Back to Journals » Infection and Drug Resistance » Volume 13
Molecular Characteristics of Carbapenem-Resistant Enterobacter cloacae in a Tertiary Hospital in China
Authors Jin C, Zhou F, Cui Q, Qiang J, An C
Received 15 March 2020
Accepted for publication 8 May 2020
Published 28 May 2020 Volume 2020:13 Pages 1575—1581
DOI https://doi.org/10.2147/IDR.S254056
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Chunmei Jin,1 Fuxian Zhou,1 Qingsong Cui,2 Jixiang Qiang,1 Changshan An3
1Department of Clinical Laboratory, Yanbian University Hospital, Yanji, People’s Republic of China; 2Department of Intensive Care Unit, Yanbian University Hospital, Yanji, People’s Republic of China; 3Department of Respiratory Medicine, Yanbian University Hospital, Yanji, People’s Republic of China
Correspondence: Changshan An Tel +86-0433-2660064
Email [email protected]
Background: Infections caused by the carbapenem-resistant Enterobacter cloacae (CREC) bring great challenges to the clinical treatment and pose a serious threat to public health. In this study, we investigated the molecular characteristics of CREC in a tertiary hospital.
Materials and Methods: A total of 12 non-duplicate CREC strains isolated during the period of November 2016 to July 2019 were subjected to automated microbial identification and antimicrobial susceptibility testing (AST) using the BD Phoenix-100 identification and antimicrobial susceptibility testing (ID/AST) system. The strains were also subjected to phenotypic screening for the detection of antibiotic resistance genes such as the carbapenemase and other β-lactamase genes, with the use of the polymerase chain reaction assay (PCR). Finally, multi-locus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE)-based homology analysis were applied.
Results: Four types of carbapenemases namely IMP-26, NDM-5, NDM-1, and KPC-2 were identified in 12 CREC strains. IMP-26 was the most prevalent type (6/12 strains, 50 %), followed by NDM-5 (3/12 strains, 25 %). The results of MLST revealed that these 12 strains could be divided into five sequence types (STs) among which ST544 was the dominant type (6/12 strains, 50 %). The PFGE results divided the 12 strains into four clusters.
Conclusion: Our study indicated that the epidemics of the IMP-26-producing E. cloacae ST544 strain did occur in the intensive care unit (ICU) of a tertiary hospital. Therefore, early surveillance and strict implementation of control measures are crucial for the prevention of nosocomial infections and transmissions in hospitals.
Keywords: Enterobacter cloacae, carbapenemase, IMP-26, ST544
Introduction
Enterobacter cloacae, which belongs to the genus Enterobacter of the family Enterobacteriaceae is widely distributed in nature and is part of the normal microbiota of warm blooded animals. However, it is also a conditional pathogen that has become one of the major nosocomial pathogens in recent years.1 E. cloacae is capable of causing infections in various organs and systems, such as the respiratory tract, urinary tract, skin and soft tissues, and blood.
E. cloacae often exhibits resistance to various antibiotics. Its mechanism of resistance is primarily conferred by the production of extended-spectrum β-lactamases (ESBLs) and AmpC β-lactamases.1 Carbapenems are a class of antibiotics possessing the broadest spectrum of activity to date with an extremely strong antibacterial effect. They are ideal antibiotics for the treatment of severe nosocomial infections caused by ESBL and/or AmpC-producing Enterobacteriaceae. Carbapenems were once considered the last-resort antibiotics for infections caused by multidrug-resistant Gram-negative bacteria. However, in recent years we have witnessed the worldwide emergence of the carbapenem-resistant E. cloacae (CREC) as a result of antibiotic selective pressure following the extensive use of carbapenem antibiotics. The emergence of CREC has brought great challenges to the clinical treatment of infections.2
The production of carbapenemases represents one of the main antibiotic resistance mechanisms in CREC. Carbapenemases are members of three (A, B, and D) out of the four molecular classes of β-lactamases. Class A carbapenemases mainly include NMC/IMI, SME, KPC, and GES. Class B carbapenemases include VIM, IMP, GIM, SPM, SIM, AIM, DIM, and NDM, while Class D carbapenemases includes OXA-48.3–5
IMP carbapenemases were first discovered in Pseudomonas aeruginosa,6 while Serratia marcescens harboring the IMP genes was first reported in Japan in 1991.7 Subsequently, IMP-producing Enterobacteriaceae were primarily detected in sporadic and epidemic cases in Japan, Taiwan, and Australia.8 IMP-positive bacteria include Klebsiella pneumoniae, S. marcescens, Escherichia coli, E. cloacae, and other Enterobacteriaceae. IMP carbapenemases are prevalent in E. cloacae include IMP-1, IMP-4, and IMP-8,3 while the IMP-26 carbapenemase is rarely found in E. cloacae. The main aim of this study was to understand the molecular characteristics of the carbapenem-resistant E. cloacae in a tertiary hospital.
