Back to Journals » Pathology and Laboratory Medicine International » Volume 6
Molecular and immunohistochemical profiling of invasive micropapillary carcinoma of the breast
Authors Thomas A, Askeland R, Guseva N, Sompallae R, Ma D
Received 6 June 2014
Accepted for publication 9 July 2014
Published 3 October 2014 Volume 2014:6 Pages 33—40
DOI https://doi.org/10.2147/PLMI.S67836
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Alexandra Thomas,1 Ryan W Askeland,2 Natalya V Guseva,2 Ramakrishna Sompallae,2,3 Deqin Ma2
1Department of Internal Medicine, 2Department of Pathology, 3Bioinformatics Division, Iowa Institute of Human Genetics, University of Iowa Hospitals and Clinics, Iowa City, IA, USA
Background: In this study, molecular and immunohistochemical profiling of invasive micropapillary carcinoma of the breast was used to identify potentially useful markers for targeted therapies with a focus on BRAF V600E mutation.
Methods: Formalin-fixed, paraffin-embedded tumor blocks from seven patients were identified from the archives at our institution and tumor registry from 1997 to 2012. Massively parallel (Next-generation) sequencing was performed using the Ion AmpliSeq™ Cancer Hotspot Panel version 2 (Life Technologies, Carlsbad, CA, USA). Mutation analysis for BRAF V600E was performed using a single nucleotide primer extension assay. Immunohistochemistry studies for estrogen receptor (ER), progesterone receptor (PR), Her2/Neu, phosphatase and tensin homolog (PTEN), and non-metastatic protein 23 homologue 1 (NM23H1) were performed using the same tumor blocks. Staining for ER, PR, and Her2/Neu was scored according to American Society of Clinical Oncology/College of American Pathologists guidelines, and a four-tier system, ie, strong homogenous, heterogeneous, positive with negative foci, reduced in more than 50%, and lost in all or majority was used for PTEN and NM23H1 staining.
Results: No pathogenic mutations were identified in the tumors by next-generation sequencing. The lack of BRAF V600E mutation was confirmed by single nucleotide primer extension assay. All tumors were positive for ER and PR, and showed no overexpression of Her2/Neu. Loss of or reduced PTEN expression was observed in six of seven cases and was associated with lymph node metastasis. Reduced NM23H1 expression was observed in three of seven cases, all of which had concurrent PTEN loss.
Conclusion: No somatic mutation was identified consistent with the reported lack of driver mutations in this tumor. The frequent loss of PTEN in invasive micropapillary carcinoma may have implications for targeted therapy towards the phosphatidylinositol-4,5-bisphosphate 3-kinase pathway in this subgroup of patients.
Keywords: invasive micropapillary carcinoma, next-generation sequencing, immunohistochemistry
Introduction
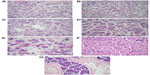
Invasive micropapillary carcinoma (IMPC) of the breast is an uncommon subtype of mammary carcinoma. Histology of IMPC characteristically shows clusters of tumor cells that are surrounded by clear stromal spaces and exhibit an “inside-out” growth pattern with the apical pole of the cells facing the stroma (Figure 1). Series reporting clinical outcomes for this subtype of breast cancer have generally found it to be more aggressive with regard to histopathologic features, stage at presentation, and clinical outcomes compared with invasive carcinoma of no special type.1–3
Micropapillary carcinomas occur at a variety of other anatomic sites including lung, bladder, thyroid, and ovary.4–7 Studies have shown that 3%–5% of lung adenocarcinomas harbor BRAF V600E mutation,8–10 and the presence of BRAF mutation was often found to be associated with tumors of a predominantly micropapillary histology.8 Melanomas with BRAF V600E mutation are responsive to targeted treatments.11 Investigations to expand the use of vemurafenib, dabrafenib, and other agents targeting the mitogen-activated protein kinase (MAPK pathway) in non-melanocytic tumors are ongoing.12
The most common drivers in breast cancers are alterations in the PIK3CA gene as well as its downstream molecules.13,14 Several drugs targeting this pathway are currently in advanced clinical trials in breast cancers (http://www.clinicaltrials.gov). Breast cancers rarely (only about 3%) have BRAF mutation or other aberrations in the MAPK pathway.14 Limited information is known about molecular drivers of IMPC due to the low incidence of this tumor type.
