isolates (86, 429, and 43). The PFGE dendrogram identified 5 clusters (A-E) that exhibited ≥85% similarity coefficient. Cluster A was predominating, as it included 12 isolates, clusters B, C, D, and E included 4, 5, 2, and 7 isolates, respectively. However, 9/30 (30%) of the isolates demonstrated a similarity coefficient ≥95%. The discriminatory power for this analysis was 0.76.
 |
Figure 4 PFGE typing of the blaOXA-48-harbouring carbapenem-resistant K. pneumoniae isolates A dendrogram was generated from the PFGE patterns of XbaI digested chromosomal DNA of 30 carbapenem-resistant K. pneumoniae clinical isolates. The comparison was performed with the BioNumerics fingerprint data software v7.62 using the UPGMA clustering method and the dice similarity coefficient with 1% optimization and 1.5% position tolerance. PFGE pulsotypes (P) were assigned to isolates according to Tenover criteria.27 The numbers written on the branches indicate the similarity percentage of the different pulsotypes. The black frame indicates the isolates that were indistinguishable and assumed to be the same clone. The different clusters are designated by letters (A–E). On the right side of the dendrogram detailed results for PFGE including the code of each isolate, pulsotypes, date of isolation, hospital wards, and screened carbapenemases and ESBLs determinants (blaOXA-48, blaNDM, blaIMP, blaVIM, blaCTX-M, blaTEM, and blaSHV). The (+) and (-) signs indicate present and absent, respectively. The alphabets indicate the type of the hospital wards; a: hematology ward, b: solid tumor ward c: surgery ward. Abbreviations: IP, in-patient; ICU, intensive care unit; stepdown unit, intermediate care unit. |
The Conjugation Experiments
The blaOXA-48 gene from the two donor strains was successfully transferred to the recipient E. coli K12. PCR results indicated that the gene was successfully transferred in both conjugations. The meropenem MIC values determined were consistent with the transfer of carbapenemase expression. As the MIC value of the recipient strains changed from ≤0.5 µg/mL to 4 µg/mL and 8 µg/mL with the donner strains 128 and 116, respectively. Accordingly, this experiment revealed that the plasmids harboring the blaOXA-48 gene in some of the investigated isolates are transferrable via conjugation and capable of conferring resistance to meropenem to E. coli K12.
Discussion
In 2017, the World Health Organization (WHO) defined CRE as one member of the “critical priority list of antibiotic-resistant pathogens”.33 In 2018, the European Centre for Disease Prevention and Control (ECDC) conducted a review to update the risk assessment of CRE, as it represents an important threat to patients in healthcare systems.34 A recent study in Egypt reported that there is a continuous increase in the prevalence of CRE in the country, with at least one CRE isolate in 64% from 72 hospitals during the study period.35 As reported in other previous studies in Egypt, our study also concluded that the predominant species among the isolated CRE was MDR K. pneumoniae, which represents a frightening causative agent of different types of infections among hospitalized pediatric cancer patients.35–38 It was reported that class D carbapenemases (blaOXA-48= 58%) are predominant in K. pneumoniae followed by the blaNDM-1 gene, in several studies conducted in different countries notably in the Mediterranean region including Spain, France, Turkey, Tunisia and Egypt.20,21 Furthermore, the majority (41.8%) of the K. pneumoniae isolates in the current study harbored one or more carbapenem resistance determinants (Figure 2) that agreed with previously reported studies.40–42 the unusual combination of the 3 carbapenemase genes (blaOXA-48, blaNDM-1, and blaIMP-1), was among our alarming findings, especially it was not reported before among isolates in Egypt. Interestingly, several studies from China, Japan, and multiple countries in the Mediterranean region including; France, Morocco, Egypt, Algeria, Tunisia, and Spain reported the presence of single blaIMP-1 or in combination with either blaNDM-1 or blaOXA-48 carbapenemase genes.24,25 However, the current triple gene-combination has not been previously reported. Ten percent of the CRKP isolates were non-carbapenemase producers, which could be explained by the overexpression of AmpC or ESBLs combined with permeability alteration (deficient expression or loss of porins) and/or other carbapenemases that were not identified.43