Materials and Methods
Specimen Source
From November 2016 to July 2019, all CREC (resistant to imipenem or meropenem) were collected from Yanbian University Hospital. All strains were non-duplicate (only the carbapenem-resistant E.cloacae strains isolated at the first instance were retained for the same patient).
Strain Identification and Antimicrobial Susceptibility Testing
Microbial identification and antimicrobial susceptibility testing (AST) were performed using the BD Phoenix-100 automated ID/AST system (Becton, Dickinson and Co., USA). E. coli ATCC25922 was used as the quality control strain.
Phenotypic Screening for Carbapenemases and the Detection of Antibiotic Resistance Genes
The phenotypic screening for carbapenemase genes in CREC was carried out according to the modified carbapenem inactivation method (mCIM) provided in the Clinical and Laboratory Standards Institute (CLSI) guideline (2017). Carbapenemase genes (blaNDM, blaKPC, blaIMP, blaVIM, and blaOXA48like) and other β-lactamase genes (blaCTX-M, blaACT, blaDHA, and blaCMY) were detected using the PCR assay,9,10 and the resulting PCR products were subjected to DNA sequencing (Beijing Tsingke Biotechnology Co., Ltd, Chian). Nucleotide sequences were compared by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Multi-Locus Sequence Typing (MLST) and Pulsed-Field Gel Electrophoresis (PFGE)
MLST was performed according to a previously described method (https://pubmlst.org/ecloacae/). The Sequence Type Analysis and Recombinational Tests2 (START2) (http://pubmlst.org/software/analysis/start2/) software was used to generate the phylogenetic tree.11 E. cloacae strains were characterized by PFGE according to the previously described by Cui et al.12 Salmonella enterica serotype H9812 was used as a marker, and the PFGE was established using the XbaI digestion. The agarose gel electrophoresis was performed for 19 h at 14°C, with switch times from 2.2 s to 54.2 s at 6 V/cm on a Bio-Rad CHEF Mapper Pulsed Field Electrophoresis System. Comparison of the PFGE patterns was performed in BioNumerics 7.6 using the Dice Similarity coefficient.
Statistical Analyses
All analyses were performed using the WHONET software (version 5.6) and the SPSS software (version 25.0).
Results
Isolation and AST of Bacterial Strains
A total of 12 non-duplicate strains of CREC were isolated from the sputum (5/12 strains, 41.7%), blood (4/12 strains, 33.3%), or wound (3/12 strains, 25.0%) of patients. The majority of these strains were isolated from patients admitted to the Intensive Care Unit (ICU) (8/12 strains, 66.7%), followed by the department of pain management (2/12 strains, 16.7%), the department of orthopaedics (1/12 strains, 8.3%), and the department of general surgery (1/12 strains, 8.3%). 10 patients were male (83.3.%), and the median age was 66.50 (60.25–71.00) years (Table 1). The AST results showed that all strains were resistant to imipenem, meropenem, and cefepime, but were susceptible to amikacin and colistin. Besides, the IMP-26-producing strains were susceptible to aztreonam, amikacin, and colistin (Table 2).
 |
Table 1 Clinical Characteristics of Carbapenem-Resistant Enterobacter cloacae Strains |
 |
Table 2 Molecular Characteristics and Antimicrobial Susceptibilities of Carbapenem-Resistant Enterobacter cloacae Strains |
Phenotypic Screening and Genotyping of Antibiotic Resistant Enzymes
All strains yielded positive mCIM results. Four types of carbapenemases, IMP-26 (6 strains, 50.0%), NDM-5 (3 strains, 25.0%), NDM-1 (2 strains, 16.7%), and KPC-2 (one strain, 8.3%), were detected in these strains. Moreover, our analysis also identified two CTX-M-3-producing strains and one DHA1-producing strain (Table 2).