Recent studies have shown that the addition of phosphatase and tensin homolog (PTEN) tumor suppressor status to histologic findings may improve the sensitivity of the Manchester scoring method in breast cancer patients,15 and the expression of NM23H1, a metastatic suppressor, was associated with reduced potential for lymph node metastasis and vascular invasion in breast cancers.16
In this study, we performed mutational profiling using massively parallel sequencing (AmpliSeq™ Cancer Hotspot Panel version 2, Life Technologies, Carlsbad, CA, USA) in a cohort of seven IMPCs with particular attention to BRAF mutations. Expression of PTEN and NM23H1 was also examined.
Materials and methods
The study was reviewed and approved by the institutional research review committee of University of Iowa (Iowa City, IA). No informed consent from patients was required.
Case selection
Formalin-fixed, paraffin-embedded (FFPE) tissue blocks from seven patients with pure or predominant IMPC were selected from the archives of the Department of Pathology at our institution and the Tumor Registry from 1997 to 2012. Each case was reviewed by two surgical pathologists and the previously documented diagnosis of IMPC was confirmed. We targeted only areas with at least 20% IMPC tumor cells. Areas with the highest percentage of IMPC tumor cells and least amount of contaminating materials (non-neoplastic cells, debris, and mucin) were marked by the pathologists. The age, anatomic location, specimen type, histologic grade, axillary lymph node status, and estrogen receptor (ER)/progesterone receptor (PR)/Her2/Neu results were acquired from the medical record.
Extraction of genomic DNA
One hematoxylin-eosin-stained slide and ten unstained sections (6 microns in thickness) were cut. Areas with the highest percentage of tumor were identified on the hematoxylin-eosin slide and corresponding areas from the unstained slides were manually microdissected using a razor blade. The paraffin flakes were placed in a 1.5 mL microcentrifuge tube, deparaffinized with 1,200 μL of xylene, vortexed, and centrifuged (16,000 g ×5 minutes). The tissue pellet was washed with 95% ethanol twice before proceeding with the extraction. Genomic DNA was isolated from the microdissected FFPE sections using a QIAamp DNA FFPE tissue kit (Qiagen, Valencia, CA, USA). The DNA concentration was measured using a Qubit dsDNA HS assay kit (Life Technologies) and the TaqMan RNase P detection reagents kit (Life Technologies).
Massively parallel sequencing
Massively parallel (next-generation) sequencing was performed using the Ion Torrent AmpliSeq Cancer Hotspot Panel version 2 (Life Technologies) according to the manufacturer’s instruction. Briefly, 10 ng of genomic DNA was used for preparation of the amplicon libraries using the Ion AmpliSeq 2.0 technology. The libraries were bar-coded, clonally amplified by emulsion polymerase chain reaction in vitro, and sequenced on an Ion 318 Chip. The data were analyzed using the Ion Torrent Suite Software (Life Technologies) followed by a laboratory developed/validated pipeline.
Mutation analysis for BRAF V600E by single nucleotide primer extension assay
BRAF V600E mutation analysis was performed by a laboratory-developed single nucleotide primer extension assay using the SNaPshot Multiplex kit (Applied Biosystems, Foster City, CA). The V600 codon was interrogated with a sense probe consisting of deoxythymidine monophosphate homopolymers of varied sizes for discrimination of different mutation products. After amplification, the product was subjected to probe annealing and addition of a single, fluorescently labeled dideoxynucleotide. The primer extension products were analyzed by capillary electrophoresis.
