Back to Journals » Infection and Drug Resistance » Volume 13
Molecular Basis for Pathogenicity of Human Coronaviruses
Authors Pourrajab F, Zare-Khormizi MR, Sheikhha MH
Received 30 March 2020
Accepted for publication 24 June 2020
Published 17 July 2020 Volume 2020:13 Pages 2385—2405
DOI https://doi.org/10.2147/IDR.S255156
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Fatemeh Pourrajab,1,2 Mohamad Reza Zare-Khormizi,3 Mohammad Hasan Sheikhha4
1Nutrition and Food Security Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran; 2Department of Biochemistry and Molecular Biology, School of Medicine, Shahid Sadoughi University of Medical Sciences, Yazd, Iran; 3School of Medicine, Shahid Sadoughi University of Medical Sciences, Yazd, Iran; 4Biotechnology Research Center, International Campus, Shahid Sadoughi University of Medical Sciences, Yazd, Iran
Correspondence: Fatemeh Pourrajab
Nutrition and Food Security Research Centre, Shahid Sadoughi University of Medical Sciences, Yazd, Iran
Email [email protected]
Mohammad Hasan Sheikhha
Biotechnology Research Center, International Campus, Shahid Sadoughi University of Medical Sciences, Yazd, Iran
Tel +983538247085
Fax +983538247087
Email [email protected]
Abstract: Over the past years, several zoonotic viruses have crossed the species barrier into humans and have been causing outbreaks of severe, and often fatal, respiratory illness. The 21st century has seen the worldwide spread of three recognized coronaviruses (CoVs) which can cause pneumonia and severe acute respiratory symptoms (SARSs), SARS, MERS, and recently SARS-CoV-2. Herein, it is raising concerns about the dissemination of another new and highly lethal pandemic outbreak. Preparing for a pandemic outbreak involves a great deal of awareness necessary to stop initial outbreaks, through recognizing the molecular mechanisms underlying virus transmission and pathogenicity. CoV spike protein S is the key determinant of host tropism and viral pathogenicity which can undergo variations and makes the CoV a highly pathogenic and diffusible virus capable of sustained human-to-human transmission and spread easily. The three mentioned CoVs exhibit some similarities in S protein whereby constitute a promising target for the development of prophylactics and therapeutics in the future.
Keywords: coronavirus, transmission, receptor binding, spike protein
Coronaviruses
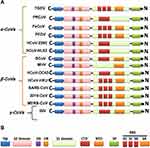
Coronaviruses (CoVs) are zoonotic viruses that enter either directly from wildlife reservoirs or indirectly via an intermediate domestic animal host. They have the largest known single-stranded RNA genome (~30 kb) and are categorized in three groups, based on phylogenetic analyses and surface antigenic characteristic: (a) alpha-CoVs, responsible for gastrointestinal disorders in human, dogs, pigs, and cats; (b) beta-CoVs, including the human severe acute respiratory syndrome virus (SRAS-CoV), the Middle Eastern respiratory syndrome (MERS-CoV), and recently the novel Cov 2019 (2019-nCoV/SARS-CoV-2) virus; (c) gamma-CoVs, which infect avian species (Table 1 and Figure 1).1–3
 |
Table 1 Coronavirus Genera, Species and Their Binding Receptors in Hosts |
Viral RNA produces both genomic and sub-genomic RNAs serving as mRNAs for the structural and accessory genes which reside downstream of the replicase polyproteins. The organization of the CoV genome is 5′-leader-UTR-replicase-S (Spike)–E (Envelope)-M (Membrane)-N (Nucleocapsid)-3′UTR-poly (A) tail with accessory genes interspersed within the structural genes at the 3′ end of the genome. The replicase gene encodes two large ORFs, rep1a and rep1b, which express two co-terminal polyproteins, pp1a and pp1ab. There are non-structural proteins (nsp) that assemble into the replicase-transcriptase complex (RTC) which is responsible for RNA replication and transcription of the sub-genomic RNAs. The ns-proteins contain certain enzymatic functions, for example, nsp12 encodes the RNA-dependent RNA polymerase (RdRp) domain; nsp13 encodes the RNA helicase domain and RNA 5′-triphosphatase activity; nsp14 encodes the exoribonuclease (ExoN) which is involved in virus replication and playing a role in genome recombination and viral mutation.3–5
Several facts indicate another animal acting as an intermediate host or mixing vessel between an animal reservoir and humans. They can infect humans and cause disease to varying degrees, from upper respiratory tract infections (URTIs) resembling the common cold, to lower respiratory tract infections (LRTIs) such as bronchitis, pneumonia, and even severe acute respiratory syndrome (SARS). They most often affect the respiratory or intestinal tract, but some representatives may also infect other tissues or organs.1–3
In late December 2019, an outbreak of pneumonia with an unknown etiology appeared in Wuhan, China, that shortly thereafter became pandemic and serious for human health.4,5 Noteworthy, the World Health Organization (WHO) had previously, in April of 2018, predicted it in a priority list of pathogens which including MERS and SARS-CoV, and notified it as a new pathogen which causing “Disease X”. Based on phylogeny, taxonomy, and sequence analysis, this novel pathogen was named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and forming a sister clade to the prototype human and bat-related SARS-CoVs. The novel SARS-CoV-2 exhibit about 35%, 79.5, and 96% sequence identity to MERS, SARS-CoV, and a bat coronavirus, SL-CoV-RaTG13, respectively (Figure 2A).3–6 Such similarity between SARS-CoV-2 and the bat-related SARS-CoV genome (>95% sequence identity) argues for a recombinant virus transmitted from bats to human hosts by the mean of an intermediate host. Herein, the novel pathogen, SARS-CoV-2, has been categorized into the BetaCoronaVirus family (group B), the 7th recognized as a human pathogen, and the 3rd causing a severe clinical syndrome after SARS and MERS-CoV.3–5
 |
Figure 2 Continued. |
 |
Figure 2 Continued. |
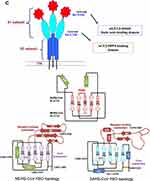 |
Figure 2 Continued. |
Despite the name and some similarities, there are differences between MERS, SARS, and SARS-CoV-2 in genetic, receptor recognition, and clinical features (Figures 1 and 2).6,7
Compared to the human representative SARS and MERS-CoVs, SARS-CoV-2 shows superior pathogenicity and inferior clinical outcomes that are addressed to the receptor recognition by the spike protein S and plasma membrane fusion capacity.4–6 SARS-CoV-2 is a high diffusible pathogen, spread by droplets, direct contact, and contact with infected objects. However, the mortality rate of the infected individuals with the complete severe respiratory syndrome disease (COVID-19) is estimated to about 0.2% in young healthy individuals wherein growing to the highest rate in people older than 80 years and with pre-existing heart disease, while SARS and MERS-CoV mortality rate reach 10% and 35%, respectively.3,5 As posing a serious threat to global public health, therefore, it is important to know the molecular mechanism of CoV pathogenicity and the differences exist to promptly develop specific anti-CoV therapeutics and prophylactics for treatment and prevention of MERS, SARS-CoV and recently SARS-CoV-2 disease, COVID-19.