MLST and PFGE
MLST divided the strains into five STs, ST544 (6 strains), ST114 (2 strains), ST78 (2 strains), ST171 (one strain), and ST97 (one strain) (Fig 1). The PFGE results showed that strains 3, 4, 5, 7, 8, and 11 (6 strains) shared 93.9% homology; strains 1, 9, and 12 shared 82.4% homology; strains 2 and 6 shared 80% homology. However, strain 10 lacked homology with the other strains (Figure 2).
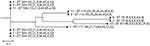 |
Figure 1 Multi-locus sequence typing (MLST) phylogenetic tree of the 12 carbapenem-resistant E. cloacae strains. |
 |
Figure 2 Dendrogram of patterns for carbapenem-resistant Enterobacter cloacae isolates obtained by PFGE. |
Discussion
Carbapenem-resistant E. cloacae strains that produce carbapenemases such as the OXA-48, KPC-3, VIM-1, NDM-1, and IMP-4, have been reported in numerous countries.13–16 It has been found that NDM-1-producing strains predominate in China.17 In this study, four types of carbapenemases were identified, the IMP-26 (6 strains, 50.0%), NDM-5 (3 strains, 25.0%), NDM-1 (2 strains, 16.7%), and KPC-2 (one strain, 8.3%), while the VIM and OXA-48 carbapenemases were not detected. In addition, there was no strain harboring more than one type of carbapenemases. Our results also identified the metallo-β-lactamases, mainly the IMP-26 carbapenemase, as the most prevalent type of carbapenemases, followed by the NDM-5 carbapenemase. This differs from the findings reported in some domestic studies.17
IMP carbapenemases are metallo-β-lactamases that can hydrolyze all β-lactam antibiotics with the exception of aztreonam. In 2010, IMP-26, which is an IMP-4 variant, was first reported by Koh et al in a clinical carbapenem-resistant isolate of P. aeruginosa in Singapore.18 Since then, there were only sporadic reports of IMP-26-producing Gram-negative bacilli, especially of bacilli belonging in the family of Enterobacteriaceae. It has been suggested that the IMP-26-expressing strains have a significantly greater resistance to meropenem than the IMP-1-expressing strains.19 In China, the IMP-8 and IMP-4 carbapenemases are the most frequently detected IMP subtypes in E. cloacae,12,17,20 while the IMP-26-producing E. cloacae isolates have been sporadically reported in Shanghai, Chongqing, and Ningxia.21–24 In this study, all IMP-26-producing strains displayed 100% susceptibility to aztreonam, amikacin, and colistin, and showed 100% resistance to meropenem, imipenem, cefepime, ceftazidime, cefotaxime, ciprofloxacin, levofloxacin, and tetracyclines. In addition, 83.3% of the IMP-26-producing strains were resistant to gentamicin, and 83.3% were susceptible to piperacillin/tazobactam. Hence, piperacillin/tazobactam, amikacin, aztreonam, and colistin, depending on the patient’s condition, can be selected as antibiotics for the treatment against IMP-26-producing E. cloacae.
