Back to Journals » Infection and Drug Resistance » Volume 15
Molecular Characterization of Carbapenem-Resistant Acinetobacter baumannii Isolates Among Intensive Care Unit Patients and Environment
Authors Hu H , Lou Y, Feng H, Tao J, Shi W, Ni S, Pan Q, Ge T, Shen P, Zhong Z, Xiao Y, Qu T
Received 29 November 2021
Accepted for publication 26 March 2022
Published 13 April 2022 Volume 2022:15 Pages 1821—1829
DOI https://doi.org/10.2147/IDR.S349895
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Hangbin Hu,1,* Yifeng Lou,1,2,* Haiting Feng,3 Jingjing Tao,1 Weixiao Shi,1 Shuangling Ni,4 Qunying Pan,3 Tianxiang Ge,3 Ping Shen,1 Zifeng Zhong,3 Yonghong Xiao,1 Tingting Qu1,3
1State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, National Medical Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, People’s Republic of China; 2Infectious Disease Department, Sanmen People’s Hospital, Taizhou, Zhejiang, People’s Republic of China; 3Infection Control Department, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, People’s Republic of China; 4Infectious Disease Department, Lishui People’s Hospital, Lishui, Zhejiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tingting Qu; Yonghong Xiao, Tel +86 571 87236673, Email [email protected]; [email protected]
Objective: Critical patients in intensive care unit (ICU) are highly susceptible to acquiring carbapenem-resistant Acinetobacter baumannii (CRAB) infection. To investigate the relationship between nosocomial infections and environmental health, we studied the distribution and homology of CRAB isolates from patients and environment and evaluated the effectiveness of infection control measures.
Methods: In the 4-month study, we conducted a monthly CRAB screening of the ICU environment prior to disinfection in a Chinese teaching hospital. The ICU underwent routine disinfection procedures twice a day. We collected samples from the environment around the patients before disinfection. Clinical specimens from patients were also screened. The samples obtained were studied for phenotype and homology via antibiotic susceptibility testing, pulsed-field gel electrophoresis (PFGE), and whole-genome sequencing (WGS).
Results: Ten specimens were sampled from ICU environments. Five were obtained in May 2020, and sputums from patient a in bed A at this time were cultured for CRAB isolates; in June 2020 another 5 environmental specimens were obtained from the same bed unit for CRAB, and sputums from patient b in bed A at this time were also cultured for CRAB isolates. Following intensive infection control measures, environmental sampling was negative in July and August. These 18 CRAB isolates all carried OXA-66 and OXA-23 genes and showed a similar resistance phenotype. WGS showed a close relationship among specimens from patients’ sputum and their surroundings, but no homology between patients.
Conclusion: The analysis of cgMLST and SNPs is more accurate for strain homology analysis. Our data confirm that CRAB isolates spread from patient to environment in ICU; however, contact isolation and disinfection measures are effective in avoiding transmission, highlighting the importance of continued education and surveillance of CRAB. WGS could provide rich information on antimicrobial resistance, which is of great value in scientific research and clinical diagnosis.
Keywords: carbapenem-resistant Acinetobacter baumannii, intensive care unit, whole genome sequencing, cgMLST, PFGE
Introduction
The spread of Carbapenem-resistant Organism (CRO), especially in intensive care units (ICUs), has become a serious problem and health threat worldwide.1–4 The carbapenems are antimicrobial agents with significant activity and potency against Gram-positive and Gram-negative bacteria.5 It has been widely used in clinical studies since the first carbapenem was authorized in the United States for the treatment of microbial infections in 1985.6 Unfortunately, since the abuse of antibiotics over the past few decades, a set of bacteria have become resistant to carbapenems. The main mechanisms are the overproduction of AmpC cephalosporinase, Extended-spectrum β-lactamases (ESBLs), and carbapenemases.7–9 Hospitalized patients are susceptible to CRAB colonization. If CRAB enters the bloodstream or reaches vital organ, it leads to longer hospital stays, fewer treatment options, and higher risks for complications, an increased morbidity.10–12 In China, the detection rate of CRAB increased substantially from 31.0% in 2005 to 71.5% in 2021 (http://www.chinets.com/Data/GermYear), which contribute to increasing healthcare costs.