Regarding, the CarbaNP and mCIM phenotypic tests results, both tests indicated that the numbers of positive isolates are higher than that detected by the MHT especially for those isolates encoding for the blaOXA-48. Hence, our findings are consistent with other previously reported results regarding the rates of detecting positive results by these three assays.47,48
The present study revealed that 49% of the recovered isolates were MDR. However, amikacin, colistin, and tigecycline were still effective against most of the K. pneumoniae isolates (Figure 1), similar to other studies conducted in Egypt and elsewhere.46 Although there is a marked increase in the resistance of amikacin, colistin, and tigecycline in recent studies in Egypt, they are still effective against carbapenem-resistant Enterobacteriaceae.37,39,47 Molecular typing is a useful tool for determining the genetic relatedness of bacteria. This is particularly important in the investigation of the epidemiology of several bacterial infectious diseases accompanied by HAI.48 To understand how blaOXA-48 is disseminated through the study hospital, two different DNA-based fingerprinting methods were used to compare the isolates at the molecular level. The dendrogram relationships of the 30 isolates for both the ERIC and PFGE analyses revealed high genetic diversity among the investigated isolates except for two pairs of isolates that were closely related. Since the 30 genotyped isolates co-harbored a diverse range of resistance determinants, therefore they exhibited resistance to carbapenems with different mechanisms that are considered an alarm for the potential dissemination of these clones among the hospital wards. According to the aforementioned results, when compared with PFGE, ERIC-PCR displayed a much lower discriminatory power. The isolates were categorized by the PFGE among a larger number of clusters compared to ERIC-PCR typing. A dominant PFGE pattern with ≥90% similarity coefficient was observed in 26 isolates, while the other isolates displayed unique PFGE patterns ranging from 85% to 90% similarity. These findings indicate the emergence and spread of these resistant isolates across the hospital wards. Moreover, they also indicate that cross-transmission is not restricted to a certain ward or time. For example, the 3 isolates (86, 429, and 430) showed the same pulsotype with 100% similarity and were isolated from the same ward but almost one year apart regarding the date of sample collection. Additionally, the latter isolates revealed the same resistance determinants (blaOXA-48, blaTEM-1, blaSHV-1, and blaCTX-M15) combination except for (blaSHV-1 and blaCTX-M15) in isolate no. 86. Another example was isolate no.134 that exhibited 99% similarity and co-harbored 5 different resistance determinants blaOXA-48, blaNDM-1, blaIMP-1 blaSHV-1, and blaCTX-M15 (Figure 4).
The two molecular typing methods revealed that 5 K. pneumoniae isolates (429, 430, and 86) and (3 and 134) were categorized as a unique subtype. Also, they exhibited MIC values between 4 µg/mL and ≥16µg/mL for meropenem and imipenem, respectively. Interestingly, we observed that the aforementioned 5 isolates (86, 429, 430, 3, and 134) were obtained from the hospital ICUs. Similar to our study, recent two studies were conducted also in Egypt that reported the prevalence of diverse clones from CRKP using the PFGE and BOX PCR as molecular typing methods, respectively.37,49
It could be inferred from using the two different typing methods that ERIC-PCR method might be less discriminatory in the evaluation of distantly related isolates. Conversely, it could be a useful tool for evaluating isolates that are closely related genetically.
Since 57% of the total K. pneumoniae isolates were recovered from the ICU, therefore, the increase in the ICU admission in our study could highlight the increase of bacterial infection with K. pneumoniae, as it can form a biofilm. Thus, allowing its adherence to different invasive procedures that are often used in that ward.50 Those results are consistent with other studies conducted in Egypt, Colombia, the United States, and Greece, among pediatric patients, admitted to the ICU.16,37,51 On the other hand, a study in the Netherlands reported that admission and long ICU stay, use of invasive medical devices and carbapenem as an empirical therapy were the most significant risk factors for CRE acquisition by hospitalized patients particularly vulnerable patients.52 Accordingly, infection control measures should focus on the ICU, where the relevant patients at risk for either colonization and/or infection with HAI causing microorganisms are common.