Immunohistochemistry studies
The same tumor blocks used for the molecular test were used for immunohistochemistry studies. Heat-induced antigen retrieval was performed with citrate buffer at pH 6.0. Immunohistochemistry was carried out with a primary antibody to PTEN (clone 6H2.1, 1:100; Dako, Carpinteria, CA, USA) and NM23H1 (clone Nm301, 1:50; Santa Cruz Biotechnology, Santa Cruz, CA, USA) incubated with tissue sections for 30 minutes at room temperature. Unbound antibodies were washed away, and the slides were incubated with the secondary antibody for 15 minutes using the Dako Rabbit Envision System (HRP #K4003112). Signal was detected after incubation with the Dako DAB Plus reagent (#K346811-2; 5 minutes) and signal intensity was enhanced with the Dako DAB Enhancer (#S1961-31-2; 3 minutes). Finally, a Harris hematoxylin counterstain was applied. The slides were evaluated and scored by two anatomic pathologists using a four-tier system for PTEN and NM23H1: P: strong homogenous; H: heterogeneous (positive with negative foci); R: reduced in more than 50%; and L: lost in all or majority of the tumor cells. Immunohistochemistry studies for ER, PR, and Her2/Neu were scored according to American Society of Clinical Oncology/College of American Pathologists guidelines.17,18
Statistical analysis
The statistical analysis was performed using Fisher’s exact test. A P-value <0.05 was considered to be statistically significant.
Results
Patient characteristics
Seven IMPC patients ranging in age from 50 to 83 years with a mean age 67 years were identified from patients seen at our hospital and the Tumor Registry over a period of 15 years. Four were treated with partial breast excision while three had total mastectomy. On microscopic examination, four patients had pure IMPC, two tumors exhibited a minor component of invasive ductal carcinoma, and the last one had a component of mucinous carcinoma. Four of the seven patients presented with axillary lymph node metastases at surgery. Six patients had histologic Ellis-Elliston grade 2 and one had grade 1. Figure 1 shows representative images of all cases. The characteristics of the cases were summarized in Table 1.
Mutational profiling by massively parallel sequencing
Massively parallel sequencing was performed on tumors from all seven patients. Four tumors gave satisfactory results, with one sample showing a benign polymorphism in the KIT gene (c.1621A>C, p.M541L, Figure 2). No pathogenic mutations were identified in any of the samples examined. Library preparation of three tumors failed and the amplifiable DNA in these samples was less than 5%, as quantified by a TaqMan RNase P detection real-time polymerase chain reaction assay (Life Technologies, data not shown). The suboptimal DNA quality was most likely due to the prolonged storage time of the blocks or suboptimal tissue fixation and processing.
The lack of BRAF V600E mutation in all seven tumors was confirmed by a laboratory-developed single nucleotide primer extension assay with an analytical sensitivity of 5%. Representative result is shown in Figure 3.
Immunohistochemistry studies
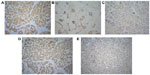
Expression of PTEN and NM23H1 was scored using the four-tier system described earlier in this section (representative images of the immunostain are shown in Figure 4).
All the tumors were strongly positive for ER and PR, with no overexpression of Her2/Neu. Loss or reduced PTEN expression was seen in six of seven cases (four with complete PTEN loss). Four patients presented with lymph node metastasis; all had complete (3/4) or partial (1/4) loss of PTEN expression and two had reduced expression of NM23H1. The third case with reduced NM23H1 expression had a metachronous urothelial carcinoma with micropapillary histology. All three tumors with reduced NM23H1 expression showed concurrent PTEN loss.
No statistically significant difference was observed between the reduced expression of PTEN and NM23H1 and lymph node metastasis (P=0.4), most likely due to the small number of samples.
Discussion
Mutation profiling by massively parallel sequencing has identified recurrent mutations or translocations in different subtypes of breast cancers.19–22 Limited information is available on IMPC due to its lower incidence in the population. Recently, Flatley et al evaluated a panel of hotspot mutations by mass spectrometry analysis in IMPCs from 19 patients.23 Aberrations in the MAPK pathway were predominant in this series, with the majority of mutations seen in the PIK3CA or AKT1 gene. KRAS mutation was only seen in one patient and no BRAF mutation was identified. Another group performed hotspot mutation analysis of 19 cancer-related genes and fusion genes that had been described in secretory or adenoid cystic carcinoma.24 No driver mutation was identified.
