Back to Journals » Infection and Drug Resistance » Volume 11
Molecular analysis of multidrug resistance in clinical isolates of Shigella spp. from 2001–2010 in Kolkata, India: role of integrons, plasmids, and topoisomerase mutations
Authors Rajpara N, Nair M, Chowdhury G , Mukhopadhyay AK, Ramamurthy T, Niyogi SK, Bhardwaj AK
Received 10 August 2017
Accepted for publication 10 October 2017
Published 12 January 2018 Volume 2018:11 Pages 87—102
DOI https://doi.org/10.2147/IDR.S148726
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Neha Rajpara,1,2 Mrinalini Nair,2 Goutam Chowdhury,3 Asish K Mukhopadhyay,3 Thandavarayan Ramamurthy,4 Swapan Kumar Niyogi,3 Ashima Kushwaha Bhardwaj1
1Department of Human Health and Diseases, Indian Institute of Advanced Research, Koba Institutional Area, Gandhinagar, 2Department of Microbiology and Biotechnology Centre, Maharaja Sayaji Rao University of Baroda, Vadodara, Gujarat, 3Department of Bacteriology, National Institute of Cholera and Enteric Diseases, Kolkata, 4Center for Human Microbial Ecology, Translational Health Science and Technology Institute, Faridabad, India
Abstract: To understand the genetic basis of high drug resistance in Shigella, 95 clinical isolates of Shigella spp. (2001–2010) were obtained from the Infectious Diseases Hospital, Kolkata, India. Ninety-three isolates were resistant to three or more antibiotics. Resistance to nalidixic acid, trimethoprim, streptomycin, and co-trimoxazole was most common in this population. Dendrogram analysis showed that S. sonnei strains were more clonally related when compared to the other Shigella species. The role of mobile genetic elements and chromosome-borne resistance factors was analyzed in detail. Integron analysis indicated the preponderance of class 2 and atypical class 1 integrons in that population. Typical class 1 integron was present in only one S. sonnei isolate and harbored trimethoprim resistance-encoding gene dfrV, while atypical class 1 integrons harbored dfrA1–aadA or blaOXA-aadA gene cassettes responsible for resistance to trimethoprim, aminoglycosides, and β-lactams. Class 2 integrons harbored either dfrA1-sat-aadA or dfrA1-sat gene cassettes. Most importantly, a novel gene cassette array InsE-InsO-dfrA1-sat was found in class 2 integron of S. sonnei NK4846. Many of the resistance traits for antibiotics such as trimethoprim, co-trimoxazole, kanamycin, ampicillin, and tetracycline were transferred from parent Shigella isolates to recipient Escherichia coli during conjugation, establishing the role of plasmids in horizontal transfer of resistance genes. Multiple mutations such as S80→I, S83→L, and D87→G/N/Y in quinolone resistance determining regions of topoisomerases from the representative quinolone-resistant isolates could explain the spectrum of minimal inhibitory concentration values for various quinolones. To the best of our knowledge, this is the first comprehensive report that describes the contribution of mobile (plasmids, integrons, and quinolone resistance genes named qnr) and innate genetic elements (mutations in topoisomerases) in determining the resistance phenotype of all the four species of Shigella over a span of ten years.
Keywords: mobile genetic element, conjugation, atypical class 1 integron, quinolone resistance, efflux pumps
Introduction
In February 2017, WHO published a global priority list of antibiotic-resistant pathogens that pose greatest threat to human health and for which new antibiotics are needed.1 Based on the urgency for development of new antibiotics, pathogens have been divided into critical, high, and medium category.1 Fluoroquinolone-resistant Shigella has been included as a medium priority pathogen in this list. This has reiterated that multidrug resistant (MDR) Shigella poses a threat to human health and underlines the need to unravel and understand the molecular basis of MDR in Shigella. Shigellosis caused by Shigella flexneri, S. sonnei, S. dysenteriae, and S. boydii mostly occurs in resource-poor communities that do not have proper sanitation.2 Annually, the number of Shigella episodes is estimated to be 80–165 million and ~74,000–600,000 deaths worldwide.3 Antibiotics are used for the treatment of shigellosis, but with increasing MDR pathogens, treatment of this disease has become complicated. There have been many reports around the world indicating the rising problem of MDR in shigellosis.4–12 Increase in resistance to fluoroquinolones, third-generation cephalosporins, and azithromycin has been reported globally.13