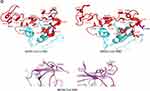
The envelope spike protein S is the key determinant of virus-host specificity, tropism, and infectivity. The graphical and high-resolution structures of the MERS, SARS, and SARS-CoV-2 S proteins partly resemble each other, except the receptor-binding domains (RBDs) and receptor binding motifs (RBMs). The RBD in the spike protein is the most variable part of the coronavirus genome. The overall structure of the SARS-CoV-2 RBD is supposed to be similar to that of the SARS-CoV RBD whose in turn shares some similarities with MERS-CoV RBD (Figure 2B–D).2,5,6,8 The overall receptor-binding mode of the SARS and SARS-CoV-2 RBD is nearly identical that can utilize ACE2 as the cell receptor. Some residues have been identified in the SARS and SARS-CoV-2 RBDs which are critical for ACE2 binding, and are either conserved or shared similar side chain properties (Figure 2D).2,4,5 According to some similarities in the structure and sequence, the literature argues for a convergent evolution between SARS and SARS-CoV-2. However, there are differences in the receptor-binding regions of the SARS-CoV-2 S protein. Additionally, the structure of SARS-CoV-2 S protein is longer than SARS-CoV S protein which may be addressed to more transmissibility and pathogenicity of SARS-CoV-2 (Figure 2B and D).2,4,5 There are reports that the core subdomains of RBDs in MERS and SARS-CoV share a high degree of structural similarities, whereas the receptor binding subdomains are notably divergent (Figure 2C and D). There have been identified several key residues in the receptor-binding subdomains that are critical for viral binding to DPP4 and ACE2 and entry into the target cell (Figure 2D) 6,8
Herein, the heavily N-linked glycosylated spike protein S (~150 kDa) is involved in virus transmission and infectivity. It makes up homotrimers and a distinctive spike structure on the surface of CoVs. The CoV S proteins underwent cleavage at two sites; S1/S2 and S2ʹ. The cleavage at the S2ʹ site releases the fusion peptide to enable the conformational changes of the spike and facilitate the viral and host membrane coalescence. The cleavage at S1/S2 site is dispensable and could be performed before viral particle budding/release by furin protease (as in MERS) in producing cells or after release by a host protease, while the S2ʹ site is pivotal to host entry and is mostly cleaved by host proteases such as TMPRSSII (as in SARS-CoV-2) at the cell surface or Cathepsin in endosome.8,9 The N-terminal S1 subunit constitutes the globular region and makes up the large receptor-binding subunit of the S protein while the stalk part is made up of the membrane-proximal S2 subunit (Figure 2B). The N-terminal S1 and C-terminal S2 of the S protein play a similar role in all CoVs, the S1 region is related to receptor binding, and the S2 domain plays a role in the membrane fusion process (Figure 2C and D).2,6,8 The amino acid sequences of S1 diverge across different genera but are relatively conserved within each genus. S1 contains two independent domains, an N-terminal domain (S1-NTD) and a C-terminal domain (S1-CTD) (Figure 2C). Either or both of these S1 domains can function as a receptor-binding domain (RBD) (Figure 1 and Table 1). Specific interactions between RBD and host receptors are the key determinants of tissue tropism, host range and cross-species infection.2,6,10,11
The S1 domains are important determinants for the host range and tissue tropism of CoVs. S1 initiates infection by binding to host cell surface molecules, either proteinaceous, sialoglycan based, or both (Figure 1 and Table 1). The S1 subunit comprises four β-rich sub-domains, designated A, B, C, and D, where sub-domain A and B acting specifically as receptor-binding domains in different CoVs. However, the transmembrane C-terminal S2 subunit is the metastable spring-loaded fusion machinery (Table 1).2,6,12 The S1A sub-domain engages host sialic acids/sugar ligands and the S1B sub-domain recognizes host membrane-bound proteins. This bivalent-binding interaction enables broad zoonotic CoV infection and host-restricted infectivity.4,6,13
Most of CoVs utilize peptidases as their membrane-bond protein receptors. It is not yet clear why peptidases are used, as entry occurs even in the absence of the enzymatic domain of these receptors. Many α-CoVs utilize aminopeptidase N (APN) as their receptor, SARS-CoV/HCoV-NL63 and the recently identified SARS-CoV-2 binds to angiotensin-converting enzyme 2 (ACE2) to gain entry into the human cells, MERS-CoV, as well as, SARS-CoV-2 (which was recently identified) use dipeptidyl-peptidase 4 (DPP4) as their receptors, and MHV enters through CEACAM1 (Table 1).8,11
Here, the high mortality rate of certain CoVs, along with their ease of transmission in humans underpins the need for more research into CoV molecular biology which can aid in the production of effective anti-CoV agents for both humans CoVs and enzootic CoVs infections.
Molecular Pathogenicity of SARS-CoV
SARS-CoV as the emerging infectious viral disease of the 2003 outbreak, is characterized by severe clinical manifestations of the lower respiratory tract where resulting in diffuse alveolar damages. SARS-CoV infection damages lung tissues owing to elevated levels of production and activation of proinflammatory chemokines and cytokines, resulting in atypical pneumonia with rapid respiratory deterioration and failure.14,15 Like MERS-CoV, SARS infects primarily type II pneumocytes in alveoli that lead to the release of many cytokines and chemokines produced by these cell types and their elevation in the serum of SARS-CoV infected patients. Virus infection and release of cytokines in the alveolar compartment with its proximity to the pulmonary capillary bed disrupt the barrier and allow the systemic spread of the virus to distant organs, especially in the context of inflammation and alveolar-capillary leak. Many cytokines and chemokines elevated in the serum of SARS-CoV infected patients, were produced by these cell types.13,16,17
Accordingly, samples isolated from patients’ respiratory tissues infected with SARS-CoV exhibited diffuse alveolar damages including desquamated epithelial cells, type II cell hyperplasia, fibrin, and collagen deposits in the alveolar space, increased mononuclear infiltrates in the interstitium, and, in some cases, the presence of multinucleated syncytial cells. Such damages reflect the combined effects of primary infection, host immune responses, and therapeutic interventions.16 In the case of epidemic and 2003, 2012 and 2019 outbreaks of SARS, MERS and novel CoV 2019, several findings point to the spread and potential binding of infectious CoVs such as MERS-CoV through the mucosa in the throat, respiratory tract and the eyes.13,18,19 SARS, MERS, and SARS-CoV-2 all cause severe pneumonia, sharing similarities in their pathogenesis. SARS-CoV and SARS-CoV-2 both spread through respiratory secretions, such as droplets, via direct person-to-person contact.1,7,14,20 Autopsies from SARS-CoV-infects patients showing severe disease and secondary complications, including respiratory failure, indicated the presence of a virus in both proximal and distal pulmonary epithelia. The epithelium mucosa of the conducting airways, the major site of respiratory droplet deposition, supports the binding, entry, and replication of SARS-CoV. Furthermore, the virus entry and spread is preferentially through the apical surface of well-differentiated cells. This apical exit pathway of virions would favor the efficacious entry and spread of infection along the respiratory tract.15,17
SARS-CoV primarily infects epithelial cells, pneumocytes, and enterocytes in the respiratory system within the lung, while being capable of entering macrophages and dendritic cells but only leads to an abortive infection.17 SARS-CoV can also infect mucosal cells of intestines, tubular epithelial cells of kidneys, epithelial cells of renal tubules, cerebral neurons, and immune cells. Infectious viral particles in patients with SARS can be excreted through respiratory secretions, stool, urine, and sweat.14,15
The productive infection of conducting airway epithelia and the apically release of SARS-CoVs may lead to virus removal by mucociliary clearance where SARS-CoVs gains access to the gastrointestinal tract. Here, diarrhea is a clinical sign commonly observed in SARS-infected patients where SARS-CoV has gained the gastrointestinal tract and infected the cells.17 Upon exposure of the host to the virus, the virus binds to cells expressing the virus receptors; ACE2 as the main receptor, and CD209L as an alternative receptor with a much lower affinity.14,15
Origin and Evolution of SARS-CoV