The MLST results showed that there were five STs; the ST544 was the dominant ST that accounted for 50% of the strains, while the remaining strains were assigned with ST114 (2 strains), ST78 (2 strains), ST171 (one strain), and ST97 (one strain). Moreover, the MLST data revealed a polymorphism between those strains, among which, ST544 was the dominant ST. The rarely reported E. cloacae ST544 was first discovered by the Taiwan scholars in 2016 and does not produce carbapenemases. Besides, the meropenem-susceptible, DHA1-producing E. cloacae ST544 has been previously found in animal specimens.25 In this study, all E. cloacae ST544 strains were found to be IMP-26-producing strains that did not produce CTX-M, DHA, ACT, and CMY β-lactamases. There were five strains isolated from patients admitted to the ICU and one strain isolated from patients admitted to the department of general surgery, who were once also admitted to the ICU, suggesting that there were small outbreaks of the ST544-IMP-26 strain in the ICU. The results of cluster analysis using the START2 software shows that the ST544 strain is closer to ST114 and ST171 strains. Our study identified one NDM-1-producing ST171 strain and two ST78 strains, one of which produced the NDM-1 carbapenemase, and the CTX-M-3 and DHA1 β-lactamases, while the other strain produced the KPC-2 carbapenemase and the CTX-M-3 β-lactamase. The ST171 strain, which primarily produces KPC carbapenemases, is a major epidemic strain in the United States.26 There have also been sporadic reports of the ST171 strain in China.27 The clonal expansion of ST171 across the United States and its subsequent local transmission suggested that this high-risk clone requires increased attention. Therefore, there is a need for enhanced surveillance to prevent the spread of high-risk clones. ST78 was first identified by the Japanese scholar Tohru Miyoshi-Akiyama in 2013. The ST78 clone has been shown to produce various β-lactamases. Previous population analyses on the Multi-drug resistance E. cloacae have demonstrated that ST78 is a widespread and globally dominant ESBL-producing clone, indicating that it is a very common clone associated with nosocomial infections with a unique ability to accept plasmids harboring antibiotic resistance genes.28 Previous studies have revealed that the NDM-1-producing E. cloacae ST78 has a higher epidemic potential and is more likely to cause severe antibiotic resistance outbreaks.24 The NDM-1-producing E. cloacae ST78 deserves clinical attention as it has been previously reported in China24,27 and has also been detected in this study. It has been demonstrated that the widespread of CREC is attributable to its higher tendency to acquire and spread multidrug resistance determinants instead of increased virulence,28 as well as its adaptability to the hospital environments. Both the NDM-5-producing ST114 strains in this study were isolated from the wound exudate of patients. Besides, our study has also identified an ST97 strain that produces NDM-5. The E. cloacae ST114 strain has been detected in France, the United States, and China,24,29,30 while sporadic cases of the E. cloacae ST97 strain have also been reported in China.27,31
The PFGE results showed that the 12 strains could be divided into four clusters. There were six strains (strains 3, 4, 5, 7, 8, and 11) that shared 93.9% homology, all of which were IMP-26-producing strains that belonged to ST544 (MLST). Strains 1, 9, and 12 shared 82.4% homology and belonged to ST114 and ST171. Strains 2 and 6 shared 80% homology and belonged to ST78. Finally, strain 10 lacked homology with the other strains. Taken together, our analysis revealed that both sporadic cases and small outbreaks did occur in the hospital.
The occurrence of carbapenem-resistant Enterobacteriaceae is related to many factors, including ICU admission, prior antimicrobial exposure, and invasive treatment.32 Research has shown that exposure to third or fourth-generation cephalosporins and carbapenems is an independent risk factor for nosocomial infection with carbapenem-resistant Enterobacteriaceae.33 Moreover, if the drug resistance gene is located on the plasmid, it can easily cause horizontal transmission of the drug resistance gene, resulting in disseminated infection in the same ward or department. In this study, small-scale prevalence of the strain IMP-26-producing E. cloacae ST544 was found. Analysis of clinical data showed that all 6 patients were only admitted or had once been admitted to the ICU. Endotracheal intubation and mechanical ventilation were used before the isolation of the strains. Owing to the severity of the infection symptoms, 5 patients were administered meropenem once to control the infection. A study by Wang et al shows that the IMP-26 gene exists on the CREC plasmid and it can be easily spread.21 Its prevalence in the hospital might be related to the above-mentioned factors. After the carbapenem-resistant Enterobacter cloacae was isolated in the hospital, several measures such as isolation of patients, enforcement of the hand hygiene practice, and nosocomial infection monitoring were put in place. Since the implementation of these measures, no other large-scale epidemic event has occurred in any department or in the hospital.