CRAB is an important cause of healthcare acquired infections (HAIs). Human-to-human transmission may be the primary route of HAIs among the Healthcare-workers (HCWs) and patients, while the contact of patients’ surroundings may also contribute to the HAIs since the environment could be contaminated by patients. Recent researcher has revealed the environmental contamination of CRAB was existed and had an epidemiological link between clinical samples.13,14 These results suggest that the environment could be a route of cross-transmission, which eventually causes HAIs among patients to be possible.
In this study, we conducted environmental surveillance in ICU wards to investigate whether the environmental contamination may contribute to CRAB transmission. Pulsed field gel electrophoresis (PFGE) and whole-genome sequencing (WGS) were performed to analyze the epidemiological relevance between environmental isolates and patient samples.
Methods
Study Design
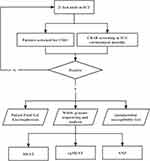
This study was conducted in the First Affiliated Hospital of Zhejiang University, China. The hospital was a teaching hospital with approximately 2500 beds. The environmental surveillance was carried out in the ICU wards from May 2020 to August 2020. The environmental sampling was conducted every month before routine disinfection. Stool specimens from patients were collected once a week for CRO screening, and other clinical specimens were sampled when the patients were suspected to be infected. Study flowchart is shown in Figure 1.
Environmental Sampling
Environmental sampling was conducted on surfaces that were frequently contacted by patients or HCWs, including bed rails, nightstand, bed curtains and cardiograph monitor panel, non-contact ear thermometer, personal digital assistant (PDA), keyboard, landline telephone, and hand-washing sink. Samples were also collected from the hands of HCWs before hand-hygiene was performed. Surfaces and hands were sampled with Custom Sponge-Stick with Neutralizing Buffer (3M, St. Paul, MN, USA). When a patient had respiratory CRAB infection or colonization, condensates in mechanical ventilator circuits were collected. All samples were sent to the laboratory immediately after collection. Swabs were inoculated onto blood agar plates (Oxoid, Altrincham, UK) and Mueller-Hinton agar plates containing 2 μg/mL imipenem and cultured for 24 hours at 37°C. For the condensate samples, membranes with 0.45 μm pore sizes were used to trap and support bacterial growth for subsequent culture and analysis. The VITEK 2 system (bioMérieux, France) was performed to identify CRAB when single colonies were obtained, and the strains were stored at −80°C for later use. Clinical specimens such as sputum, blood, and urine were cultured, and CRAB isolates were also stored for further study. The isolation times of the 18 CRAB strains are shown in Figure 2.
 |
Figure 2 Months of isolation of 18 CRAB strains. The upper one is the time of clinical specimen isolation, and the lower Ea-1~Eb-5 stand for the samples collected from environment. |
Antimicrobial Agents Susceptibility Testing
All CRAB isolates were tested for antibiotic susceptibility using agar dilution and broth dilution methods for 14 commonly used antimicrobial agents (ceftazidime, cefepime, piperacillin-tazobactam, imipenem, meropenem, amikacin, ciprofloxacin, ampicillin, gentamicin, tetracycline, trimethoprim/sulfadioxazole, levofloxacin, tigecycline, and polymyxin B). While tigecycline results were interpreted based on the Food and Drug Administration (FDA) criteria,15 other antibiotics results were interpreted according to the Clinical and Laboratory Standards guidelines (CLSI 2020).16
Genomic DNA Extraction
Genomic DNA of CRAB strains was extracted using a QIAamp DNA MiniKit (Qiagen, Valencia, CA, USA) under the manufacturer’s protocol.17 After amplified, the monoclonal strains were lysed and purified by adding reagents to obtain DNA.
Genome Sequencing and Annotation
The genome was sequenced by the HiSeq X Ten platform (Illumina, San Diego, CA, USA) with 2 × 150-bp paired-end reads. Fasta files were assembled using Shovill 0.9.0. The genomes were annotated using Prokka v 1.14.5 (https://github.com/tseemann/prokka). Acquired resistance genes were detected by ResFinder 4.1 tool on the CGE Server (https://cge.cbs.dtu.dk/services/ResFinder/), and multilocus sequence typing was obtained using MLST 2.0 tool on the same server (https://cge.cbs.dtu.dk/services/MLST/).