Evidence suggests that human SARS-CoVs may have transmitted from bats to human hosts where civets were found to be the intermediate host. Civet SARS-CoVs was found more similar to bat SARS-related CoVs (bat-SR-CoVs) than any other virus identified to date. The genome sequences of human SARS-CoVs were almost identical to the genomes of civet SARS-CoVs. However, two genes in human SARS-CoVs showed major variation with civet SARS-CoVs where the first detected variable region was the accessory gene orf8.9,15,18 On the basis of orf8 variation, the human transmission of SARS-CoV 2002–2003 outbreak was divided into three phases. The early phase was characterized by a limited number of localized cases. The viral genomes from early- phase patients contain two genotypes of orf8, one with a complete orf8 (369 nucleotides) and the other containing an 82-nucleotide deletion. The middle phase was during which a super-spreader event occurred in the hospital. Most of the genomes from middle-phase patients contain a split orf8 (orf8a and orf8b) owing to a 29-nucleotide deletion; with two exceptions containing the early phase deletion in orf8 and the other with the whole orf8 deleted, completely. Finally, the late phase was initiated with international spread, where viral genomes from the late-phase patients were like as most of the middle-phase genomes. The human isolates from 2004 and all civet SARS-CoV genomes have a complete orf8 except one civet strain with an 82-nucleotide deletion. These data indicate that orf8 genes underwent adaptations during transmission from animals to humans during the SARS epidemic. ORF8a protein is not essential for SARS-CoV replication.10
The second major variation between human SARS-CoVs and civet SARS-CoVs was seen the spike protein S. Molecular analysis and structural comparisons of S1-CTD from different SARS-CoV strains and its interactions with ACE2 from different host species have revealed the molecular mechanisms by which SARS-CoV do cross-species transmission and transmit from animals to humans and caused the SARS epidemic.11,14
In SARS-CoV S protein, S1-CTD functions as the RBD and is responsible for binding to ACE2 and entering cells.10 The RBD is composed of amino acids 318–510 where tyrosine-rich residues 424–494 make complete interactions with the ACE2 receptor, and create receptor-binding motif (RBM). In RBM, 14 residues are in direct contact with ACE2 and six of them are tyrosine, since representing both the hydroxyl group and the hydrophobic ring. The RBD region also contains multiple cysteine residues that are linked by disulfide bonds. These disulfide bonds are stabilizing the structure of RBD and important in RBD-ACE2 interaction (Figure 2C).15 Substitution in RBM residue Lys479 to Asn479 showed an important role in inducing the binding affinity of civet SARS-CoV RBD for human ACE2 and the civet-to-human transmission of SARS-CoV.10
Additionally in the ACE2, at the interface of RBD and human ACE2, two virus-binding hot spots: Lys31 (hot spot 31) and Lys353 (hot spot 353) have been identified make favorable interactions with the residues 479 and 487 at the RBD-human ACE2 interface. Interactions at at the RBM with ACE2, provide significant energy to enhance viral binding to human ACE2, and played a critical role in the civet-SARS-CoV transmission to human. Both of these virus-binding hot spots consist of a salt bridge (Lys31 with Glu35 and Lys353 with Asp38) that is buried in a hydrophobic pocket and contribute a substantial amount of energy to RBD–ACE2 binding as well as filling voids at the RBD–ACE2 interface. Notable, all of the naturally selected viral mutations found in SARS-CoV and SARS-CoV-2 RBM surrounded these two hot spots, with a significant impact on the structure of RBM, the ACE2 binding affinity, and the host-immune responses.10,11 One of the naturally selected RBM mutations was K479N, which facilitated the palm civets-SARS-CoV transmission to humans. Another viral naturally selected mutation was S487T, facilitated the human-to-human transmission of SARS-CoV and makes more infectious spread if virus. These two mutations contributed significantly to the SARS epidemic from 2002 to 2003. Interestingly, these two positions at the S1-CTD of bat-related SARS-CoV (corresponding to residues 479 and 487 in human SARS-CoV strains), contain two Asn (N). The interaction between human ACE2 and the first Asn is favorable, while the second one is less favorable. Thus, the bat-related SARS-CoV recognizes human ACE2 but less well than the human SARS-CoV strains do.9,14,15 Generally, three substitutions Arg/Lys/Asn479 have been found in the palm civets SARS-CoV wherein all fit well into the interface between the RBD and civet ACE2, and infect civet cells efficiently. Between them, Lys479 is incompatible with human ACE2, while Arg479 provides the most favorable interaction between civet SARS-CoV RBD for human ACE2 where forming a salt bridge with ACE2 residue Asp38. In sum, strains that contain Asn479/Arg479 substitutions recognize human ACE2 well and preferentially infect human cells, whereas strains that contain Lys479 show less affinity for human ACE2 and infect human cells inefficiently. Hence, Asn479 and Arg479have been defined as viral adaptations to human ACE2, whereas; Lys479 was accounted for as a viral adaptation to civet ACE2.10,15 Mutation in RBM residue 487 was accounted to have an important role in the SARS-CoV human-to-human transmission. The residue The487 recognized human ACE2 efficiently and was transmitted between humans during the 2002–2003 SARS epidemics. The487 makes favorable interaction with the human ACE2-hot spot 353 and provides stacking support for the formation of the salt bridge between Lys353 and Asp38. By contrast, in the other strains isolated from humans and civets, there was Ser487 that cannot provide stacking support to the hot spot 353, and hence recognize human ACE2 inefficiently and cannot transmit between humans.10,11,15
Thereby, two naturally selected mutations at the positions 479 and 487, determined SARS-CoV tropism (host range from civet to human), and epidemic (human-to-human transmission). Any residue changes in these two positions might, therefore, affect cross-species and/or intra-species transmission of SARS-CoVs. In the other word, changes of only a few amino acids in the specific positions of the civet SARS-CoV RBD that are responsible for binding to the peptidase domain of ACE2, can enhance viral affinity for the human receptor and causing the civet SARS-CoV transmission from animals (including civets, mice, and rats) to human.
Most importantly in SARS RBD, two disulfide bonds are linking C323 to C348 and C467 to C474 which stabilizing the RBD structure and preserving its interaction with ACE2 (Figure 2C). One disulfide bond linking two residues C467 and C474 is located in RBM, where directly involved in the interaction between RBD-ACE2 and another linking between residues C323 and C348 is positioned in the core structure of RBD.15
Infectivity and Pathogenicity of SARS-CoV
S1-CTD of both MERS-CoV and SARS-CoV contains a core structure and an RBM which recognizes and binds the host receptor (Figure 2C). In MERS-CoVs, S1-CTD and NTD both participate to recognize host receptors and make a complex with them where SB-SD sub-domains are included in the core structure and RBM (interacting with protein receptor) and the S1-NTD makes S1A sub-domain (interacting with α-2,3/2,6-linked SiA receptors, Table 1). Like as MERS-CoV, the SARS-CoV core structure consists of a five-stranded anti-parallel β-sheet (β1–β4 and β7) connecting with three short α-helices (αA-αC). The core structures of both CoVs are highly similar to each other, but their RBMs are markedly different, leading to different receptor specificities. SARS-CoV has a two-stranded β-sheet (β5 and β6), loop-dominated RBM in RBD, while MERS-CoV RBM mainly consists of a four-stranded β-sheet.
Herein, like as that is known in MERS-CoV, S1-NTD may participate to create S1A sub-domain which in combination with the core and RBM interact and recognize specific receptors in the host (Figure 2C and Table 1).8,11,15,22
SARS-CoV receptor ACE2 is a membrane-associated aminopeptidase converting Ang I to Ang II, expressed in vascular endothelia, renal and cardiovascular tissues, and epithelia of the small intestine and testes, balancing the renin-angiotensin system (RAS).7,17 In the respiratory tract, ACE2 is widely expressed on the epithelial cells of alveoli, trachea, bronchi, bronchial serous glands, and alveolar monocytes and macrophages. Furthermore, as a membrane-bond molecule, ACE2 is providing a variety of susceptible cells to SARS-CoV infection. For example, it is diffusely localized on the endothelia of arteries and veins, the mucosal cells of the intestines, tubular epithelia of the kidneys, epithelial cells of the renal tubules, and cerebral neurons and immune cells.14,15 Further, in diseased conditions, ACE2 can be highly expressed in alveolar cells of the lung, esophagus upper and stratified epithelial cells, absorptive enterocytes from ileum and colon, cholangiocytes, myocardial cells, kidney proximal tubule cells, and bladder urothelial cells.17,23,24 The most abundant expression of ACE2 was observed in well-differentiated primary airway epithelia with the strongest and signal intensity on the apical rather than the basolateral surface (Figure 2). Furthermore, the intensity of ACE2 expression was depended on the state of cellular differentiation. Thereby, SARS-CoVs infect well-differentiated cells from the apical surface and preferentially exits from the apical side wherein easily is spread by respiratory droplets and contact and enters the host through the mucosa of the respiratory tract and the eyes. The epithelium of the conducting airways is the major site of respiratory droplet deposition and supports the replication of SARS-CoV,17 In Acute respiratory distress syndrome, the RAS is crucial in maintaining oxygenation as widespread lung injury would otherwise result in a complete pulmonary shutdown. In patients with pneumonia or lung injury, locally increased Ang II production is triggering for maintaining oxygenation. Ang II induces pulmonary vasoconstriction in response to hypoxia, which is important in preventing shunting in patients with pneumonia or lung injury, where subsequently increasing vascular permeability facilitating pulmonary edema. Thus, ACE2 is also highly expressed in the lung. Pulmonary ACE2 appears here in regulating the balance of circulating Ang II/Ang 1–7 levels. Further, ACE2 plays a crucial role in elderly people by regulating the RAS via opposing the actions of Ang II, because it has a beneficial role in many diseases such as hypertension, diabetes, and cardiovascular disease.10,24