Resistance to antibiotics in bacteria has been attributed to chromosome-borne inherent and mobile genetic factors. For examples of chromosome-borne factors, an organism may lack the target of antibiotics (mutations in target genes) or export antibacterial agents before they exert their effect (efflux pumps) or may restrict an antibiotic from entering into the bacteria (porins). Mobile genetic elements (MGEs) include plasmids, integrons, and integrating conjugative elements (ICEs), which are potent vectors for acquisition and dissemination of antibiotic resistance genes among the bacterial populations.14–19 Plasmids evolved as an essential part of the bacterial genome, providing resistance genes for carbapenem resistance, extended spectrum beta-lactamase and quinolone resistance proteins that can be easily exchanged among bacteria by horizontal gene transfer.18 Integrons are gene-capture elements and till date five classes of integrons have been well characterized based on their integrase sequences.17 A typical class 1 integron consists of two conserved segments (CS) at their 5′ and 3′ ends, separated by a variable region that usually comprises of one or more extraneous gene cassettes. The 5′ CS region contains the integrase gene, the integration site, and a promoter region that allows the expression of any number of gene cassettes inserted at the attI1 site in a suitable orientation.17 The 3′ CS region usually comprises of qacEΔ1 encoding resistance to quaternary ammonium compounds and sul1 encoding resistance to sulphonamides.17 While the atypical class 1 integron consists of 5′ CS of class 1 integron and a variable region, it consists of an IS1 element at 3′ end in place of qacEΔ1 and sul1.16 Atypical class 1 integron was first found on the pathogenicity island carrying Shigella resistance locus, on chromosome of S. flexneri 2a strain YSH6000 where it harbored two resistance determinants for chloramphenicol and tetracycline.15,16 The presence of atypical class 1 integron in Shigella isolates has commonly been reported from Asia and has also now been reported from Africa.20–23 SXT elements are the best studied examples of ICE, which harbor the sulphonamide, trimethoprim, streptomycin, and chloramphenicol resistance genes.14
Shigellosis is endemic in India and is one of the major causes of diarrhea in this country 2,13 Due to varied subgroups, varied distribution of different subgroups in various geographical locations, and different location-specific antibiograms in Shigella infections, the problem of treatment of Shigella infection is multidimensional. Therefore, detailed analysis of the species/serogroup-specific antibiotic resistance patterns and the genetic factors governing these resistance phenotypes is important to understand these bacterial infections. For this purpose, 95 clinical isolates of Shigella were analyzed for their antibiotic resistance profiles. Various genetic factors borne on chromosomes or plasmids or integrons were deciphered as reasons for the observed MDR phenotypes.
Materials and methods
Bacterial strains and DNA isolation
Patients who took any antibiotic were not enrolled in this study. Confirmed isolate from each patient was included for further analysis. Only one isolate was taken from each patient. Stool specimens collected from patients with dysentery were analyzed for common enteric pathogens using standard microbiological technique.24 Subsequently, the identity of the Shigella isolates was confirmed by biochemical tests24 and they were serotyped with an antiserum kit (Denka Seiken Co. Ltd, Tokyo, Japan). These Shigella isolates were representation of a large number of Shigella cases that were treated at the hospital during the ten-year period. As performing detailed molecular analysis for such a large number of isolates was not feasible, only representative population was taken without any selection bias. Therefore, 95 Shigella isolates (S. flexneri [n=42], S. sonnei [n=42], S. dysenteriae [n=5], and S. boydii [n=6]) were collected from patients with bloody diarrhea admitted to the Infectious Diseases Hospital, Kolkata, India, from 2001 to 2010. The patients provided written informed consent for participating in the study, and in the case of children, written informed consent was obtained from their parents. The consent procedure was approved by the Institutional Ethical Clearance Committee of NICED. The study was approved by the Institutional Biosafety Committee of Indian Institute of Advanced Research, Gandhinagar, and the Review Committee on Genetic Manipulation governed by the guidelines laid down by the Department of Biotechnology, Government of India. Genomic and plasmid DNA were prepared as described previously.25 Plasmid DNA was prepared using alkaline lysis method.25,26 The plasmid DNA samples were electrophoresed in 1% agarose gel prepared in 1× Tris-Acetate-EDTA (TAE) using 1× TAE running buffer at 6–7 V/cm. 1 kb DNA ladder and λ Hind III marker were used to determine the molecular size of plasmids.
Pulsed field gel electrophoresis (PFGE) analysis
Genomic DNA for PFGE was prepared in agarose plugs using the method described earlier.27 PFGE of NotI- or XbaI-digested genomic DNA was performed using a CHEF-Mapper (Bio-Rad Laboratories) according to the PulseNet standardized protocol for subtyping of Shigella species. PFGE images were captured using a gel documentation system (Vilber Lourmat, Marne-la-Vallée, France). PFGE profiles were analyzed using the BioNumerics version 4.0 software (Applied Maths, Sint-Martens-Latem, Belgium). The tagged image file formats were normalized by using the universal Salmonella enterica serotype Braenderup (H9812) size standard on each gel against the reference in the database. In the dendrogram analysis, the PFGE profiles were matched using the Dice coefficient and unweighted pair group method using arithmetic averages. Clustering of PFGE profiles was made using 1.5% band position tolerance window and 1.5% optimization.