Homology Analysis
These CRAB isolates belonging to the same species were characterized by PFGE as described previously.18 After obtaining whole-gene sequence data, the genomic relationships of CRAB isolates belonging to the same species were analyzed core genome multilocus sequence typing (cgMLST) separately using Ridom SeqSphere+ Version 5.1.0 (Ridom, Münster, Germany) with default parameter. Core gene single nucleotide polymorphism (SNP) analysis was further performed for strains belonging to the same ST using Snippy (v4.4.5; https://github.com/tseemann/snippy).
Disinfection
Routine disinfection procedures were performed 3 times a day in the ICU ward. Surfaces of objects and facilities are wiped using disposable towels with 500 mg/L chlorinated disinfectant, and waited for 30 minutes. The cleaning and disinfection procedures of floors were performed in the same way. Visible contaminants should be removed thoroughly at any time with sanitary wipes (Clinell Universal Wipes; GAMA Healthcare Ltd, London, UK) before disinfection.
Results
Environment Surveillance and Clinical Sampling
Between May 2020 and August 2020, the environmental sampling was performed every month in the ICU. The CRAB was detected from patient a’s surroundings (surfaces; Table 1) in May 12th, while the other environmental samples were all negative for CRAB detection. Patient a was admitted to the ICU on April 4th and he was in bed A during the hospitalization. The CRAB was isolated from his sputum specimen on April 16th. Patient b was admitted to the hospital on May 14th and transferred to ICU on May 23th. He was also in bed A during the hospitalization in ICU. CRAB was isolated from his sputum specimen on June 6th. Patient c was admitted to the ICU on May 23th. The CRAB was isolated from his sputum specimen on June 12th. Patient c was in bed B during the hospitalization. He was transferred to the hepatobiliary surgical ward on June 16th. The environmental samples of patient c were all negative for CRAB detection.
 |
Table 1 CRAB Isolates from the ICU Settings |
Antimicrobial Agents Sensitivity Results and Distribution of Drug Resistance Genes
All of the 18 CRAB strains presented similar resistance phenotypes to commonly used antimicrobials in clinic, including ceftazidime, cefepime, piperacillin tazobactam, imipenem, meropenem, amikacin, ciprofloxacin, ampicillin, tetracycline, trimethoprim/sulfadioxazole, and levofloxacin (Figure 3). Eb-4 was resistant to polymyxin B (MIC 4 mg/L), the other 17 strains were intermediate (MIC 1~2 mg/L). Most isolates (88.9%, 16/18) were sensitive to tigecycline. According to the Oxford scheme, the sequence type (ST) of A. baumannii isolates was divided into ST369 and ST208. Except for the earliest sputum sample obtained (Pa-1), which belonged to ST208, patient a’s sputum specimens with his surrounding environmental specimens belongs to ST369. Patient b’ sputum specimens and his surrounding environmental specimens are ST208. The CRAB strains all contained OXA-23, OXA-25 and OXA-66 (Figure 3).
Homology Analysis Based on PFGE and Whole Genome Sequencing (WGS)
The 18 CRAB isolates were divided into three clusters according to the PFGE fingerprints (Figure 4A): environment of patient a, sputum of patient a, b and c, and environment of patient c. Isolates in a same cluster differed only slightly from each other. Homology analysis of the genetics between isolates by cgMLST showed different results (Figure 4B). Sputum specimens from patient a (excluding Pa-1) and all samples from his environment formed the same cluster, while sputum specimens from patient b and isolates from his environment formed another cluster. Isolates Pa-1 and Pc-1 did not belong to any cluster.
In addition, SNP mutations were widespread and polymorphism-rich in respective populations of ST208 and ST369. The SNPs distribution of ST208 isolates ranged from 11 to 196, whereas the SNP distribution of ST369 ranged from 5 to 2891 (Figure 4C and D).