A recent study reported that the Asian donors had variants with much higher ACE2 expression cell ratio than white and African American donors. In this study, data analysis for allele frequency (AF) of expression quantitative trait loci (eQTLs), reported potential functional coding variants in ACE2 among East Asian participants, which may be involved in the epidemic occurrence, spreading and distribution of 2019-nCoV/SARS-CoV-2. According to the report, higher AFs in the eQTL variants may be associated with higher ACE2 expression in tissues (Figure 1C), which may cause different susceptibility or response to 2019-nCoV/SARS-CoV-2. In this study, 6 variants (rs4646127, rs2158082, rs5936011, rs6629110, rs4830983, and rs5936029) of ACE2 were introduced to be expressed higher than 95% in EAS population, whereas the AFs of these variants in European population was reported to be much lower (52–65%).25
Molecular Pathogenicity of MERS-CoV
Three SARS-CoV (2003), MERS-CoV (2012), and SARS-CoV-2 (2019) are CoVs that are known to cause severe pneumonia in humans and share some common features in clinical manifestations and similarities in their genomic organization and glycoprotein S structures (Figure 2). The primary receptor for MERS-CoV is a multifunctional cell surface protein DPP4, also known as the immune-modulator factor CD26. DPP4 is widely expressed on epithelia of the kidney, alveoli, small intestine, liver, and prostate, and on activated leukocytes. Thereby, MERS-CoV can infect cells of several human organs including the lower respiratory tract, kidney, and intestine. MERS-CoV infection appears with various manifestations including clinical symptoms fever, cough, sore throat, myalgia, chest pain, diarrhea, vomiting, and abdominal pain (but not restricted to), as well as, acute, highly lethal pneumonia and renal dysfunction. The highly expressed DPP4 on the kidney cells causes renal dysfunctions by either hypoxic damage or direct infection of the epithelia. In animal models of MERS-CoV lung infection, alveolar edema and infiltration of neutrophils and macrophages is occurred.18,26 Remarkably, MERS-CoV can invade the immune system by infecting the human T and dendritic cells, and macrophages. There may stimulate T-cell apoptosis and evade antiviral T-cell responses. MERS-CoV may dysregulate immune responses through diminishing innate immune responses and delayed proinflammatory cytokines.14,27,28
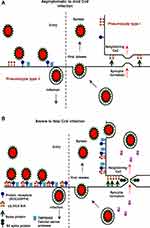
In dromedary camels as the natural host, MERS-CoV spreads more efficiently than in humans. It transmits easily between the animals but causes a mild infection. In dromedaries, MERS-CoV mostly replicate in the nasal epithelium upon infection, while in humans, MERS-CoV mainly replicates in the lower respiratory tract, particularly in the bronchiolar and alveolar epithelia. Inter-species and intra-species differences observed in the infectivity and transmissibility of the virus highlight the role of host receptors in the MERS-CoV pathogenesis (Figure 3). These differences in transmissibility and pathogenicity are attributed to the different tropism of MERS-CoV in these two species. These differences can be found in SARS-CoV and SARS-CoV-2 infection, and can be addressed to the localization and distribution of receptors in the respiratory tract (with some exceptions) or other organs.1,10,13
Origin and Evolution of MERS-CoV
To enhancing the understanding of CoV evolution, cross-species transmission, and host range, should identify the key structures involved in S-receptor interaction and complex formation.9,11 Herein, similar to SARS-CoV infection, the MERS-CoV S protein is cleaved into a receptor-binding subunit S1 and a membrane-fusion subunit S2, during the infection process (Figure 1A). The MERS-CoV and SARS-CoV RBDs differ from each other although they share a high similarity in the structure of the S1-CTD and the core structure.14,15 Despite the high similarity in the core, MERS and SARS-CoV RBMs and S1-NTDs seem to be markedly different, leading to different receptor specificities (Figure 2D). The RBM of MERS-CoV S1-CTD mainly consists of a four-stranded antiparallel β-sheet connecting to the core via loops. Like the VBM for SARS-CoV on ACE2, the VBM for MERS-CoV is also located on the outer surface of DPP4, away from the peptidase active site. The conserved core structure of SARS-CoV and MERS-CoV S1-CTDs suggests a common evolutionary origin and the different RBM of two S1-CTDs indicate a divergent evolutionary pathway that has led to their recognition of different host receptors and transmission.8,9,11 In MERS-CoV, the core structure of RBD (RBM) is stabilized by three disulfide bonds, while in SARS-CoV it was done by just one disulfide bond (Figure 2D).8,14,15 Particularly, the residues 484–567 of RBM take charge of directly interacting with the extracellular β-propeller domain of DPP4, however, the fusion core formation of MERS-CoV resembles that of SARS-CoV.13 Despite differences in the S1-CTDs and S1-NTDs, MERS-CoV and the highly related bat-Cov HKU4 recognize DPP4 in very similar ways and appear a close evolutionary relationship between the two viruses.8,9,11
Infectivity and Pathogenicity of MERS-CoV
By focusing on β-CoV MERS-CoV, the virus particles show dual binding activity and specificity via sialoglycans and proteinaceous molecules (Figures 1 and 3). The receptor binding is facilitated by distinct sub-domains of the S1 subunit, S1A-D. In particular, the S1B sub-domain located in S1-CTD binds DPP4. The peptidase receptor is a functional receptor of MERS-CoV that have a major influence on viral host range and tropism since its tissue localization varies between species.15,26 Despite the interaction with DPP4, MERS-CoV host restriction, and tissue tropism is defined by binding to cell glycotopes sialic acids (SiA) (Figure 1 and Table 1).13,19
MERS-CoV targets the respiratory tract both in humans and in its natural host, the dromedary camel, via the initial binding to α2,3/2,6-linked glycotopes. The nasal epithelium of dromedary camels and type II pneumocytes in human lungs express high levels of α2,3/2,6-linked SiA but not the nasal epithelium of pigs and rabbits. The Sia-binding activity is assigned to the sub-domain S1A with a preference for α2,3-linked over α2,6-linked SiA. The primary distribution of α2,3-linked SiA in the upper and lower respiratory tracts correlates with the predominant sites of MERS-CoV replication in camels and humans, respectively.18,26
In dromedaries, MERS-CoV replicates predominantly in the nasal epithelium upon the animal infections, while in humans, MERS-CoV mainly replicates in the lower respiratory tract, particularly in the bronchiolar and alveolar epithelia. Consistent with this, the distribution of α2,3-linked SiA is mainly in the nasal epithelium and upper respiratory tract, in dromedary camels. In contrast, α2,3-linked SiA is present mainly in the lower respiratory tract, in humans. Thereby, MERS-CoV spread more efficiently in dromedary and transmits easily, in contrast to humans. This difference in transmissibility is addressed to the different tropism of MERS-CoV in these two species. The α2,3-linked SiA-binding of MERS-CoV is specified by tissue localization of these glycotopes which varies between various tissues in susceptible species.13,26,27
Dromedary camels, pigs, and rabbits, all exhibit high levels and localization of DPP4 in their respiratory tract but pigs and rabbits are hardly infected by MERS-CoV or unable to transmit the virus by contact or airborne droplets. Most likely, the reason is that both pigs and rabbits, unlike the dromedary camels, express low levels of α2,3/2,6-linked SiA, in their respiratory tract where they shed low/no levels of the infectious virus upon MERS-CoV inoculation. Samples from bat species not only support these findings but also present insectivorous bats as the original host of MERS-CoV, besides indicating the importance of intestinal tropism and fecal-oral transmission of MERS-like-CoV in these insectivorous bats.13,26