Antimicrobial susceptibility tests and minimal inhibitory concentration (MIC) assays
Shigella isolates were tested for their susceptibility to ampicillin (10 mg), azithromycin (15 mg), ceftriaxone (30 mg), cefuroxime (30 mg), chloramphenicol (30 mg), co-trimoxazole (1.25 mg trimethoprim/23.75 mg sulfamethoxazole), ciprofloxacin (CIP; 5 mg), gentamicin (10 mg), streptomycin (10 mg), trimethoprim (5 mg), tetracycline (30 mg), nalidixic acid (NAL; 30 mg), norfloxacin (NOR; 10 mg), kanamycin (30 mg), and ofloxacin (OFX; 5 mg) by disk diffusion method using commercial disks (HiMedia, Mumbai, India) in accordance with the criteria recommended by the Clinical and Laboratory Standards Institute (CLSI) standards.28 For the interpretation of antibiotic susceptibility, CLSI chart for enterobacteriaceae was used. MDR was defined as resistance acquired to at least one drug in three or more antimicrobial categories.29
MIC assays were performed for quinolones using commercial strips (EzyMIC strip; HiMedia) as per the manufacturer’s protocol. Escherichia coli ATCC 25922 was used for quality control in both the experiments. Experiments were performed at least three times.
Ascertaining the role of efflux pumps in drug resistance
To ascertain the role of efflux pumps in imparting resistance to drugs, synergy test was carried out as described earlier.30–32 The test was performed using various antibiotics, i.e., ampicillin, azithromycin, chloramphenicol, and CIP to which a particular clinical isolate was resistant. The efflux pump inhibitor carbonyl cyanide-m-chlorophenyl hydrazone (CCCP) was added on Mueller-Hinton Agar at 4 mg/L concentration. Susceptibility testing for antibiotics by MIC strip was performed as described in earlier section, both in the presence and absence of CCCP. A decrease in the MIC of the isolates in the presence of CCCP indicated the role of efflux pumps in reducing the concentration of that drug inside the cell by throwing it out.
Polymerase chain reaction (PCR)
Presence of factors such as integrons, SXT elements, quinolone resistance genes, and topoisomerases in Shigella isolates was established by PCR using primers specific for each element. Genomic DNA (200 ng) were used as templates in PCR with the primers and conditions mentioned in Table 1 for the screening of class 1, class 2, and class 3 integrons and SXT integrase. PCR was carried out with an initial denaturation at 95°C for 4 min. Subsequent to this, 25–30 amplification cycles were performed, each consisting of an initial denaturation at 95°C for 0.5 min followed by annealing and extension steps (Table 1). The final polymerization was carried out at 72°C for 10 min. Recombinant Taq polymerase (Thermo Scientific) was used along with appropriate buffers containing ammonium sulfate (1×) and magnesium chloride (2 mM).
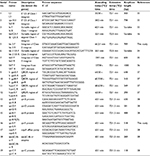  | Table 1 Primers used in the present study Abbreviations: CS, conserved segments; QRDR, quinolone resistance determining regions. |
All the quinolone-resistant isolates (n=85) were analyzed for plasmid-mediated quinolone resistance (PMQR) genes (qnrA, qnrD, qnrB, qnrS, oqxA, aac-(6′)-Ib-cr, and qnrC) by multiplex PCR using the conditions described earlier (Table 1).38 The positive controls for PMQRs were taken from the bacterial lysates derived from Enterobacter cloacae (qnrA), Klebsiella pneumoniae, and S. enterica Newport (qnrB), S. enterica Saintpaul (qnrS), S. enterica Typhimurium (aac(6′)Ib-cr), E. coli transformant pHSII (qnrC), and E. coli transformant p2007057 (qnrD).39–44 These bacterial lysates were procured from Technical University of Denmark (courtesy Dr Lina Cavaco). K. pneumoniae MTCC 7028 for oqxA was obtained from Microbial Type Culture Collection, Chandigarh, India.
Based on their quinolone resistance profiles, two or more representative isolates from each of the species were used to study the effect of topoisomerase mutations in quinolone resistance. A quinolone-sensitive isolate from each of the Shigella species was included as a control. The quinolone resistance determining regions (QRDRs) of topoisomerases (gyrA, gyrB, parC, parE) were amplified using the primers and conditions described in Table 1.37 PCR amplicons were sequenced and these sequences were compared with the standard or control Shigella topoisomerase sequences to detect the mutations.