The 30 genotyped CPKP isolates were ESBLS producers and showed significant resistance to ampicillin, cephalosporins, fluoroquinolones, and carbapenems, but exhibited differential susceptibility to amikacin (1%) and colistin (2.3%). These findings highlighted the risk of clonal dissemination of blaOXA-48-harboring K. pneumoniae among the hospital wards similar to a study conducted in the United Arab of Emirates, which reported comparable results.53 However, in Japan, Khalifa and co-workers reported no clonal relationship among carbapenemase-producing isolates.40 Furthermore, a study in Italy reported that there is a relationship between the hospital environment, and the increased prevalence of resistant bacteria causing infections often encountered in the ICU setting, followed by non-ICU hospital wards and outpatient hospital settings.54 In 2016, a study in France reported the persistence of OXA-48-producing K pneumoniae for 20 months in a hospital ICU environment.55
The dissemination of some clonally-related isolates among the hospital’s different wards during the study period is an indication of the probability of cross-transmission of these resistant isolates in the hospital. The spread of the carbapenemases is linked mainly to mobile genetic elements such as plasmids, transposons, and integrons that carry not only resistance but might also harbor virulence determinants. Thus, facilitating transmission of resistant determinants between the same and different bacterial species including the bacteria present in the human microbiome.56 Consequently, this could affect the bacterial ability for colonization and infection within the host.57 The blaOXA-48 like genes are always carried on a broad range of plasmids such as IncH, IncA/C, IncX3, and ColKP3.58 The genes encoding for these resistance mechanisms are transferable between bacteria. Hence, there is a high risk of dissemination and potential healthcare-associated outbreaks of Enterobacteriaceae with these resistance mechanisms.57 In the current study, the conjugation experiment for the blaOXA-48 harboring plasmids from representative isolates was successful consistent with other studies.59–61
Based on our molecular typing results, both methods can be used for the determination of the clonal relationships of K. pneumoniae strains. Although ERIC-PCR is a more rapid and easy method compared to PFGE, its discriminatory power seemed to be less than that of PFGE.
The current study presents the molecular epidemiological situation of the MDR CRKP spread among hospitalized pediatric cancer patients in a major hospital in Egypt. It is notable from our findings that there might be dissemination of clonally-related strains among the ICU wards in the hospital. Therefore, the implementation of strict control measures for CRKP among the healthcare wards requires reliable and rapid identification methods. Additionally, hospital settings should have a protocol for effective screening for CPE, especially in high-risk patients. Further studies are required to investigate the sequence type of the disseminating clones in the hospital settings and consider the carbapenemases’ impact on the fitness and virulence in Enterobacteriaceae.
Ethics Approval
The study approval was obtained from the Research Ethics Committee of the Faculty of Pharmacy, Cairo University (approval no. MI-1203).
Acknowledgments
We would like to thank the lab members of the Clinical Pathology Department in the Children’s Cancer Hospital for facilitating isolates collection. Also, we would like to thank the staff and team members of the Biotechnology Centre in the Faculty of Pharmacy, Cairo University for assistance in the PFGE work and providing resources.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work”.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Perdikouri EIA, Arvaniti K, Lathyris D, et al. Infections due to multidrug-resistant bacteria in oncological patients: insights from a five-year epidemiological and clinical analysis. Microorganisms. 2019;7(9):1–13. doi:10.3390/microorganisms7090277
2. WHO. The fight against antimicrobial resistance is significant for cancer prevention and treatment. 2021. Available from: https://apps.who.int/iris/bitstream/handle/10665/337511/WHO-EURO-2020-1628-41379-56382-eng.pdf.
3. De Araújo Jácome PRL, Alves LR, Jácome-Júnior AT, et al. Detection of blaSPM-1, blaKPC, blaTEM and blaCTX-M genes in isolates of Pseudomonas aeruginosa, Acinetobacter spp. And Klebsiella spp. from cancer patients with healthcare-associated infections. J Med Microbiol. 2016;65(7):658–665. doi:10.1099/jmm.0.000280
4. Satlin MJ, Cohen N, Ma KC, et al. Bacteremia due to carbapenem-resistant Enterobacteriaceae in neutropenic patients with hematologic malignancies. J Infect. 2016;73(4):336–345. doi:10.1016/j.jinf.2016.07.002
5. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019. https://www.cdc.gov/drugresistance/biggest_threats.html.
6. Papanicolas LE, Gordon DL, Wesselingh SL, Rogers GB. Not just antibiotics: is cancer chemotherapy driving antimicrobial resistance? Trends Microbiol. 2018;26(5):393–400. doi:10.1016/j.tim.2017.10.009
7. Rapoport B, Klastersky J, Raftopoulos H, et al. The emerging problem of bacterial resistance in cancer patients; proceedings of a workshop held by MASCC B neutropenia, infection, and myelosuppression ^ study group during the MASCC Annual Meeting Held in Berlin on 27 – 29 June 2013. Supportive Care Cancer. 2016;24:2819–2826. doi:10.1007/s00520-016-3183-5
8. Codjoe FS, Donkor ES. Carbapenem Resistance: a review. Med Sci. 2017;6(1). https://www.ncbi.nlm.nih.gov/pubmed/29267233.