In this study, we performed mutation profiling using targeted next-generation sequencing that could simultaneously detect over 2,800 mutations in 50 cancer-related genes. No pathogenic mutation was identified consistent with the findings in the literature.19,20 The p.M545L mutation in the KIT gene has been reported in a variety of tumor types including breast cancer13,14 as well as normal tissue of our patient (Case 5). This variant therefore represents a benign polymorphism.
Mutations in the BRAF gene have been reported in a subset of lung adenocarcinomas, and the presence of BRAF V600E mutation has been shown to be associated with micropapillary morphology in lung cancer.8–10 We confirmed the lack of BRAF mutation in these tumors using a laboratory-developed, sensitive (analytical sensitivity 5%) single nucleotide primer extension assay. Unlike what has been observed in lung cancer, BRAF mutation does not appear to be associated with breast carcinoma with micropapillary histology. A recent study by Vasan et al25 screened 52 breast cancers for mutations in 3,230 exons in 182 cancer-related genes and translocations in 14 genes and found the most common actionable alternations were in the PIK3CA gene and no BRAF mutation was identified. Another study of 65 triple-negative breast cancers also showed that this subtype lacked BRAF V600 mutation.26 Based on the reports in the literature and our findings, it seems that BRAF and other components of the MAPK pathway is a less common target for mammary tumors.
Studies by Natrajan et al27 showed that IMPCs were heterogeneous but significantly enriched for luminal B breast cancer mutations, consistent with the findings described in The Cancer Genome Atlas.14 Using microarray analysis, Horlings et al reported that five of six IMPC tumors included in their study clustered with higher grade ER-positive invasive carcinoma of no special type.28 In our study cohort, all tumors were strongly positive for ER (3+; >95%) and PR (3+; 30->98%), and with no overexpression of Her2/Neu (0–1+). The majority of the tumors (6/7) had Ellis-Elliston grade 2 and four of seven had lymph node metastasis. Our findings add to the body of evidence that IMPC generally has a molecular signature that can be grouped with higher grade, luminal B type breast cancer.
PTEN is a known tumor suppressor which is mutated in multiple human cancers.29,30 Patients with PTEN loss in their tumors may benefit from targeted therapy against the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) pathway.31,32 In our study, loss of or reduced expression of PTEN was observed in six of seven IMPCs and was associated with lymph node metastasis. Since no PTEN mutation was identified, the reduced expression at the protein level was most likely due to post-transcriptional modifications or large deletions or truncation of the gene.
Metastasis accounts for most of the morbidity and mortality associated with breast cancer.33 NM23H1 is the first metastatic suppressor identified which controls metastasis across several tumor types including breast, thyroid, and melanoma.34 A direct correlation between PTEN and NM23H1 expression was observed in four of seven cases. All three cases with reduced NM23H1 expression showed concurrent PTEN loss. In two of the three cases with reduced NM23H1 expression, patients developed lymph node metastasis, and interestingly, the third patient had a metachronous urothelial carcinoma with micropapillary histology. The potential molecular mechanism underlying concurrent NM23H1 and PTEN loss could be that the tumor suppressor and metastatic suppressor work synergistically to promote tumorigenesis and tumor progression, which might explain the aggressiveness of IMPC.
A limitation of our study is the small number of cases. Over a period of 15 years, only seven cases were identified from archives in our department and the tumor registry. Next-generation sequencing failed in some cases, and was most likely due to prolonged storage and a suboptimal fixation process of the specimens. Although no statistically significant difference was achieved due to our small study cohort, we found that IMPCs had no driver mutations in the hotspots of the 50 cancer-related genes. The lack of BRAF and RAS mutations and frequent loss of PTEN expression in these tumors indicate that the PI3K pathway, rather than the MAPK pathway, plays a more important role in the pathogenesis of IMPC. The identification of frequent PTEN loss in these tumors further supports a role for targeted therapy using PI3K-mammalian target of rapamycin inhibitors in IMPC, which is currently a very active area of breast cancer drug development.35–39 The correlation of reduced NM23H1 expression and PTEN loss may play a role in explaining the clinical behavior of the invasive nature of IMPC.