Conjugation
Based on antibiograms, agarose gel analysis of plasmid preparations, and integron analysis, representative isolates of Shigella were selected to study transferability of their resistance traits to the recipient by conjugation.36,45 E. coli XL1-BLUE (resistant to tetracycline and NAL) or E. coli J53 KACC 16628 (resistant to sodium azide) was used as a recipient. Briefly, the recipient and donor were mixed in a ratio of 1:1 on a sterile 0.45 µm nylon membrane (Nytran N, Whatman) and incubated overnight for mating on Luria-Bertani agar at 37°C. Transconjugants were selected on MacConkey agar containing appropriate antibiotics and tested for acquired resistance traits by determination of their antibiotic susceptibility profiles.
Sequence analysis
DNA sequencing was performed by Sanger’s chain termination method using DNA sequencer (Applied Biosystems; 3730/3730xl DNA analyzer). All the amplicons except topoisomerases were cloned in pDrive (Qiagen) or pBAD (Invitrogen) TA cloning vectors. The amplicons (>1.0 kb) were sequenced by primer walking, and the sequences were assembled and analyzed by Basic Local Alignment Search Tool (BLAST). The ORF (open reading frame) finder tool at the NCBI website (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to predict all the possible ORFs in these sequences. Sixty seven assembled sequences were submitted to GenBank (accession numbers KX583660-KX583675, KX817766-KX817795, KX768278-KX768283, KX536824-KX536827, KX463270-KX463271, KX781232-KX781233, KX777251-KX777252, KX792556, and KX863674-KX863676).
Results
Bacterial isolates and their resistance profiles
Ninety five clinical isolates of Shigella included 42 S. flexneri, 42 S. sonnei, 6 S. boydii, and 5 S. dysenteriae isolates. Among S. flexneri isolates, serotype 2a was most common (22/42), followed by serotype 3a (11/42). Rest of the nine isolates consisted of serotypes 1a (2/42), 1b (3/42), 2b (2/42), and one each of serotypes 4a and 6. S. boydii population had representations from serotypes 1, 2, 4, 11 (2/6), and 12. S. dysenteriae consisted of serotypes 3 (2/5), 9, 5, and 12.
Out of the 95 isolates, 93 were resistant to three or more antibiotics. The percentage of Shigella isolates resistant to each of the antibiotics is shown in Table 2. Intermediate resistance and complete resistance were together considered to be a resistance trait. More than 90% of S. flexneri isolates showed resistance to streptomycin, trimethoprim, and tetracycline, and 69–85% showed resistance to NAL, chloramphenicol, co-trimoxazole, and CIP (Table 2). Above 80% of S. sonnei isolates showed resistance to NAL, kanamycin, trimethoprim, co-trimoxazole, streptomycin, and azithromycin. S. dysenteriae isolates chiefly showed resistance to co-trimoxazole, trimethoprim, tetracycline, NAL, and streptomycin. In S. boydii isolates, resistance to trimethoprim and streptomycin was prominent, i.e., above 80%.
  | Table 2 Percentage of drug resistance in Shigella isolates Notes: Complete resistant and intermediate resistant traits were considered to be resistant traits. |
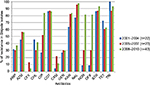
Year-wise analysis of the aforementioned antibiotic susceptibility profiles (Figure 1) revealed that the MDR status of these isolates persisted through all these years (Figures 2 and 3). Most importantly, during ten years, resistance to fluoroquinolone drugs such as CIP, NOR, and OFX was remarkably increased as compared to NAL (Figure 1). Resistance to either of the four quinolone drugs was 89.5%, while resistance to all the four quinolone drugs together was 47.4%. An interesting pattern was observed for resistance to cephalosporins. A bell-shaped pattern showed the maximal resistance to them (13% for ceftriaxone and 21.7% for cefuroxime) from the years 2005 to 2007. This resistance decreased in later years.
Clonality in Shigella isolates
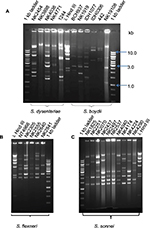
The genotypes obtained by PFGE were analyzed separately according to Shigella species: S. sonnei (Figure 2), S. flexneri (Figure 3), S. boydii (Figure S1), and S. dysenteriae (Figure S2). PFGE analysis of XbaI-digested chromosomal DNA of the 42 S. sonnei strains yielded 21 reproducible PFGE patterns. As shown in Figure 2, two PFGE patterns were shared by nine isolates (21%) from the years 2002, 2004, 2006, and 2009, and eight isolates (19%) from the years 2007 and 2008. The similarity index for all S. sonnei isolates was 92–100%, suggesting that they were clonally related to each other. PFGE analysis of S. flexneri after digestion with NotI revealed serotype-specific clusters, with ~80–100% similarity within each serotype. S. flexneri exhibited three distinct clades (Figure 3). Clade A and B belonged to S. flexneri 1a, 1b, and 4a and S. flexneri 3a isolates, respectively. Clade C consisted of S. flexneri 2a and 2b. PFGE analysis revealed that S. boydii isolates belonging to different serotypes had ~80% similarity and S. dysenteriae isolates were also observed to be 80% clonally related. As could be observed from Figures 2 and 3, where antibiogram profiles and PFGE profiles are aligned, there was no correlation between clonality and MDR status of the isolates belonging to the same clade.