9. Tamma PD, Simner PJ. Phenotypic detection of carbapenemase-producing organisms from clinical isolates. J Clin Microbiol. 2018;56(11):1–13. doi:10.1128/JCM.01140-18
10. Sekyere JO, Govinden U, Essack S. The molecular epidemiology and genetic environment of carbapenemases detected in Africa. Microb Drug Resist. 2016;22(1):59–68. doi:10.1089/mdr.2015.0053
11. Montazeri EA, Khosravi AD, Saki M, Sirous M, Keikhaei B, Seyed-Mohammadi S. Prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae causing bloodstream infections in cancer patients from southwest of Iran. Infect Drug Resist. 2020;13:1319–1326. doi:10.2147/IDR.S254357
12. Yazdansetad S, Alkhudhairy MK, Najafpour R, et al. Preliminary survey of extended-spectrum β-lactamases (ESBLs) in nosocomial uropathogen Klebsiella pneumoniae in north-central Iran. Heliyon. 2019;5(9):e02349. doi:10.1016/j.heliyon.2019.e02349
13. Mohamed ER, Aly SA, Halby HM, Ahmed SH, Zakaria AM, El-Asheer OM. Epidemiological typing of multidrug-resistant Klebsiella pneumoniae, which causes paediatric ventilator-associated pneumonia in Egypt. J Med Microbiol. 2017;66(5):628–634. doi:10.1099/jmm.0.000473
14. Duman Y, Ersoy Y, Gursoy NC, Toplu SA, Otlu B. A silent outbreak due to Klebsiella pneumoniae that co-produced NDM-1andOXA-48 carbapenemases, and infection control measures. Iran J Basic Med Sci. 2020;23(1):46–50. doi:10.22038/IJBMS.2019.35269.8400
15. Baranovsky S, Romano-Bertrand S, Dumont Y, et al. Tracking carbapenemase-producing bacteria by molecular typing: population diversity and sampling pitfall. Infect Genet Evol. 2018;65:104–106. doi:10.1016/j.meegid.2018.07.020
16. Darda VM, Iosifidis E, Antachopoulos C, et al. Risk factors for carbapenem resistance and outcomes when treating bloodstream infections in a pediatric intensive care unit. Acta Paediatr Int J Paediatr. 2019;108(10):1923–1924. doi:10.1111/apa.14923
17. Hudzicki J. Kirby-bauer disk diffusion susceptibility test protocol. Am Soc Microbiol. 2009;1–13. https://www.asm.org/Protocols/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Pro.
18. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-seventh informational supplement. CLSI document M100-S27. 27th ed. 2017; Available from: www.clsi.org.
19. Ragheb SM, Tawfick MM, El-Kholy AA, Abdulall AK. Phenotypic and genotypic features of klebsiella pneumoniae harboring carbapenemases in Egypt: OXA-48-like carbapenemases as an investigated model. Antibiotics. 2020;9(12):852. doi:10.3390/antibiotics9120852
20. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement CLSI. Vol. 32; 2015.
21. Georgios M, Egki T, Effrosyni S. Phenotypic, and molecular methods for the detection of antibiotic resistance mechanisms in gram-negative nosocomial pathogens. In: Saxena SK, ed. Trends in Infectious Diseases. Rijeka: IntechOpen. 2014:139–162. doi:10.5772/57582
22. Xia Y, Liang Z, Su X, Xiong Y. Characterization of carbapenemase genes in Enterobacteriaceae species exhibiting decreased susceptibility to carbapenems in a university hospital in Chongqing, China. Ann Lab Med. 2012;32(4):270–275. doi:10.3343/alm.2012.32.4.270
23. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi:10.1016/S0022-2836(05)80360-2
24. Bilung LM, Pui CF, Su’Ut L, Apun K. Evaluation of BOX-PCR and ERIC-PCR as molecular typing tools for pathogenic leptospira. Dis Markers. 2018;2018:1–9. doi:10.1155/2018/1351634
25. Sneath PHA, Sokal RR. 1975. Numerical taxonomy. the principles and practice of numerical classification. In: Peter HAS, Sokal RR, editors. The Quarterly Review of Biology. Vol. 50. Taylor & Francis. 525–526. doi:10.1086/408956
26. CDC (Centers for Disease Control and Prevention). Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexner. Available from: https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf. 2013.
27. Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi:10.1128/jcm.33.9.2233-2239.1995
28. Manohar P, Leptihn S, Lopes BS, Ramesh N. Dissemination of Carbapenem-resistance and Plasmids-encoding Carbapenemases in Gram-negative Bacteria Isolated from India. bioRxiv. 2020:102434. doi:10.1101/2020.05.18.102434
29. Khajuria A, Praharaj AK, Kumar M, Grover N. Carbapenem resistance among enterobacter species in a tertiary care hospital in central India. Chemother Res Pract. 2014;2014:6. doi:10.1155/2014/972646
30. Gong X, Zhang J, Su S, et al. Molecular characterization and epidemiology of carbapenem non-susceptible enterobacteriaceae isolated from the eastern region of heilongjiang province, China. BMC Infect Dis. 2018;18(1):1–10. doi:10.1186/s12879-018-3294-3
31. Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26(11):2465–2466. doi:10.1128/JCM.26.11.2465-2466.1988
32. Magiorakos A, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
33. World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 2017. Available from: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/.
34. European Centre for Disease Prevention and Control. Carbapenem-resistant Enterobacteriaceae; 2018. Available from: https://ecdc.europa.eu/sites/portal/files/documents/RRA-Enterobacteriaceae-Carbapenems-European-Union-countries.
35. Kotb S, Lyman M, Ismail G, et al. Epidemiology of Carbapenem-resistant Enterobacteriaceae in Egyptian intensive care units using national healthcare-associated infections surveillance data, 2011–2017. Antimicrob Resist Infect Control. 2020;9(1):1–9. doi:10.1186/s13756-019-0639-7
36. Dandachi I, Chaddad A, Hanna J, Matta J, Daoud Z. Understanding the epidemiology of multi-drug resistant gram-negative bacilli in the Middle East using a one health approach. Front Microbiol. 2019;10(AUG). doi:10.3389/fmicb.2019.01941
37. El-Badawy MF, El-Far SW, Althobaiti SS, Abou-Elazm FI, Shohayeb MM. The first Egyptian report showing the co-existence of blaNDM-25, blaOXA-23, blaOXA-181, and blaGES-1 among carbapenem-resistant K. pneumoniae clinical isolates genotyped by BOX-PCR. Infect Drug Resist. 2020;13:1237–1250. doi:10.2147/IDR.S244064
38. Tawfick MM, Alshareef WA, Bendary HA, ElMahalawy H, Abdulall AK. The emergence of carbapenemase blaNDM genotype among carbapenem-resistant Enterobacteriaceae isolates from Egyptian cancer patients. Eur J Clin Microbiol Infect Dis. 2020;39(7):1251–1259. doi:10.1007/s10096-020-03839-2
39. Awad S, Ghanem S, Helal M, et al. Phenotypic and genotypic characteristics of community-acquired and hospital-acquired carbapenem-resistant Enterobacteriaceae in patients with liver cirrhosis at the National Liver Institute of Egypt. Can J Infect Control. 2019;34(2):100–103. doi:10.36584/cjic.2019.011
40. Khalifa HO, Soliman AM, Ahmed AM, et al. High carbapenem resistance in clinical gram-negative pathogens isolated in Egypt. Microb Drug Resist. 2017;23(7):838–844. doi:10.1089/mdr.2015.0339
41. David S, Reuter S, Harris SR, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4(11):1919–1929. doi:10.1038/s41564-019-0492-8
42. Touati A, Mairi A. Epidemiology of carbapenemase-producing Enterobacterales in the Middle East: a systematic review. Expert Rev Anti Infect Ther. 2020;18(3):241–250. doi:10.1080/14787210.2020.1729126
43. Vanegas JM, Parra OL, Jiménez JN. Molecular epidemiology of carbapenem-resistant Gram-negative bacilli from infected pediatric population in tertiary-care hospitals in Medellín, Colombia: an increasing problem. BMC Infect Dis. 2016;16(1):1–10. doi:10.1186/s12879-016-1805-7
44. Jing X, Zhou H, Min X, et al. The simplified carbapenem inactivation method (sCIM) for simple and accurate detection of carbapenemase-producing gram-negative bacilli. Front Microbiol. 2018;9(October):1–7. doi:10.3389/fmicb.2018.02391
45. Zhong H, Wu ML, Feng WJ, Huang SF, Yang P. Accuracy and applicability of different phenotypic methods for carbapenemase detection in Enterobacteriaceae: a systematic review and meta-analysis. J Glob Antimicrob Resist. 2020;21:138–147. doi:10.1016/j.jgar.2019.10.010
46. Jeong SH, Kim HS, Kim JS, et al. Prevalence and molecular characteristics of carbapenemase-producing Enterobacteriaceae from five hospitals in Korea. Ann Lab Med. 2016;36(6):529–535. doi:10.3343/alm.2016.36.6.529
47. Zafer MM, El-Mahallawy HA, Abdulhak A, Amin MA, Al-Agamy MH, Radwan HH. Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann Clin Microbiol Antimicrob. 2019;18(1):1–8. doi:10.1186/s12941-019-0339-4