Notably, an actionable mutation in one solid tumor does not guarantee response in another. Recently, vemurafenib, which is highly effective in BRAF V600E-mutated melanoma, was found to have only limited activity in BRAF-mutated colon cancers.40 Linking molecular signatures with active therapy will require us to further understand the complex interactions within a tumor’s genetic profile as well as between the tumor and the patient. Continuing to address these questions will allow us to move forward to an era of truly personalized, precise, and effective therapy.
Acknowledgments
This study was supported by the Breast Cancer Research Group for the Holden Comprehensive Cancer Center of University of Iowa Hospitals and Clinics. The authors thank the molecular pathology and core histology research laboratories in the Department of Pathology at our institution for their technical support.
Disclosure
The authors report no conflicts of interest in this work.
References
Chen L, Fan Y, Lang RG, et al. Breast carcinoma with micropapillary features: clinicopathologic study and long-term follow-up of 100 cases. Int J Surg Pathol. 2008;16(2):155–163. | |
Yu JI, Choi DH, Park W, et al. Differences in prognostic factors and patterns of failure between invasive micropapillary carcinoma and invasive ductal carcinoma of the breast: matched case-control study. Breast. 2010;19(3):231–237. | |
Pettinato G, Manivel CJ, Panico L, Sparano L, Petrella G. Invasive micropapillary carcinoma of the breast: clinicopathologic study of 62 cases of a poorly recognized variant with highly aggressive behavior. Am J Clin Pathol. 2004;121(6):857–866. | |
Kuroda N, Hamaguchi N, Ohara M, Hirouchi T, Miyzaki E, Mizuno K. Intracytoplasmic lumina in invasive micropapillary carcinoma of the lung. Diagn Cytopathol. 2006;34(3):224–226. | |
Wang J, Wang FW. The natural history, treatment pattern, and outcomes of patients with micropapillary bladder carcinoma. Am J Clin Oncol. September 21, 2013. [Epub ahead of print.] | |
Motosugi U, Murata S, Nagata K, Yasuda M, Shimizu M. Thyroid papillary carcinoma with micropapillary and hobnail growth pattern: a histological variant with intermediate malignancy? Thyroid. 2009;19(5):535–537. | |
May T, Virtanen C, Sharma M, et al. Low malignant potential tumors with micropapillary features are molecularly similar to low-grade serous carcinoma of the ovary. Gynecol Oncol. 2010; 117(1):9–17. | |
Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29(26):3574–3579. | |
Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29(15):2046–2051. | |
Li H, Pan Y, Li Y, et al. Frequency of well-identified oncogenic driver mutations in lung adenocarcinoma of smokers varies with histological subtypes and graduated smoking dose. Lung Cancer. 2013;79(1):8–13. | |
McArthur GA, Chapman PB, Robert C, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 2014;15(3):323–332. | |
Planchard D, Mazieres J, Riely JG, et al. Interim results of phase II study BRF113928 of dabrafenib in BRAF V600E mutation-positive non-small cell lung cancer patients. J Clin Oncol. 2013;31 Suppl:Abstr 8009. | |
Santarpia L, Qi Y, Stemke-Hale K, et al. Mutation profiling identifies numerous rare drug targets and distinct mutation patterns in different clinical subtypes of breast cancers. Breast Cancer Res Treat. 2012;134(1):333–343. | |
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. | |
Phuah SY, Looi LM, Hassan N, et al. Triple-negative breast cancer and PTEN (phosphatase and tensin homologue) loss are predictors of BRCA1 germline mutations in women with early-onset and familial breast cancer, but not in women with isolated late-onset breast cancer. Breast Cancer Res. 2012;14(6). | |