Presence of integrons
Out of 95, 42 isolates were found to be positive for 5′ CS of class 1 integrase by PCR. When analyzed for the presence of 3′ CS, only one S. sonnei isolate IDH0734 yielded the expected 0.8 kb amplicon indicating that rest of the 41 isolates harbored atypical class 1 integron. Since IDH0734 harbored atypical class 1 integron with 5′ CS and 3′ CS, it was analyzed for its variable region where it yielded ~750 bp amplicon. Atypical class1 integrons were found in 38 out of 42 S. flexneri, two out of five S. dysenteriae, and one out of six S. boydii isolates (Table 3). For analysis of atypical class 1 integrons, their variable regions were amplified with the primer pair intI1CA F/IS1CA R in all the 41 isolates. Gel analysis of PCR products showed a major band with mobility of 2.4 kb in S. flexneri (2a, 1a, 1b, and 2b) and S. dysenteriae, while 2.0 kb band was observed in S. flexneri 3a and S. boydii.
  | Table 3 Presence of integrons in Shigella isolates and the gene cassettes found in the variable regions of integrons |
In PCR, 83.2% isolates (79/95) were found to be positive for class 2 integrase (Table 3). Integrase 2-positive isolates were subsequently used to amplify variable regions using the primers specific for 5′ and 3′ conserved sequences (Int2VA F/Int2VA R primers) of class 2 integrons. Amplicons with varying sizes (1.4 kb or 2.4 kb or 2.2 kb) were obtained. Of the class 2 integrase–positive S. flexneri isolates, 28 out of 29 harbored 2.2 kb variable region and H20145 harbored 1.4 kb variable region. Of the S. sonnei isolates, 41 out of 42 were positive for class 2 integrase and all of them except two carried 1.4 kb variable region. One of these two harbored 2.4 kb variable region, while the other one did not show any amplification. All the five S. dysenteriae and four out of six S. boydii isolates were positive for class 2 integrase, and these harbored either 1.4 kb or 2.2 kb variable regions. Class 3 integron and SXT element were absent in these isolates.
Presence of both atypical class 1 and class 2 integrons in 28 out of 42 S. flexneri and two out of five S. dysenteriae isolates (Table 3) underlined the important role of integrons in imparting drug resistance to these isolates, also later proved by sequence analysis. In the S. sonnei population, only IDH0734 carried both typical class 1 and class 2 integrons (Table 3).
Sequence analysis of integrons
Sequences corresponding to 5′ CS and 3′ CS described in the earlier section were analyzed and submitted to GenBank (KX768278-KX768283 and KX777252). As mentioned earlier, these Shigella isolates carried variable regions with a major band of 750 bp for typical class 1 integron: bands of 2.4 kb and 2.0 kb for atypical class 1 integrons. Variable regions of 1.4 kb, 2.4 kb, and 2.2 kb were obtained in class 2 integrons. For each species, a sample representative of each band size was used for the sequence analysis. For example, two band sizes were found in variable regions of class 2 integrons in S. flexneri isolates, so representative 1.4 kb and 2.2 kb variable regions were sequenced.
Sequence of the variable region of typical class 1 integron from S. sonnei IDH0734 revealed that it encoded dfrV (KX777251) responsible for trimethoprim resistance. The 2.0 kb variable region of atypical class 1 integron in S. flexneri 593 and S. boydii NK1919 (Table 3) showed the cassettes dfrA1-aadA (KX817770 and KX817771). The 2.4 kb variable region of atypical class 1 integron from S. flexneri 102 and S. dysenteriae 1244 carried blaoxa-aadA (KX817769 and KX951422, respectively) gene cassettes (Table 3). Therefore, atypical class 1 integrons carried resistance traits for trimethoprim (dfrA1), beta-lactams (blaoxa), and aminoglycosides (aadA).