48. Liu D, ed. Molecular Detection of Human Bacterial Pathogens. New York: CRC Press; 2011. doi:10.1201/b10848.
49. Ghaith DM, Zafer MM, Said HM, et al. Genetic diversity of carbapenem-resistant Klebsiella pneumoniae causing neonatal sepsis in intensive care unit, Cairo, Egypt. Eur J Clin Microbiol Infect Dis. 2020;39(3):583–591. doi:10.1007/s10096-019-03761-2
50. Martelius T, Jalava J, Kärki T, Möttönen T, Ollgren J, Lyytikäinen O. Nosocomial bloodstream infections caused by Escherichia coli and Klebsiella pneumoniae resistant to third-generation cephalosporins, Finland, 1999–2013: trends, patient characteristics, and mortality. Infect Dis. 2016;48(3):229–234. doi:10.3109/23744235.2015.1109135
51. Chiotos K, Han JH, Tamma PD. Carbapenem-resistant enterobacteriaceae infections in children. Curr Infect Dis Rep. 2017;18(1):139–148. doi:10.1016/j.physbeh.2017.03.040
52. van Loon K, Vos M. A systematic review and meta-analyses of the clinical. Antimicrob Agents Chemother. 2018;62(e01730–17):1–18. doi:10.1128/AAC.01730-17
53. Moubareck CA, Mouftah SF, Pál T, et al. Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int J Antimicrob Agents. 2018;52(1):90–95. doi:10.1016/j.ijantimicag.2018.03.003
54. Ripabelli G, Tamburro M, Guerrizio G, et al. Tracking multidrug-resistant klebsiella pneumoniae from an Italian hospital: molecular epidemiology and surveillance by PFGE, RAPD, and PCR-based resistance genes prevalence. Curr Microbiol. 2018;75(8):977–987. doi:10.1007/s00284-018-1475-3
55. Pantel A, Richaud-Morel B, Cazaban M, Bouziges N, Sotto A, Lavigne JP. Environmental persistence of OXA-48-producing Klebsiella pneumoniae in a French intensive care unit previous presentations: presented to the Congress of French Society of Hygiene, June 2015, Tours, France. Am J Infect Control. 2016;44(3):366–368. doi:10.1016/j.ajic.2015.09.021
56. European Centre for Disease Prevention and Control. Outbreak of Carbapenemase-Producing (NDM-1 and OXA-48) and Colistin-Resistant Klebsiella Pneumoniae ST307. North-East Germany: ECDC: Stockholm; 2019.
57. Li B, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9(9):1071–1081. doi:10.2217/fmb.14.48
58. Shankar C, Mathur P, Venkatesan M, et al. Rapidly disseminating blaOXA-232 carrying Klebsiella pneumoniae belonging to ST231 in India: multiple and varied mobile genetic elements. BMC Microbiol. 2019;19(1):1–8. doi:10.1186/s12866-019-1513-8
59. David S, Cohen V, Reuter S, et al. Genomic analysis of carbapenemase-encoding plasmids from Klebsiella pneumoniae across Europe highlights three major patterns of dissemination. bioRxiv. 2019:1–36. 10.1101/2019.12.19.873935.
60. Solgi H, Nematzadeh S, Giske CG, et al. Molecular epidemiology of OXA-48 and NDM-1 producing enterobacterales species at a University Hospital in Tehran, Iran, between 2015 and 2016. Front Microbiol. 2020;11(May):1–13. doi:10.3389/fmicb.2020.00936
61. Hamprecht A, Sommer J, Willmann M, et al. Pathogenicity of clinical OXA-48 isolates and impact of the OXA-48 IncL plasmid on virulence and bacterial fitness. Front Microbiol. 2019;10(November):1–12. doi:10.3389/fmicb.2019.02509
62. Versalovic J, Koeuth T, Lupski R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19(24):6823–6831. doi:10.1093/nar/19.24.6823
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.