Disscussion
Genus acinetobacter are considered to be ubiquitous in nature because they can be recovered from soil and water samples.19 Acinetobacter baumannii is found in most of the hospital environment, particularly in ICUs, burn wards, and surgical wards.6 It can be isolated from various clinical specimens, such as sputum, bloodstream, wounded skin and tissues, and feces. Nosocomial infections in the healthcare system caused by CRAB are increasing in recent years, especially in ICUs.20,21 Due to the increasing drug resistance pattern of Acinetobacter baumannii and the limitation of drugs that can be used for treatment, the CRAB has been considered as a global priority by the World Health Organization.22
Recent studies proved that CRAB can survive for long periods in dry environments and be isolated from medical settings, medical device, health-care workers’ hands and dander from colonized patients.23 Organisms may further migrate and interact with environmental strains, eventually leading to the formation of a reservoir for various antibiotic resistance genes.24 In addition, the difficulty in eradicating CRAB from the environment contributed to the increasing prevalence of HAIs, due to its persistence on surfaces and reduced susceptibility to disinfectant.25 In this study, we proved that the environmental contamination of CRAB was existed in the ICU. The CRAB isolates were detected from the surroundings of patient a and patient b, both of whom stayed in the same bed A unit at different time points. And CRAB isolate was also detected in the sputum specimen of patient c who stay in the bed B which was positioned next to bed A.
A total of 18 CRAB isolates were divided into 2 sequence types, 10 ST208 and 8 ST369, all belonging to the 92 clonal complex (CC92).26,27 In China, ST208 is the predominant ST of CRAB,28 while ST369 is a high-risk clone.26 These CRAB isolates all carried OXA-66 and OXA-23 genes, the two main causes of bacterial carbapenem resistance, which were reported to be highly correlated with the outbreak of A. baumannii CC92 as previous studies.27,29 According to the WGS and cgMLST results, homology analysis of CRAB strains isolated from one patient and his corresponding environmental samples were proved to be in the same cluster, and the strains in environmental samples were isolated after that were found in sputum specimen of the same patient. These results suggested that the environmental contamination could be attributable to patients with CRAB colonization/infection. A number of researchers have demonstrated that hands, gloves and gowns of healthcare workers may get contaminated when direct or indirect contact with patients, and it would mediate the transmission of CRAB from patient to environment or from patient to patient.30,31 In our study, the CRAB strains were also isolated from ventilator condensate samples when patients’ sputum specimens were positive for CRAB detection, suggesting that CRO had the potential to transmit through aerosol, as described in the previous study.27 Therefore, single-room isolation is strongly recommended according to World Health Organization (WHO) guidelines for preventing the cross-transmission of CRO (eg, CRAB).32 The contamination by CRAB was detected from monitor screens, keyboards, PDAs, bed rails and bedside tables around bed units, which highlights the importance of environmental disinfection.
In this study, the PFGE pattern suggested that sputum samples cultured from three patients were highly related, indicating a probability of cross-infections and horizontal transmission. However, cgMLST and SNP analysis based on WGS technique indicated that the sputum samples from three patients did not belong to the same cluster, which contradicted the result from PFGE. Operator errors may occur in identifying bands, especially when PFGE gel image analysis is shifted or weak.33 PFGE used to be considered as the golden standard for molecular epidemiology of bacterial strains.34 However, with the development of science and technology, WGS has become a new technique for pathogen identification with its superior resolution than PFGE, and it could distinguish isolates that could not be distinguished by PFGE.35 WGS-based cgMLST had higher sensitivity and accuracy in results when compared with PFGE.36 These studies all highlighted that WGS technology had a higher discriminatory power in determining the relevance of isolates compared to traditional phenotypic methods including PFGE. In addition, it was interesting that the first CRAB isolate from sputum specimen belonged to ST369 and latter ones belonged to ST208 in patient A, both of which could be detected from patient’s surroundings. These results indicated that multiple clones coexisted in the same patient, or the dominant clone evolved in response to the interaction of antibiotic treatment with host immunity, which needed further investigation.37,38