Conclusion
Our study provided evidence that IMP-26-producing E. cloacae ST544 is a major epidemic strain in a tertiary hospital in China. Early detection and surveillance can prevent the spread of the bacteria.
Ethics Statement
This study was approved by the research ethics board at Yanbian University Hospital.
Acknowledgments
The authors specially thank professor Hui Wang for her guidance and support.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wang S, Xiao SZ, Gu FF, et al. Antimicrobial susceptibility and molecular epidemiology of clinical Enterobacter cloacae bloodstream isolates in Shanghai, China. PLoS One. 2017;12(12):e0189713. doi:10.1371/journal.pone.0189713
2. Liu C, Qin S, Xu H, et al. New Delhi Metallo-beta-Lactamase 1(NDM-1), the dominant carbapenemase detected in carbapenem-resistant Enterobacter cloacae from Henan Province, China. PLoS One. 2015;10(8):e0135044. doi:10.1371/journal.pone.0135044
3. Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi:10.1128/CMR.00001-07
4. Bush K. Carbapenemases: partners in crime. J Glob Antimicrob Resist. 2013;1(1):7–16. doi:10.1016/j.jgar.2013.01.005
5. Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25(4):682–707. doi:10.1128/CMR.05035-11
6. Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35(1):147–151. doi:10.1128/AAC.35.1.147
7. Osano E, Arakawa Y, Wacharotayankun R, et al. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994;38(1):71–78. doi:10.1128/AAC.38.1.71
8. Matsumura Y, Peirano G, Motyl MR, et al. Global molecular epidemiology of IMP-producing enterobacteriaceae. Antimicrob Agents Chemother. 2017;61:4. doi:10.1128/AAC.02729-16
9. Lewis JS
10. Yang Q, Wang H, Sun H, Chen H, Xu Y, Chen M. Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: results from large hospital-based surveillance studies in China. Antimicrob Agents Chemother. 2010;54(1):573–577. doi:10.1128/AAC.01099-09
11. Jolley KA, Feil EJ, Maiden M-SC. Sequence type analysis and recombinational tests (START)\n. Bioinformatics. 2001;12:12.
12. Cui L, Zhao J, Lu J. Molecular characteristics of extended spectrum beta-lactamase and carbapenemase genes carried by carbapenem-resistant Enterobacter cloacae in a Chinese university hospital. Turk J Med Sci. 2015;45(6):1321–1328. doi:10.3906/sag-1407-62
13. Fernández J, Montero I, Martínez Ó, et al. Dissemination of multiresistant Enterobacter cloacae isolates producing OXA-48 and CTX-M-15 in a Spanish hospital. Int J Antimicrob Agents. 2015;46(4):469–474. doi:10.1016/j.ijantimicag.2015.07.003
14. Kiedrowski LM, Guerrero DM, Perez F, et al. Carbapenem-resistant enterobacter cloacae isolates producing KPC-3, North Dakota, USA. Emerg Infect Dis. 2014;20(9):1583–1585. doi:10.3201/eid2009.140344
15. Sidjabat HE, Townell N, Nimmo GR, et al. Dominance of IMP-4-producing enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob Agents Chemother. 2015;59(7):4059–4066. doi:10.1128/AAC.04378-14
16. Villa J, Viedma E, Branas P, Orellana MA, Otero JR, Chaves F. Multiclonal spread of VIM-1-producing Enterobacter cloacae isolates associated with In624 and In488 integrons located in an IncHI2 plasmid. Int J Antimicrob Agents. 2014;43(5):451–455. doi:10.1016/j.ijantimicag.2014.02.006
17. Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistant enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012-2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi:10.1093/cid/ciy660
18. Koh TH, Khoo CT, Tan TT, et al. Multilocus sequence types of carbapenem-resistant Pseudomonas aeruginosa in Singapore carrying metallo-beta-lactamase genes, including the novel bla(IMP-26) gene. J Clin Microbiol. 2010;48(7):2563–2564. doi:10.1128/JCM.01905-09
19. Tada T, Nhung PH, Miyoshi-Akiyama T, et al. Multidrug-resistant sequence type 235 pseudomonas aeruginosa clinical isolates producing IMP-26 with increased carbapenem-hydrolyzing activities in Vietnam. Antimicrob Agents Chemother. 2016;60(11):6853–6858. doi:10.1128/AAC.01177-16
20. Zhao Q, Jia X, Pang F, Li Y. Study on genotype and clinical characteristics of infection of carbapenemase-producing Enterobacter cloacae. Med J China. 2015;95(40):3264–3268.