Data revealed that the virus particles of CoVs bind to target cells in humans in the two-step process. In the early phase of attachment, the CoV first adheres to SiA linkages where then a subsequent engagement with protein receptors occurs during infectious cell entry. Accordingly, binding to SiA glycotopes promotes infection and also supports the intercellular expansion of CoV through syncytial development (Figure 3B). Additionally, the subsequent receptor interaction of DPP4 is required for an efficient virus infection. Notable, in absence of Sia linkages MERS-CoV binding and infectivity are hampered.12,18
Herein, viruses may spread/transmit in the absence of the prototype protein receptors whereby adaptive mutations in the S1A sub-domain facilitated the virus attachment and increased the intercellular expansion of CoV.19 During virus natural selection, adaptive mutations in the SiA-binding domains of spike protein increases the intercellular expansion process. In experiments, SiA acted as a host receptor to help CoV entry without the need for protein receptors and sufficiently facilitated the later stages of virus spread through cell-cell membrane fusion, without requiring protein receptors. Overall findings propose a key role for SiA in the early virus-host interaction, viral-directed cell-cell fusion, and easy spread of infection. Accordingly, the lectin-like properties of S1-NTD contribute to facile zoonotic transmission of CoVs and intercellular spread within infected organisms.12,13,19 According to the findings, MERS-CoV infection can involve several human organs including lower respiratory tract, kidney, intestine, and liver where causes acute, highly lethal pneumonia and renal dysfunction with various clinical symptoms.18,26
Differences observed between individuals in the behavior of virus infectivity and pathogenicity are attributed to the certain risk factors up-regulating spike protein receptors DPP4/ACE2 and α2,3/2,6-SiA linkages. For example, in chronic obstructive pulmonary disease (COPD), there is an increased expression of these receptors in the lungs that contributing to the development of SARS or severe MERS-CoV infection.21,28,29 In healthy human lungs, DPP4/ACE2 is almost exclusively expressed in type II pneumocytes that can regenerate alveolar epithelium upon injury and α2,3-SiA linkages are present mainly in the lower reparatory tract (Figure 3A), however, in the lungs of smokers and COPD patients, unlike in healthy human lungs, DPP4/ACE2 would be up-regulated in both type I and II pneumocytes and α2,3/2,6-SiA linkages is highly present both in the upper and lower reparatory tract, indicating fatal MERS/SARS-CoV. When both types I and II pneumocytes became infected and damaged by the CoV, that leads to diffuse alveolar damage and the production of more infectious virus. In fatal MERS/SARS-CoV patients, both types I and II pneumocytes became infected and activated (Figure 3A and B). In the case of fatal MERS-CoV infection, DPP4/ACE2 is widely expressed on the epithelia of the kidneys, alveoli, small intestine, liver, and prostate, and activated leukocytes. The highly expressed DPP4 (CD26)/ACE2 in the kidneys can cause renal dysfunctions by either hypoxic damage or direct infection of the epithelia.13,14,26,28
Molecular Pathogenicity of SARS-CoV-2
The SARS epidemic that emerged in 2019 was named as novel CoV (2019-nCoV/SARS-CoV-2), causing symptoms in humans similar to those caused by the SARS-CoV outbreak in 2003 in addition to high human-to-human transmissibility. The novel CoV was first identified in Wuhan, China, in December 2019, and known as the causative pathogen for the epidemic of viral pneumonia outbreak which becoming pandemic, soon after. The SARS-CoV-2 shows clinical and molecular characteristics similar to the highly pathogenic SARS-CoV, where it has been also named SARS CoV-2 and the related pandemic disease as COVID-19. Genome analysis of SARS-CoV-2 isolates obtained from patients samples revealed evidence both for human-to-human (HTHT) and animal-to-human (ATHT) transmission. Despite mutations arising during every replication cycle, the RNA sequences of SARS-CoV-2 isolated from samples of different patients were almost identical, with greater than 99·9% sequence identity, which exhibited direct HTHT.1–3 At the level of whole-genome sequencing of samples isolated from patients airway epithelial cells, SARS-CoV-2 showed close similarity to bat-SARS-Like-CoVs (~87% identity), however, at the receptor interactions, the characteristics interface of S protein-receptor complex and the structure of external sub-domains of S1 subunit, RBD and RBM, was found highly similar to that of SARS-CoV 2003. The high similarity observed between the RBD domain of 2003 SRAR-CoV and 2109-nCoV (SARS-CoV-2) S-protein supported the same targets and strong interaction with human ACE2, which was following the similar symptoms observed in 2019 outbreak and infection by the novel SARS-CoV.1,23,30
Recently published literature exhibited some new aspects of SRAS-CoV-2 entry and host infection. Data confirmed that SARS-CoV-2 engages ACE2 as the entry receptor and employs the cellular serine protease TMPRSS2 for S protein priming and entry into the target cells. The s2ʹ site in the S protein that is pivotal to host entry, is mostly cleaved by host protease TMPRSSII at the cell surface.
Indeed, the cell/tissue tropism of SARS-CoV-2 was found not only determined by the distribution of ACE2 but also by that of TMPRSSII (Figure 3B). In addition to SARS-CoV-2, TMPRSS2 is a host cell factor that is critical for the spread of several clinically relevant viruses, including influenza A viruses and CoVs. The protease inhibitors might constitute a treatment option whereby blocking the viral entry and spread in the infected host and prevent pathogenesis (Figure 3A and B). Furthermore, data indicate that the SARS-CoV-2 S1 domain is able to interact with the human CD26 (DPP4), a key immune-modulator for hijacking and virulence (Figure 3B). Studies also report unique N- and O-linked glycosylation sites of SARS-CoV-2 spike glycoprotein that distinguish it from the SARS and MERS spike proteins and underline shielding and camouflage of pathogen from the host-defense system. Indeed, the cell/tissue tropism of SARS-CoV-2 may be not only determined by the distribution of ACE2 but also by that of TMPRSSII, glycan shields, and CD26. These findings accentuate the potential features of SARS-CoV-2 in host infectivity.31,32,44,45
SARS-CoV-2 genome was reported to encode a highly complex transcriptome owing to numerous discontinuous transcription events. Frequent fusion events were detected to occur in the transcripts which may provide a basis for a variant generation. SARS-CoV-2 expresses nine canonical subgenomic RNAs (S, 3a, E, M, 6, 7a, 7b, 8, and N) together with the genomic RNA. Transcriptome analysis showed that the N RNA is the most abundantly expressed transcript, followed by S, 7a, 3a, 8, M, E, 6, and 7b. In addition to the canonical genomic and 9 subgenomic RNAs, SARS-CoV-2 produces transcripts encoding unknown ORFs with fusion, deletion, and frame-shifts which might be a result of noncanonical “‘splicing’” events. There were identified at least 41 RNA modification sites on viral transcripts, with the most frequent motif, AAGAA. Modified RNAs (eg ORF1ab and S) have shorter poly(A) tails than unmodified RNAs, suggesting a link between the modification and the poly(A) tail. Like other RNA viruses, SARS-CoV-2 undergoes frequent recombination (a basis for variant generation, immune invasion, and pathogenesis), which may allow rapid evolution to change their host/tissue specificity and drug sensitivity. It was proposed that the new ORFs may participate as accessory proteins that contribute to viral replication and invade host immune response. The RNA modifications may also contribute to viral survival and immune evasion in infected tissues as the innate immune system is known to be less sensitive to RNAs with nucleoside modification.33
Additionally, mutation counts occurring in the viral genome represent a positive correlation with the case fatality and are strengthened with time and virus transmission. Single nucleotide substitutions (SNVs) are the most common mutations occurred in the virus genome which notifies an important consideration in virus detection, clinical treatment, drug design, and vaccine development to avoid target shifting. Mutations have critical impacts on the natural selection and the adaptation of viral strains to the local environment and can alter viral transmissibility and pathogenicity, disease manifestation, and the efficacy of treatment and vaccination. One study identified at least six phylogenetic clusters of the SARS-CoV −2 strains, which also exhibit a geographic preference in different continents. According to the public database GISAID and according to genome analysis of SARS-CoV-2 specimens scattered across the globe, three major clades of SARS-CoV-2 has been identified, that have been subsequently named as clade G (a variant of the spike protein S-D614G), clade V (a variant of the ORF3a coding protein NS3-G251), and clade S (variant ORF8-L84S). In particular, clade G is prevalent in Europe. The most prevalent mutation in sequenced genomes worldwide is a substitution of adenosine with guanosine (A23403G), defining as G-clade of SARS-CoV-2 genomes, prevalent in Europe, Oceania, South America, and Africa. The most frequent mutation that has been identified in Asia is mutation G11083T, causing L37F in Non-structural protein 6. Apart from the S-D614G mutation in the Spike protein, there has been identified a silent mutation (F106F) in NSP6, a proline-to-leucine in NSP12b (P314L) and a mutation in the 5ʹUTR at genomic coordinates 241 that are almost always co-occurring with the S-D614G mutation and determining the strongest signature for the G clade of SARS-CoV-2 genomes. The signature is prevalent in Europe, Africa, the Americas, and Oceania sequenced genomes.34,35