Duenas-Gonzalez A, Abad-Hernandez MM, Garcia-Mata J, Paz- Bouza JI, Cruz-Hernandez JJ, Gonzalez-Sarmiento R. Analysis of nm23-H1 expression in breast cancer. Correlation with p53 expression and clinicopathologic findings. Cancer Lett. 1996;101(2):137–142. | |
Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med. 2010;134(7):e48–e72. | |
Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138(2):241–256. | |
Desmedt C, Voet T, Sotiriou C, Campbell PJ. Next-generation sequencing in breast cancer: first take home messages. Curr Opin Oncol. 2012;24(6):597–604. | |
Uji K, Naoi Y, Kagara N, et al. Significance of TP53 mutations determined by next-generation “deep” sequencing in prognosis of estrogen receptor-positive breast cancer. Cancer Lett. 2014;342(1):19–26. | |
Ozcelik H, Shi X, Chang MC, et al. Long-range PCR and next-generation sequencing of BRCA1 and BRCA2 in breast cancer. J Mol Diagn. 2012;14(5):467–475. | |
Edgren H, Murumagi A, Kangaspeska S, et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12(1):R6. | |
Flatley E, Ang D, Warrick A, Beadling C, Corless CL, Troxell ML. PIK3CA-AKT pathway mutations in micropapillary breast carcinoma. Hum Pathol. 2013;44(7):1320–1327. | |
Natrajan R, Wilkerson PM, Marchio C, et al. Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast. J Pathol. 2014;232(5):553–565. | |
Vasan N, Yelensky R, Wang K, et al. A targeted next-generation sequencing assay detects a high frequency of therapeutically targetable alterations in primary and metastatic breast cancers: implications for clinical practice. Oncologist. 2014;19(5):453–458. | |
Grob TJ, Heilenkotter U, Geist S, et al. Rare oncogenic mutations of predictive markers for targeted therapy in triple-negative breast cancer. Breast Cancer Res Treat. 2012;134(2):561–567. | |
Natrajan R, Wilkerson PM, Marchio C, et al. Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast. J Pathol. 2014;232(5):553–565. | |
Horlings HM, Weigelt B, Anderson EM, et al. Genomic profiling of histological special types of breast cancer. Breast Cancer Res Treat. 2013;142(2):257–269. | |
Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. | |
Liaw D, Marsh DJ, Li J, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16(1):64–67. | |
Ghayad SE, Cohen PA. Inhibitors of the PI3K/Akt/mTOR pathway: new hope for breast cancer patients. Recent Pat Anticancer Drug Discov. 2010;5(1):29–57. | |
Morgan TM, Koreckij TD, Corey E. Targeted therapy for advanced prostate cancer: inhibition of the PI3K/Akt/mTOR pathway. Curr Cancer Drug Targets. 2009;9(2):237–249. | |
Smith SC, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer. 2009;9(4):253–264. | |
Marino N, Nakayama J, Collins JW, Steeg PS. Insights into the biology and prevention of tumor metastasis provided by the Nm23 metastasis suppressor gene. Cancer Metastasis Rev. 2012;31(3–4):593–603. | |
Britschgi A, Radimerski T, Bentires-Alj M. Targeting PI3K, HER2 and the IL-8/JAK2 axis in metastatic breast cancer: which combination makes the whole greater than the sum of its parts? Drug Resist Updat. 2013;16(3–5):68–72. | |
Ciruelos E, Cortes-Funes H, Ghanem I, Manso L, Arteaga C. Role of inhibitors of mammalian target of rapamycin in the treatment of luminal breast cancer. Anticancer Drugs. 2013;24(8):769–780. | |
Saini KS, Loi S, de Azambuja E, et al. Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat Rev. 2013;39(8):935–946. | |
Zhang X, Li XR, Zhang J. Current status and future perspectives of PI3K and mTOR inhibitor as anticancer drugs in breast cancer. Curr Cancer Drug Targets. 2013;13(2):175–187. | |
Cidado J, Park BH. Targeting the PI3K/Akt/mTOR pathway for breast cancer therapy. J Mammary Gland Biol Neoplasia. 2012;17(3–4):205–216. | |
Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.