Sequences of class 2 integrases from each species were analyzed by ORF finder tool and submitted to GenBank (KX536824-KX536827, KX463270-KX463271). Results revealed that these were non-functional class 2 integrase genes carrying an internal stop codon TAA. This was in accordance with earlier observation that the gene encoding class 2 integrase contains a nonsense mutation in codon 179 (ochre 179) and thereby it yields a non-functional protein which can be recovered by a single mutation.46 Sequences of variable regions of class 2 integrons were analyzed from the representative isolates of each species (Table 3). The 1.4 kb band from S. flexneri H20145, S. boydii 442, and S. sonnei L1137 harbored dfrA1-sat (KX817766, KX817768, and KX781233, respectively) cassettes, while 2.2 kb band from S. flexneri 102 and S. dysenteriae 1244 carried dfrA1-sat-aadA gene cassettes (KX817767 and KX792556, respectively). S. sonnei NK4846 harbored a new cassette array InsE-InsO-dfrA1-sat (KX781232) of 2.4 kb band size on class 2 integron (Table 3). Therefore, class 2 integrons in this population of Shigella isolates carried the resistance traits for trimethoprim (dfrA1), streptothricin (sat), and aminoglycosides (aadA). In addition, the genes for transposases or insertion sequences (InsE, InsO) were also observed in one of the cassettes.
Quinolone resistance
For deciphering quinolone resistance due to mutations, sequences from the QRDRs of topoisomerases were analyzed from the quinolone resistant/sensitive isolates derived from each Shigella species as described in section “Materials and methods”. These sequences from the four topoisomerases, i.e., GyrA, GyrB, ParC, and ParE, were submitted to GenBank (KX583660-KX583675 and KX817772-KX817795). These isolates were also analyzed for their MICs to four quinolones (NAL, CIP, OFX, and NOR) in order to correlate the effect of mutations on their quinolone resistance phenotypes. A variety of mutations were detected in these isolates (Table 4). S. flexneri NK2640 (sensitive to all the tested quinolones) carried mutation in GyrA at V196→A outside the QRDR region. S. flexneri 2a isolate 102 highly resistant to all the four quinolones carried multiple mutations in GyrA (S83→L, D87→N, and H211→Y) and ParC (S80→I and E84→G). This was also evident in their high MIC values: >256 µg/mL for CIP, NAL, and NOR and >32 µg/mL for OFX. Two mutations D87→Y (inside the QRDR) and V196→A (outside the QRDR) were found in GyrA of NAL-resistant S. sonnei NK4219, while three mutations S83→L, D87→G, and V196→A were observed in GyrA of S. sonnei IDH1694 (resistant to all the quinolones). S. sonnei IDH1694 also harbored mutation S80→I in ParC region. S. sonnei IDH0734 which carried two mutations in GyrA (S83→L, D87→G) and one mutation in ParC (S80→I) showed much higher MIC values (48–192 folds) and resistance to all four quinolones as compared to S. sonnei NK4846 (Table 4). S. dysenteriae 1244 and S. boydii 442 harbored a mutation S83→L in GyrA. These mutations were not observed in control isolates S. dysenteriae NK4036 and S. boydii NK1919. No mutations were observed in GyrB and ParE from all these isolates. Therefore, as clearly depicted in Table 4 and discussed earlier, the increase in the number of mutations directly correlated with increase in resistance to spectrum of quinolones.
Results of a multiplex PCR for seven PMQR genes revealed that they were absent in these isolates except S. flexneri isolate M11560 that harbored a qnrS gene. The monoplex PCR for each PMQR gene was standardized using the controls described in section “Materials and methods”. Subsequently, multiplex PCR for all the seven genes was performed with Shigella isolates.
Transfer of resistance by conjugation
Shigella isolates harbored multiple plasmids ranging from 1.0 kb to 23 kb as observed by agarose gel (Figure S3). Based on antibiotic susceptibility, plasmid profile, and presence of integrons, six isolates were selected from the present population to examine the resistance traits transferable through conjugation (Table 5). The choice of recipient E. coli was based on the resistance profile of donor isolates. Transconjugants of S. boydii 442 were selected on trimethoprim and tetracycline, while transconjugants from rest of the Shigella isolates were selected either on trimethoprim+sodium azide or streptomycin+sodium azide. The transconjugants were obtained successfully with conjugation efficiencies ranging from 10−6 to 10−7 transconjugants per recipient cell (Table 5). These experiments showed the transferability of resistance traits such as ampicillin, azithromycin, chloramphenicol, CIP, co-trimoxazole, kanamycin, NAL, OFX, streptomycin, tetracycline, and trimethoprim to recipient E. coli, establishing the role of plasmids in horizontal gene transfer (Table 5). Most commonly, the resistance traits for co-trimoxazole and trimethoprim were transferred in all the cases, confirming the carriage of trimethoprim resistance gene on plasmids.