The antibiotic treatment was not given to patient c since bacterial colonization was considered. Patients a and b were considered CRAB lung infections and received combination therapy: one with polymyxin B combined with amikacin nebulized therapy; the other with polymyxin B combined with meropenem. Despite the use of active treatment, both patients obtained a poor clinical prognosis. Based on expert consensus, a combination of effective antibiotics is recommended to treat CRAB infections.39,40 When the MIC of carbapenems ≤8 mg/L, a combination therapy with carbapenem showed a comparable outcome than tigecycline, polymyxin B, or aminoglycoside monotherapy. Nebulized inhalation of aminoglycoside or polymyxin B is recommended to increase the local drug distribution which was increased and also decreased systemic side effects.41 However, prevention of CRAB infection is key, and antimicrobial restriction system antimicrobial restriction systems can help reduce and contribute to the appropriate use of antibiotics, especially carbapenems.42
Continuous education on infection control and prevention, environmental disinfection, and the usage of antibiotics were conducted for ICU staff in June. In addition, measures for patient unit with multi-drug resistant bacteria colonization/infection in this ICU were improved by enhancing the frequency of daily disinfection, air disinfection, ventilator condensate management, and avoidance of dust operations. The environmental samples detected in July and August were all negative: no carbapenem-resistant organisms were isolated. As contaminated surfaces around patients may increase the incidence of multidrug-resistant bacterial infections, hand hygiene, patient isolation, and strict environmental disinfection measures should be applied to prevent CRO outbreaks.32,43
There were still some limitations in this study. First, the sample size was small. Second, we did not collect the air samples for CRO screening, so evidence of airborne long-distance transmission of non-contact environmental contamination by patients was lacking. How CRAB spreads from patients to the environment by aerosol transmission requires further study.
In conclusion, environmental contamination of CRAB is common in ICU, which could cause hospital-acquired infection especially in immunocompromised patients. The CRAB contamination in patient surroundings could be transmitted from patients to environment via direct or indirect contact. No cross-infection of CRAB was detected among the three patients in the ICU. Our findings highlight the widespread use of CRAB on the object surfaces of the ICU ward and emphasize the importance of disinfection and patient isolation in preventing bacteria spreading.
Ethics Approval
This study was approved by the Institutional Review Board of the First Affiliated Hospital of Zhejiang University, China (approval number: IIT20210762A). Due to the nature of the study, the review committee waived informed consent. The study follows the guidelines of the Helsinki Declaration.
Acknowledgments
The author thanks all patients who participated in this study and all members of our institution for their help.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval for the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
None of the authors have any conflicts of interest in this work.
References
1. Ripabelli G, Sammarco ML, Scutella M, Felice V, Tamburro M. Carbapenem-resistant kpc- and tem-producing Escherichia coli st131 isolated from a hospitalized patient with urinary tract infection: first isolation in Molise region, central Italy, July 2018. Microb Drug Resist. 2020;26(1):38–45. doi:10.1089/mdr.2019.0085
2. Sarshar M, Behzadi P, Scribano D, Palamara AT, Ambrosi C. Acinetobacter baumannii: an ancient commensal with weapons of a pathogen. Pathogens. 2021;10(4):387. doi:10.3390/pathogens10040387
3. Hu H, Zhang Y, Zhang P, et al. Bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing p. Aeruginosa sequence type 463, associated with high mortality rates in China: a retrospective cohort study. Front Cell Infect Microbiol. 2021;11:756782. doi:10.3389/fcimb.2021.756782
4. Ahmadi M, Ranjbar R, Behzadi P, Mohammadian T. Virulence factors, antibiotic resistance patterns, and molecular types of clinical isolates of Klebsiella pneumoniae. Expert Rev Anti Infect Ther. 2021;20(3):463–472. doi:10.1080/14787210.2022.1990040
5. Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: past, present, and future. Antimicrob Agents Chemother. 2011;55(11):4943–4960. doi:10.1128/AAC.00296-11
6. Nguyen M, Joshi SG. Carbapenem resistance in acinetobacter baumannii, and their importance in hospital-acquired infections: a scientific review. J Appl Microbiol. 2021;131(6):2715–2738. doi:10.1111/jam.15130
7. Behzadi P, Garcia-Perdomo HA, Karpinski TM, Issakhanian L. Metallo-ss-lactamases: a review. Mol Biol Rep. 2020;47(8):6281–6294. doi:10.1007/s11033-020-05651-9