Origin and Evolution of SARS-CoV-2
Novel CoV 2019 or SARS-CoV-2 is most closely related to bat-SL-CoVs. Whole-genome sequence analysis exhibited more than 87% identity with bat-SL CoVs that suggesting events of inter-species transmissions. Notable, five gene regions (E, M, 7, N, and 14) in the virus genome exhibited sequence identities greater than 90%, with the highest being 98·7% in the E gene. The S gene of SARS-CoV-2 exhibited the lowest sequence identity with bat-SL-CoV, at only around 75%.1,10,20 The SARS-CoV-2 S2 protein showed around 93% sequence identity with bat-SL-CoVs, however, the S1 domain had only around 68% identity with related bat-derived CoVs.1,2,10
Despite the high homology found in the Spike-RBD sequence between SARS-CoV-2 and bat-SL-CoVs (as high as 95%),30 several facts, for example, the sequence identity < 99% from close relative bat-SL-CoVs, indicate another animal acting as an intermediate host or mixing vessel between bats and humans for the outbreak of SARS-CoV-2, where bats are not direct ancestors.1,2,29 Both SARS-CoV-2 S1-CTD and NTD can bind to bat host receptors. Among all CoVs from humans, SARS-CoV exhibited the highest genome sequence identity to SARS-CoV-2 (~80% identity), while MERS-CoV showed just 50% identity with SARS-CoV-2.1,2,20 The overall sequence similarities between SARS-CoV-2 spike and SARS-CoV spike protein (isolated from human, civet, or bat) were obtained about 76–78% for the whole protein, about 73–76% for the RBD, and 50–53% for the RBM.1,10,24
Importantly, papain-like protease (PLpro), main protease 3CLpro (also named 3-chymotrypsin-like protease), RNA-dependent RNA polymerase (RdRp) were found highly conserved between the two human SARS-CoV and SARS-CoV-2, especially in functional regions.30 In comparison, human MERS-CoV and bat MERS-like CoV HKU4 share lower sequence similarities in their spikes, RBDs, or RBM, and yet they recognize the same receptor DPP4, although in different species they may show a different affinity for sialoglycans. Thus, sequence similarities existing between SARS-CoV-2 and SARS-CoV spikes can confirm that they could share the same receptor ACE2. Importantly, SARS-CoV-2 RBM does not contain any deletion or insertion when it was compared to SARS-CoV RBM (except one-residue insertion on the RBM loop). Furthermore, among the 14 ACE2-contacting residues in the RBD of SARS-CoV, 9 are fully conserved in SARS-CoV-2 and 4 are partially conserved among SARS-CoV-2.23,36
Although by molecular analysis, SARS-CoV-2 and SARS-CoV were put in different groups, they still possessed around 50 conserved amino acids in the S1 subunit, whereas most of the bat-derived viruses displayed mutational differences in this position interacting with ACE2. Most of the mutational differences between SARS-CoV-2 and bat-SL-CoVs were positioned in the S1-CTD, where the RBD of ß-CoVs is located and directly engages the protein receptor as in SARS-CoV from lineage B and MERS-CoV from lineage C (Figure 2). In comparison to SARS-CoV-2, several deletions, including positions 455–457, 463–464, and 485–497, existed in the RBD of S protein obtained from bat-derived strains.1,3,10,23 Despite the high homology in spike sequence, there were differences in the five most important amino acids (L465, L495, Y502, D510, and H514) in bat-SL-CoV RBD from SARS-CoV-2 and SARS-CoV RBD, exhibited natural selections in SARS-CoV and SARS-CoV-2 which played critical roles in the cross-species transmission of SARS-CoV-2 and SARS-CoV to humans.23,30 Several key residues responsible for the binding of the SARS-CoV RBM to the ACE2 receptor were variable in the SARS-CoV-2 RBM (including Ala426/Asn439, Thr487/Asn501, Asp479/Gln493, Gly485, and Leu472/Phe486; SARS-CoV/SARS-CoV-2 numbering).1,23 Critical residues in SARS-CoV-2 RBM (particularly Gln493) provide favorable interactions with human ACE2, consistent with SARS-CoV-2’s capacity for human cell infection. Other variable residues in SARS-CoV-2 RBM Asn501 is compatible with, but not ideal for, binding human ACE2, suggesting that SARS-CoV-2 has mutated and acquired some capacity for human-to-human transmission. Potentially recognizing ACE2 from a diversity of animal species (except mice and rats), suggests a bat origin of SARS-CoV-2 and implicates these animal species as possible intermediate hosts or animal models for SARS-CoV-2 infections.23,36 Data indicate that the core structure of SARS-CoV-2 and SARS-CoV S1 are highly similar to each other, but their S1-NTD and RBM are different, leading to different receptor specificities.1,6,30 Like as SARS-CoV residues 472–487, SARS-CoV-2 residues 486–501 make up RBM and take charge of interacting with two extracellular hot spots in the VBM of ACE2, one centering on ACE2 residue Lys31 and the other centering on ACE2 residue Lys353.10,23,36 Residue 493 and 501 in SARS-CoV-2 RBD correspond to residue 479 and 487 in SARS-CoV, respectively. Overall, Leu455, Phe486, and Ser494 of SARS-CoV-2 RBD, respectively are corresponding to residues Tyr442, Leu472, and Asp480 in SARS-CoV, and provide more favorable interactions with hotspots (K31 and K353) in VBM of human ACE2. More specifically, Tyr442 of human SARS-CoV RBD does not provide favorable interactions with hot-spot 31 on human ACE2 and has been mutated to Leu455 in the SARS-CoV-2 RBD which provides favorable support for hot-spot 31, hence enhancing the viral binding to human ACE2 and civet-to-human transmission. In particular, Leu472 of human and civet SARS-CoV RBDs that shows favorable interactions with the hot-spot 31 on human ACE2, has been mutated to Phe486 in the SARS-CoV-2 RBD which provides even more support for hot-spot 31, hence also enhancing the viral binding to human ACE2 and civet-to-human transmission. Asp480 of human and civet SARS-CoV RBDs provides favorable interactions with hot-spot 353 on human ACE2 with the help of a neighboring tyrosine (this residue remains as an Asp in the optimized RBD); Ser494 in the SARS-CoV-2 RBD still provides favorable interactions with the hot-spot 353, but the support is not as favorable as provided by Asp480.1,6,23
Sialic Acids (SiA) in Host Infectivity and Pathogen Transmission
Data indicate that sialic acid (SiA) derivatives are involved in the cross-species transmission of zoonotic viruses and host specificity.18,28,32 CoVs and avian influenza viruses (AIVs), as prototypes of zoonotic viruses, become able to cross-species transmission via changes in SiA-binding domains which facilitating infection and supporting the intercellular expansion of virus through the cellular syncytial formation. Adaptive mutations in the SiA-binding domains increase the virus avidity for host and intercellular expansion processes which leads to increased infectivity and pathogenicity. Findings raise the idea that the lectin-like properties of zoonotic viruses contribute to facile cross-species transmission and intercellular spread within infected organisms.37–40 SiA distribution and localization in the human respiratory tract determine infectivity and pathogenicity of AIV and CoVs.13,38,41 The α2,3-linked and α2,6-linked SiA are mainly expressed in the human lower and upper respiratory tract, respectively. Epithelial cells in the nasal mucosa, paranasal sinuses, pharynx, trachea, and bronchi primarily express α2,6-linked SiA, whereas the cells that line the alveolar walls express α2,3-linked SiA.39 The α2,3-linked SiA is a good receptor for human MERS-CoVs, as well as, AIVs, while α2,6-linked Sia is preferential receptors for human influenza A viruses. This can explain the low frequency of human infection by avian-type influenza viruses and the current inability of these viruses to efficiently transmit among humans.39,42 For example, highly pathogenic avian H5N1 influenza viruses and MERS-CoVs preferentially infected the human lower respiratory tract and caused severe pneumonia that often progresses rapidly to the acute respiratory distress syndrome.28,38 However, in conventional human influenza viruses, H3N2 or H1N1, the upper (rather than lower) respiratory tract is the major target for virus replication wherein leading to common flu. Additionally, efficient human to human transmission requires that the CoVs and AIVs acquire mutations and recognize α2,6-linked SiA in the upper reparatory