Agarose gel analysis of the transconjugants showed that multiple plasmids were transferred during conjugation (Table 5). Therefore, associating the resistance traits to any particular plasmid was not possible. In addition, the comparison of plasmid profiles from the parent isolates and their transconjugants showed that most of the plasmids visualized on agarose gel were transferred during conjugation except for the ones shown in bold (Table 5).
Role of efflux pumps in drug resistance phenotype
The role of efflux pumps in the antibiotic resistance phenotype of the isolates used in the aforementioned studies was assessed using synergy test. MIC for different group of antibiotics was evaluated with or without efflux pump inhibitor CCCP and change in MIC was observed. The test was carried out with the antibiotics for which these Shigella isolates were resistant. S. flexneri 129 that was sensitive to all the antibiotics tested was included as a control. A reduction of 0.65- to 2.0-fold in MIC was observed on addition of CCCP as compared to the CCCP-free control in all the tested Shigella isolates (Table 6). This fold reduction in MIC was also observed in the control strain S. flexneri 129 that was sensitive to all these drugs. In the same assays, the other control Vibrio fluvialis isolates showed 1.33- to 5.33-fold changes in MIC. Therefore, these results revealed that there was no significant change in MIC in the presence of CCCP, indicating that efflux pumps did not play a major role in the resistance to these drugs (Table 6). It was also observed that the S. flexneri 2a isolate 102 had very high MICs in all the drugs tested.
Discussion
Antibiotic resistance within wide range of pathogenic bacteria is a growing public health concern globally. It hampers the effective prevention and treatment of infectious agents. Therefore, to mitigate the problem of MDR, it becomes pertinent to decipher various factors/mechanisms involved in antibiotic resistance of these infectious bacteria. This study was carried out to determine the patterns of antimicrobial resistance in 95 clinical isolates of Shigella (from 2001 to 2010) from Kolkata, India, and to unravel the possible genetic factors responsible for the observed resistance phenotypes. In this population of Shigella isolates, there was a predominance of S. flexneri and S. sonnei. Except one or two, all the isolates were resistant to three or more antibiotics out of the 15 antibiotics tested. This enormity of MDR was in accordance with the WHO reports published recently.1,11 These reports cautioned that the danger of increasing MDR was resulting in treatment failure that could lead to mortality even in the case of minor injuries and common infections in the post-antibiotic era.11 A WHO report published in February 2017 has included Shigella in the list of priority pathogens for which new antibiotics are urgently needed.1 Our study presented here clearly shows the prevalence of MDR Shigella, which as specified by WHO, could be a matter of serious concern. In the present study, resistance to quinolone drugs used for treatment of diarrheal diseases was observed to increase markedly through years. Emerging fluroquinolone resistance has also been earlier reported in Shigella spp. from India.5,47 Increase in fluoroquinolone resistance in these isolates could be attributed to an increase in the clinical prescription or over-the-counter sale and use of these drugs.48,49 Interestingly, the resistance for ampicillin, azithromycin, and cephalosporins (ceftriaxone and cefuroxime) was either not remarkably increased or reduced with years, a desirable feature for drug management of the disease. There are large numbers of reports on emergence of extended spectrum beta-lactamase producers from many countries.50 Generally, most of the shigellae remained susceptible to cephalosporins as this group of antibiotics are less used in Kolkata for the treatment of acute diarrhea/dysentery. Clonality analysis using PFGE revealed that the clonality of these isolates could not be correlated with their antibiograms.
Molecular analysis of the genetic factors that could be responsible for the observed MDR phenotypes indicated a major role of integrons, plasmids (MGEs), and topoisomerase mutations (chromosome-borne). Most strikingly, integrons of various classes, i.e., class 1 integrons, class 2 integrons, and atypical class 1 integrons were present in the majority (90/95) of the isolates, where the gene cassettes harbored by their variable regions conferred drug resistance traits on the parent isolates. Out of 95, 31 isolates harbored both class 1 and class 2 integrons. In addition to integrons, presence of multiple plasmids in all the isolates could also be the possible source of drug resistance as supported by conjugation experiments. Predominantly, the genes for trimethoprim and aminoglycoside resistance were associated with these three classes of integrons. Typical class 1 integron was present in only one S. sonnei isolate, while atypical class 1 integron was found in S. flexneri, S. dysenteriae, and S. boydii. Class 2 integron was highly prevalent in these Shigella isolates (83%). Class 2 integrases are non-functional proteins due to an internal stop codon at 179th position of the protein sequence.46,51 Therefore, the majority of the cassette arrays on class 2 integrons are usually constant. S. sonnei NK4846 harbored a new cassette array InsE-InsO-dfrA1-sat on class 2 integron with insertion elements and resistance genes for trimethoprim and streptothricin. In an earlier report, satI gene cassette was interrupted with the IS911 element on class 2 integron of S. sonnei isolates.52 In another report, a class 2 integron with an IS630 element was found in a S. flexneri isolate, and a third report described a class 2 integron with an IS1 in an E. coli.34,53 S. sonnei NK4675 harboring the class 2 integrase did not show amplification with primers specific to the variable region of class 2 integron suggesting either the presence of null integron or mutations in the regions where primers anneal for amplifying the variable regions. Therefore, in this study, MGEs such as plasmids, class 1 integron, atypical class 1 integrons, and class 2 integrons seemed to play an important role in dissemination of drug resistance in these isolates.