8. Reyes J, Aguilar AC, Caicedo A. Carbapenem-resistant Klebsiella pneumoniae: microbiology key points for clinical practice. Int J Gen Med. 2019;12:437–446. doi:10.2147/IJGM.S214305
9. Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18(5):263–272. doi:10.1016/j.molmed.2012.03.003
10. Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29(12):1099–1106. doi:10.1086/592412
11. Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–582. doi:10.1128/CMR.00058-07
12. Lemos EV, de la Hoz FP, Einarson TR, et al. Carbapenem resistance and mortality in patients with acinetobacter baumannii infection: systematic review and meta-analysis. Clin Microbiol Infect. 2014;20(5):416–423. doi:10.1111/1469-0691.12363
13. Catho G, Martischang R, Boroli F, et al. Outbreak of pseudomonas aeruginosa producing vim carbapenemase in an intensive care unit and its termination by implementation of waterless patient care. Crit Care. 2021;25(1):301. doi:10.1186/s13054-021-03726-y
14. Wei L, Wu L, Wen H, et al. Spread of carbapenem-resistant Klebsiella pneumoniae in an intensive care unit: a whole-genome sequence-based prospective observational study. Microbiol Spectr. 2021;9(1):e0005821. doi:10.1128/Spectrum.00058-21
15. Kim DH, Jung SI, Kwon KT, Ko KS. Occurrence of diverse abgri1-type genomic islands in acinetobacter baumannii global clone 2 isolates from South Korea. Antimicrob Agents Chemother. 2017;61(2). doi:10.1128/AAC.01972-16
16. CLSI. Performance standards for antimicrobial susceptibility testing.
17. Hu H, Mao J, Chen Y, et al. Clinical and microbiological characteristics of community-onset carbapenem-resistant Enterobacteriaceae isolates. Infect Drug Resist. 2020;13:3131–3143. doi:10.2147/IDR.S260804
18. Seifert H, Dolzani L, Bressan R, et al. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of acinetobacter baumannii. J Clin Microbiol. 2005;43(9):4328–4335. doi:10.1128/JCM.43.9.4328-4335.2005
19. Baumann P, Doudoroff M, Stanier RY. A study of the Moraxella group. II. Oxidative-negative species (genus acinetobacter). J Bacteriol. 1968;95(5):1520–1541. doi:10.1128/jb.95.5.1520-1541.1968
20. Lee CR, Lee JH, Park M, et al. Biology of acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017;7:55. doi:10.3389/fcimb.2017.00055
21. Hsu LY, Apisarnthanarak A, Khan E, Suwantarat N, Ghafur A, Tambyah PA. Carbapenem-resistant acinetobacter baumannii and Enterobacteriaceae in south and Southeast Asia. Clin Microbiol Rev. 2017;30(1):1–22. doi:10.1128/CMR.masthead.30-1
22. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the who priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi:10.1016/S1473-3099(17)30753-3
23. Bernards AT, Frenay HM, Lim BT, Hendriks WD, Dijkshoorn L, van Boven CP. Methicillin-resistant staphylococcus aureus and acinetobacter baumannii: an unexpected difference in epidemiologic behavior. Am J Infect Control. 1998;26(6):544–551. doi:10.1053/ic.1998.v26.a84555
24. Marti E, Variatza E, Balcazar JL. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014;22(1):36–41. doi:10.1016/j.tim.2013.11.001
25. Meschiari M, Lopez-Lozano JM, Di Pilato V, et al. A five-component infection control bundle to permanently eliminate a carbapenem-resistant acinetobacter baumannii spreading in an intensive care unit. Antimicrob Resist Infect Control. 2021;10(1):123. doi:10.1186/s13756-021-00990-z
26. Bian X, Liu X, Zhang X, et al. Epidemiological and genomic characteristics of acinetobacter baumannii from different infection sites using comparative genomics. BMC Genomics. 2021;22(1):530. doi:10.1186/s12864-021-07842-5
27. Jiang M, Liu L, Ma Y, et al. Molecular epidemiology of multi-drug resistant acinetobacter baumannii isolated in Shandong, China. Front Microbiol. 2016;7:1687. doi:10.3389/fmicb.2016.01687
28. Deng M, Zhu MH, Li JJ, et al. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of acinetobacter baumannii from a Chinese university hospital. Antimicrob Agents Chemother. 2014;58(1):297–303. doi:10.1128/AAC.01727-13
29. Chen Y, Gao J, Zhang H, Ying C. Spread of the blaoxa-23-containing tn2008 in carbapenem-resistant acinetobacter baumannii isolates grouped in cc92 from China. Front Microbiol. 2017;8:163. doi:10.3389/fmicb.2017.00163
30. Lerner A, Adler A, Abu-Hanna J, Meitus I, Navon-Venezia S, Carmeli Y. Environmental contamination by carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2013;51(1):177–181. doi:10.1128/JCM.01992-12
31. Yan Z, Zhou Y, Du M, et al. Prospective investigation of carbapenem-resistant Klebsiella pneumonia transmission among the staff, environment and patients in five major intensive care units, Beijing. J Hosp Infect. 2019;101(2):150–157. doi:10.1016/j.jhin.2018.11.019
32. World Health Organization. Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter Baumannii and Pseudomonas Aeruginosa in Health Care Facilities. Geneva: World Health Organization; 2017.