tract. Consistent with this is the early influenza virus isolates from the 1918, 1957, and 1968 pandemics that preferentially recognized α2,6-linked SiA.41,42 Even more, the avian and human H5N1 viruses causing the “bird flu” outbreak in Hong Kong in 1997 and 2003 had an affinity for binding to both avian-like α2,3-linked SiA and human-like α2,6-linked SiA.38 Herein, pigs have been regarded as a hypothetical “mixing” vessel where re-assortment of avian and human viruses can take place, potentially leading to the emergence of pandemic influenza. Pigs contain a respiratory epithelium that contains both “avian-virus” binding with α2,3-linked and “human-virus” binding with α2,6-linked SiA and can be infected with both human and avian influenza viruses and act as intermediate host.38,39 Additionally, the expression profile of α2,6- and α2,3-linked SiA on respiratory tract cell surfaces increases during developmental differentiation and aging as the lung matures. Furthermore, the sialylation increases if cells are exposed to inflammation, oxidative stress, and tumor necrosis factor.29,43 The neonatal pneumocytes and bronchus primarily express α2,3-linked SiA and the respiratory tract of young children show mainly α2,3-linked SiA with a lower level of α2,6-linked SiA expression than adult tissues. This may, in part explain why children and young are more susceptible to avian influenza H5N1 in the recent outbreaks in East Asia. In adults, the α2,3-SiA linkages are mainly present in the lower respiratory tract.28,38,39
Evidence indicates that sialoglycan binding pockets are conserved domains among CoVs and in IVs. For instance, IVA/B hemagglutinin (HA), IVC/D hemagglutinin-esterase (HAE), and CoV HE has distinct architectures compared with those of CoV S protein, common rules of ligand engagement emerge. These rules also appear to extend to the interactions of sialoglycans with adenoviruses and reoviruses. The conserved topology of sub-domain S1A among CoV S proteins indicates that it derived during viral evolution and adaptation and thus leads to the use of distinct binding residues on the same domain putatively to acquire a ligand-binding specificity. For example, favorite binding occurs between HCoV-OC43 S1 with 9-O-Ac-Me-Sia, BCoV HE with 5-N-acetyl-4,9-di-O-acetyl-neuraminic acid α-methyl glycoside, IVC HEF complexes with 9-N-Ac-Sia. Modulation of attachment to sialoglycans can, therefore, have profound effects on zoonotic transmission, tropism, and virulence of many viruses. For instance, a single point mutation in the highly pathogenic AIV H5N1 HA was proposed to account for most of the preference switch from avian enteric tract receptors (α2,3-linked SiA) to human respiratory tract receptors (α2,6-linked SiA). Similarly, these mutations may occur in ligand-binding pockets identified in MERS-CoV or likewise in SARS-CoVs and SARS-CoV-2 but involve different interactions.6–8,12,44 The spike protein of SARS-CoV or SARS-CoV-2 has not been clearly defined to interact with α2,3- or 2,6-linked Sialic acids yet, however, some reports exhibit the possibility of such interaction. For example, inhibition of ACE2 sialylation (the sialylated N-linked glycan profiles of ACE2) impairs its ability to support the transduction of SARS-CoV. Or, a recent study reported a new type of ganglioside-binding domain at the tip of S1-NTD of the SARS- CoV-2 S protein that was reported to be fully conserved among SARS-CoV-2 specimens scattered across the globe. The domain was reported to improve the attachment of the virus to cellular lipid rafts and facilitate contact with the ACE-2 receptor. The conserved domain contains residues 111–158 and was found fully conserved among clinical isolates worldwide.44,45
The β-sandwich architecture of sub-domain S1A is conserved among CoVs recognizing similar silicides and some of them feature a duplication of this sub-domain at the S1-NTD. Based on the evidence, sialoglycan binding-site is observed in CoVs like as MERS-CoV (β-coronavirus), IBV (γ-coronavirus), PRDV (α-coronavirus) and TGEV (α-coronavirus) all of which have been described to bind to sialoglycans during host cell infection (Table 1).12,22
Sterilizing Immunity, Based on the CoV S Protein
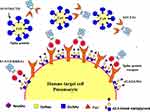
An efficient strategy recommended against virus infectivity is sterilizing immunity against virus spike protein, wherein protein and SiA receptors are reduced on target cells and innate immunity against virus infection is induced in the target organ. In sterilizing immunity, pre-infection with intranasal inoculation of low-dose virus can induce efficient antigen-specific T cell response in the lungs, where prevents subsequent effective infection of a challenge with the lethal dose of the same strain of virus or reduced infectivity by a lethal dose of homologs virus. In sterilizing immunity, local antigen-specific host immune response is induced to block virus infection and clear it before the establishment of effective infection. Only intranasal inoculation induces sterilizing immunity, whereas intramuscular injection does not block subsequent infection but may provide protection with enhanced virus clearance (Table 2).14,15,46 For example, intranasal vaccination with a low-dose CoV S protein can generate memory CD4 T cells in the airway which would be induced and mediate protection following a CoV challenge. These cells could induce anti-viral innate responses at an early stage of infection by stimulating dendritic cell migration whereby could facilitate CD8 T-cell responses. The stimulation of memory CD4 T cells in the airway has been indicated as an essential part of any strategy mentioning CoV vaccine development. Accordingly, these memory CD4 T cells target a conserved epitope within the S protein that can cross-react with some other CoVs.14,46 Another example is intranasal pre-infection with the nH1N1 influenza virus which induces sterilizing immunity whereby decreases susceptibility to the next homologous influenza virus challenge in the host. Intranasal pre-infection with nH1N1 influenza virus induced virus receptor reduction and antigen-specific T cell immune response in the lungs of mice. Receptor reduction decreases susceptibility across different strains of the influenza virus, but sterilization only takes effect with the challenge of the same strain of influenza virus (Table 2).46 Similar results have been reported after intranasal vaccination with MERS or SARS-CoV, while intravenously inoculated vaccines may induce harmful immune responses that may lead to the liver damaging in vaccinated animals or enhanced infection by a following homologous SARS-CoV challenge. Accordingly, intranasal inoculation of transgenic mice with a low-dose of SARS or MERS-CoV would result in the production of neutralizing antibodies protecting the animals from following lethal challenges with the virus. These results could probably reflect the situation in infected humans during an epidemic disease. However, there may be nasal turbinate in the upper respiratory tract and a high titer of virus replication in the lungs of the lower respiratory without any signs of morbidity or mortality in the animal models of SARS-CoV infection.14,46 Accordingly, intranasal vaccinations based on the S1 protein or RBD could induce antibodies to block virus-receptor interaction and membrane fusion or neutralize the infectious virus (Table 2). The S protein is known to be the main antigenic component among all structural proteins of MERS, SARS and SARS-CoV-2, that is responsible for inducing host immune responses, neutralizing antibodies, and/or protective immunity against virus infection. In infection by SARS-CoV strains, neutralizing antibodies against S protein raised against amino acids 485–625 in S1 or 1029–1192 in S2.14,15,22 Although full-length S protein-based SARS vaccines can induce neutralizing antibody responses against SARS-CoV infection but should mention harmful immune responses induced by full-length S protein that cause liver damage of the vaccinated animals or enhanced infection after challenge with homologous SARS-CoV, which here raising concerns about the safety and ultimate protective efficacy of vaccines that contain the full-length SARS-CoV S protein. Immunization of mice and rabbits with RBD-Fc induces long-term protective immunity against the next challenge with homologous SARS-CoV strain. The RBD-Fc induced highly potent neutralizing antibodies against SARS-CoV with great neutralizing titer in rabbits.14,15,48,49 Reports show that produced antibodies effectively cross-neutralized infection by SARS pseudoviruses that bear S proteins of both homologous and heterologous SARS-CoV isolates, including the representative strains of human 2002–2003 and 2003–2004 SARS-CoV and palm civet SARS-CoV. Herein, the fastest strategy to develop a treatment and sterilization immunity is to use a fusion protein that contained the RBDs (S1-CTD and S1-NTD) linked to human IgG1 Fc fragment (designated S1-CTD-NTD-Fc), as an immunogen successfully induced highly potent neutralizing antibodies (Figure 4), as well as, blocking virus-binding receptor (eg ACE2/DPP4). For instance, administration of the key domain RBD that binds the ACE2 receptor during entry, 193 amino acids in size, effectively blocked the entry of SARS in cell cultures.14–16,48,49