Previously, Shigella spp. were susceptible to co-trimoxazole, but on emergence of resistance to this antimicrobial, treatment recommendations were shifted to quinolone group of antibiotics and azithromycin.11 Eventually, these bacteria also developed quinolone resistance.11,13,37,54 In this study, majority of the isolates were resistant to NAL, and resistance to other quinolone antibiotics such as CIP, NOR, and OFX was higher in S. flexneri as compared to S. sonnei. Resistance to quinolones is generally caused due to mutations in topoisomerase genes, efflux pump activity, and qnr and aac(6′)-Ib-cr genes.19 In this study, an interesting array of mutations was observed in the QRDR regions of topoisomerases. Clearly, mutations in GyrA S83 or D87 positions were chiefly responsible for resistance to NAL. Mutations in GyrA have been shown to be a major reason for resistance to quinolone in various organisms such as Vibrios, Shigella, and Salmonella.13,19,30,36,45,55 An earlier study from the Democratic Republic of the Congo revealed that these mutations in Salmonella enterica serover Typhi were also responsible for the decreased CIP susceptibility.55 Wherever mutations were detected in both the topoisomerases GyrA and ParC, wider spectrum of resistance to multiple quinolone drugs was observed concomitant with high MIC values. In addition, a V196→A mutation outside the QRDR of GyrA did not appear to contribute toward quinolone resistance.
In synergy test, the isolates resistant to the tested drugs did not show a significant decrease in MIC after disruption of efflux pumps with CCCP. This indicated that efflux pumps did not play a role in mediating resistance to these drugs. Some other laboratories have shown a 64–256 times decrease in MICs for some of the mutants of Shigella, but parent isolates showing this substantial decrease in MIC have not been reported.30
Present study was not a surveillance study but strictly aimed at understanding the genetic factors responsible for emergence and dissemination of MDR. Therefore, though the number of isolates used in this study was generally small, it was sufficient to lend an insight into the genetic basis of observed drug resistance phenotypes in this geographical location. Another limitation of the study was that the patients were not followed up to get additional information on duration of shedding of MDR Shigella isolates.
To summarize, the study has indicated the prevalence of highly drug-resistant pathogens belonging to the genus Shigella from the region of Kolkata. Interplay of large number of genetic factors such as plasmids, integrons, and multiple mutations accounted for the extensive drug resistance found in these isolates.
Acknowledgments
The authors are grateful to Prof VK Chaudhary and Ms Shilpi, University of Delhi, Delhi, for sequencing of the DNA fragments. The authors are thankful to Dr Lina Cavaco, Technical University of Denmark, for sending positive controls for qnr genes and Korean Agricultural Culture Collection for providing the E. coli J53 strain. The authors also thankfully acknowledge the Puri Foundation for Education in India for providing infrastructure facilities. The technical support and advice provided by Mr Priyabrata Mohanty, Mr K Vinothkumar, Ms Aneri Shah, and Mr Shaileshkumar Bhalara are thankfully acknowledged.
The laboratory is supported by the grants from Indian Council of Medical Research, New Delhi, India (AMR/49/11-ECDI), and Gujarat State Biotechnology Mission, Department of Science and Technology, Government of Gujarat (GSBTM/MD/PROJECTS/SSA/1535/2013-14). NR is a recipient of Senior Research Fellowship from Indian Council of Medical Research, New Delhi, India.
Disclosure
The authors report no conflicts of interest in this work.
References
Supplementary materials
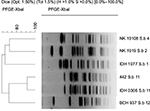  | Figure S1 Dendrogram of XbaI-digested pulsed-field gel electrophoresis profiles of the clinical isolates of Shigella boydii isolates. Scale bar indicates degree of similarity. |
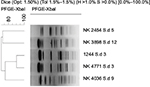  | Figure S2 Dendrogram of XbaI-digested pulsed-field gel electrophoresis profiles of the clinical isolates of Shigella dysenteriae isolates. Scale bar indicates degree of similarity. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.