33. Di Tella D, Tamburro M, Guerrizio G, Fanelli I, Sammarco ML, Ripabelli G. Molecular epidemiological insights into colistin-resistant and carbapenemases-producing clinical Klebsiella pneumoniae isolates. Infect Drug Resist. 2019;12:3783–3795. doi:10.2147/IDR.S226416
34. Lomonaco S, Verghese B, Gerner-Smidt P, et al. Novel epidemic clones of listeria monocytogenes, United States, 2011. Emerg Infect Dis. 2013;19(1):147–150. doi:10.3201/eid1901.121167
35. Xu H, Zhang W, Zhang K, et al. Characterization of salmonella serotypes prevalent in asymptomatic people and patients. BMC Infect Dis. 2021;21(1):632. doi:10.1186/s12879-021-06340-z
36. Xu Q, Fu Y, Zhao F, Jiang Y, Yu Y. Molecular characterization of carbapenem-resistant serratia marcescens clinical isolates in a tertiary hospital in Hangzhou, China. Infect Drug Resist. 2020;13:999–1008. doi:10.2147/IDR.S243197
37. Markogiannakis A, Fildisis G, Tsiplakou S, et al. Cross-transmission of multidrug-resistant acinetobacter baumannii clonal strains causing episodes of sepsis in a trauma intensive care unit. Infect Control Hosp Epidemiol. 2008;29(5):410–417. doi:10.1086/533545
38. Wheatley R, Diaz CJ, Kapel N, et al. Rapid evolution and host immunity drive the rise and fall of carbapenem resistance during an acute pseudomonas aeruginosa infection. Nat Commun. 2021;12(1):2460. doi:10.1038/s41467-021-22814-9
39. Katip W, Oberdorfer P. Clinical efficacy and nephrotoxicity of colistin alone versus colistin plus vancomycin in critically ill patients infected with carbapenem-resistant acinetobacter baumannii: a propensity score-matched analysis. Pharmaceutics. 2021;13(2):162. doi:10.3390/pharmaceutics13020162
40. Katip W, Uitrakul S, Oberdorfer P. A comparison of colistin versus colistin plus meropenem for the treatment of carbapenem-resistant acinetobacter baumannii in critically ill patients: a propensity score-matched analysis. Antibiotics. 2020;9(10). doi:10.3390/antibiotics9100647
41. Katip W, Uitrakul S, Oberdorfer P. Clinical efficacy and nephrotoxicity of the loading dose colistin for the treatment of carbapenem-resistant acinetobacter baumannii in critically ill patients. Pharmaceutics. 2021;14(1):31. doi:10.3390/pharmaceutics14010031
42. Wanla W, Katip W, Supakul S, Apiwatnakorn P, Khamsarn S. Effects of an antimicrobial restriction system on appropriate carbapenem use in a hospital without infectious diseases consultation. Int J Gen Med. 2017;10:443–449. doi:10.2147/IJGM.S145133
43. Chen W. Host innate immune responses to acinetobacter baumannii infection. Front Cell Infect Microbiol. 2020;10:486. doi:10.3389/fcimb.2020.00486
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.