21. Wang S, Zhou K, Xiao S, et al. A multidrug resistance plasmid pIMP26, carrying blaIMP-26, fosA5, blaDHA-1, and qnrB4 in Enterobacter cloacae. Sci Rep. 2019;9(1):10212. doi:10.1038/s41598-019-46777-6
22. Xia Y, Liang Z, Su X, Xiong Y. Characterization of carbapenemase genes in Enterobacteriaceae species exhibiting decreased susceptibility to carbapenems in a university hospital in Chongqing, China. Ann Lab Med. 2012;32(4):270–275. doi:10.3343/alm.2012.32.4.270
23. Dai W, Sun S, Yang P, Huang S, Zhang X, Zhang L. Characterization of carbapenemases, extended spectrum beta-lactamases and molecular epidemiology of carbapenem-non-susceptible Enterobacter cloacae in a Chinese hospital in Chongqing. Infect Genet Evol. 2013;14:1–7. doi:10.1016/j.meegid.2012.10.010
24. Cai Y, Chen C, Zhao M, et al. High prevalence of metallo-beta-lactamase-producing Enterobacter cloacae from three Tertiary Hospitals in China. Front Microbiol. 2019;10:1610. doi:10.3389/fmicb.2019.01610
25. Harada K, Shimizu T, Mukai Y, et al. Phenotypic and molecular characterization of antimicrobial resistance in Enterobacter spp. isolates from companion animals in Japan. PLoS One. 2017;12(3):e0174178. doi:10.1371/journal.pone.0174178
26. Gomez-Simmonds A, Hu Y, Sullivan SB, Wang Z, Whittier S, Uhlemann AC. Evidence from a New York City hospital of rising incidence of genetically diverse carbapenem-resistant Enterobacter cloacae and dominance of ST171, 2007-14. J Antimicrob Chemother. 2016;71(8):2351–2353. doi:10.1093/jac/dkw132
27. Jin C, Zhang J, Wang Q, et al. Molecular characterization of carbapenem-resistant Enterobacter cloacae in 11 Chinese cities. Front Microbiol. 2018;9:1597. doi:10.3389/fmicb.2018.01597
28. Gomez-Simmonds A, Annavajhala MK, Wang Z, et al. Genomic and geographic context for the evolution of high-risk carbapenem-resistant enterobacter cloacae complex clones ST171 and ST78. mBio. 2018;9:3. doi:10.1128/mBio.00542-18
29. Izdebski R, Baraniak A, Herda M, et al. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother. 2015;70(1):48–56. doi:10.1093/jac/dku359
30. Kanamori H, Parobek CM, Juliano JJ, et al. A prolonged outbreak of KPC-3-producing enterobacter cloacae and klebsiella pneumoniae driven by multiple mechanisms of resistance transmission at a large academic burn center. Antimicrob Agents Chemother. 2017;61:2. doi:10.1128/AAC.01516-16
31. Miao M, Wen H, Xu P, et al. Genetic diversity of Carbapenem-Resistant Enterobacteriaceae (CRE) clinical isolates from a Tertiary Hospital in Eastern China. Front Microbiol. 2018;9:3341. doi:10.3389/fmicb.2018.03341
32. Li S, Guo FZ, Zhao XJ, et al. Impact of individualized active surveillance of carbapenem-resistant enterobacteriaceae on the infection rate in intensive care units: a 3-year retrospective study in a teaching hospital of People’s Republic of China. Infect Drug Resist. 2019;12:1407–1414. doi:10.2147/IDR.S201644
33. Wang Q, Zhang Y, Yao X, et al. Risk factors and clinical outcomes for carbapenem-resistant Enterobacteriaceae nosocomial infections. Eur J Clin Microbiol Infect Dis. 2016;35(10):1679–1689. doi:10.1007/s10096-016-2710-0
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