 |
Table 2 A Summary of the Vaccine Development/Design, Administration Routes and Corresponding in vitro/in vivo Evaluation Results |
On the other side, a potentially more promising strategy has been proposed to create double protection where two antibody-like molecules would bind to both the virus spike protein and its receptor in the restricted host, simultaneously (Figure 4). Accordingly, another fastest strategy to develop a treatment for the next zoonotic infection, as well as, SARS-CoV-2, could be a soluble version of the viral receptor (eg ACE2). This strategy could be resistant to any mutations the virus may have in the future. This receptor should be fused to the immunoglobulin Fc fragment (ACE2-Fc). The engineered protein would then provide a neutralizing antibody with maximal breath to avoid any viral escape, as well as, helping to recruit the immune system and to build a lasting immunity. As a third mechanism of action, the ACE2-Fc therapy is proposed to supplement decreased ACE2 levels in the lungs during infection, thereby directly treating acute respiratory distress pathophysiology (Figure 4).14,16 It is well within reason that both ACE2-Fc and S1-NTD-RBD-Fc could be given to humans through the intranasal route, thereby blocking both virus-binding sites on target cells, preventing infection, as well as, inducing innate and efficient antigen-specific T cell response (Figure 4).11,14,16
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi:10.1016/S0140-6736(20)30251-8
2. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi:10.1126/science.abb2507
3. Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus. J Med Virol. 2020;92(5):522–528. doi:10.1002/jmv.25700
4. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi:10.1038/s41591-020-0820-9
5. Ceccarelli M, Berretta M, Venanzi Rullo E, Nunnari G, Cacopardo B. Differences and similarities between severe acute respiratory syndrome (SARS)-coronavirus (CoV) and SARS-CoV-2. Would a rose by another name smell as sweet? Eur Rev Med Pharmacol Sci. 2020;24(5):2781–2783. doi:10.26355/eurrev_202003_20551
6. Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi:10.1038/s41586-020-2180-5
7. van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382(16):1564–1567. doi:10.1056/NEJMc2004973
8. Wang N, Shi X, Jiang L, et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23(8):986–993. doi:10.1038/cr.2013.92
9. Yuan P, Yang Z, Song H, et al. Three main inducers of alphacoronavirus infection of enterocytes: sialic acid, proteases, and low pH. Intervirology. 2018;61(2):53–63. doi:10.1159/000492424
10. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–182. doi:10.1038/s41579-018-0118-9
11. Li F, Goff SP. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89(4):1954–1964. doi:10.1128/JVI.02615-14
12. Tortorici MA, Walls AC, Lang Y, et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019;26(6):481–489. doi:10.1038/s41594-019-0233-y
13. Widagdo W, Sooksawasdi S, Ayudhya N, Hundie GB, Haagmans BL. Host determinants of MERS-CoV transmission and pathogenesis. Viruses. 2019;11(280):2–14. doi:10.3390/v11030280
14. Song Z, Xu Y, Bao L, et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(59):1–29. doi:10.3390/v11010059
15. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226–236. doi:10.1038/nrmicro2090
16. Kruse RL. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China [version 2; peer review: 2 approved]. F1000Research. 2020;9(72):1–14. doi:10.12688/f1000research.22211.2
17. Jia HP, Look DC, Shi L, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79(23):14614–14621. doi:10.1128/JVI.79.23.14614-14621.2005
18. Li W, Hulswit RJG, Widjaja L, et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. PNAS. 2017;114(40):E8508–E8517. doi:10.1073/pnas.1712592114
19. Qing E, Hantak M, Perlman S, Gallagher T, Denison MR. Distinct roles for sialoside and protein receptors in coronavirus infection. mBio. 2020;11(1):e02764–19. doi:10.1128/mBio.02764-19
20. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi:10.1038/s41586-020-2012-7
21. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23.
22. Song EW, Gui M, Wang X, Xiang Y, Heise MT. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14(8):e1007236. doi:10.1371/journal.ppat.1007236
23. Wan Y, Shang J, Graham R, Baric RS, Li F, Gallagher T. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol. 2020;94(7). doi:10.1128/JVI.00127-20
24. Tikellis C, Thomas MC. Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin systemin health and disease. Int J Pept. 2012;2012:1–8. doi:10.1155/2012/256294
25. Cao Y, Li L, Feng Z, et al. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Disc. 2020;6(11):1–4.
26. Widagdo W, Okba NMA, Li W, et al. Species-specific colocalization of Middle East respiratory syndrome coronavirus attachment and entry receptors. J Virol. 2019;93(16):e00107–19. doi:10.1128/JVI.00107-19
27. Park YJ, Walls AC, Wang Z, et al. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat Struct Mol Biol. 2019;26(12):1151–1157. doi:10.1038/s41594-019-0334-7
28. Schwegmann-Webels C, Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconj J. 2006;23(1–2):51–58. doi:10.1007/s10719-006-5437-9
29. Wielgat P, Trofimiuk E, Czarnomysy R, Holownia A, Braszko JJ. Sialylation pattern in lung epithelial cell line and siglecs expression in monocytic THP-1 cells as cellular indicators of cigarette smoke-induced pathology in vitro. Exp Lung Res. 2018;44(3):167–177. doi:10.1080/01902148.2018.1461959
30. Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020. doi:doi.10.1016/j.apsb.2020.02.008
31. Vankadari N, Wilce JA. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg Microbes Infect. 2020;9(1):601–604. doi:10.1080/22221751.2020.1739565
32. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi:10.1016/j.cell.2020.02.052
33. Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921. doi:10.1016/j.cell.2020.04.011
34. Mercatelli D, Giorgi FM. Geographic and Genomic Distribution of SARS-Cov-2 Mutations. Preprints; 2020:2020040529. doi:10.20944/preprints202004.0529.v1
35. Yang HC, Chen CH, Wang JH, et al. Genomic, geographic and temporal distributions of SARS-CoV-2 mutations. bioRxiv. 2020. doi:10.1101/2020.04.22.055863.
36. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562–569. doi:10.1038/s41564-020-0688-y
37. Wasik BR, Barnard KN, Ossiboff RJ, et al. Distribution of O-acetylated sialic acids among target host tissues for influenza virus. mSphere. 2017;2(5):e00379–16. doi:10.1128/mSphere.00379-16
38. Nicholls JM, Bourne AJ, Chen H, Guan Y, Peiris JSM. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res. 2007;8(73):1–10. doi:10.1186/1465-9921-8-73
39. Neumann G, Shinya K, Kawaoka Y. Molecular pathogenesis of H5N1 influenza virus infections. Antivir Ther. 2007;12(4 Pt B):617–626.
40. Neumann G, Chen H, Gao GF, Shu Y, Kawaoka Y. H5N1 influenza viruses: outbreaks and biological properties. Cell Res. 2010;20(1):51–61. doi:10.1038/cr.2009.124
41. Li YH, Hu CY, Wu NP, Yao HP, Li LJ. Molecular characteristics, functions, and related pathogenicity of MERS-CoV proteins. Eng. 2019;5(5):940–947. doi:10.1016/j.eng.2018.11.035
42. HU W. Molecular features of highly pathogenic avian and human H5N1 influenza a viruses in Asia. Comput Mol Biosci. 2012;2(02):45–59. doi:10.4236/cmb.2012.22005
43. Hanisch F, Weidemann W, Großmann M, et al. Sialylation and muscle performance: sialic acid is a marker of muscle ageing. PLoS One. 2013;8(12):e80520. doi:10.1371/journal.pone.0080520
44. Fantini J, Di Scala C, Chahinian H, Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents. 2020;55(5):105960. doi:10.1016/j.ijantimicag.2020.105960
45. Zhao X, Guo F, Comunale MA, et al. Inhibition of endoplasmic reticulum-resident glucosidases impairs severe acute respiratory syndrome coronavirus and human coronavirus NL63 spike protein-mediated entry by altering the glycan processing of angiotensin I-converting enzyme 2. Antimicrob Agents Chemother. 2015;59(1):206–216. doi:10.1128/AAC.03999-14
46. Dutta A, Huang C-T, Lin C-Y, et al. Sterilizing immunity to influenza virus infection requires local antigen-specific T cell response in the lungs. Sci Rep. 2016;6(1):1–14. doi:10.1038/srep32973
47. Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16(69):2–22. doi:10.1186/s12985-019-1182-0
48. He Y, Zhou Y, Liu S, et al. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324(2):773–781. doi:10.1016/j.bbrc.2004.09.106
49. He Y, Li J, Li W, Lustigman S, Farzan M, Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J Immunol. 2006;176(10):
50. Zhao J, Zhao J, Mangalam AK, et al. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44(6):1379–1391. doi:10.1016/j.immuni.2016.05.006
51. Tai W, Wang Y, Fett CA, et al. Recombinant receptor-binding domains of multiple middle east respiratory syndrome coronaviruses (MERS-CoVs) induce cross-neutralizing antibodies against divergent human and camel MERS-CoVs and antibody escape mutants. J Virol. 2016;91(1):e01651–16. doi:10.1128/JVI.01651-16
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.