Back to Journals » International Journal of Nanomedicine » Volume 13
Modification of the surface of titanium with multifunctional chimeric peptides to prevent biofilm formation via inhibition of initial colonizers
Authors Zhang X, Geng H, Gong L, Zhang Q, Li H, Zhang X, Wang Y, Gao P
Received 11 April 2018
Accepted for publication 19 June 2018
Published 12 September 2018 Volume 2018:13 Pages 5361—5375
DOI https://doi.org/10.2147/IJN.S170819
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Linlin Sun
Xi Zhang,1 Hongjuan Geng,1 Lei Gong,2 Qian Zhang,1 Hongjie Li,1 Xu Zhang,1 Yonglan Wang,1 Ping Gao1
1School and Hospital of Stomatology, Tianjin Medical University, Tianjin 300070, China; 2Department of Esophageal Cancer, Tianjin’s Clinical Research Center for Cancer and Key Laboratory of Cancer Prevention and Therapy, National Clinical Research Center for Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin 300070, China
Background: Prevention of bacterial colonization remains a major challenge in the field of oral implant devices. Chimeric peptides with binding, antimicrobial, and osteogenesis motifs may provide a promising alternative for the inhibition of biofilm formation on titanium (Ti) surfaces.
Methods: In this study, chimeric peptides were designed by connecting an antimicrobial sequence from human β-defensin-3 with a Ti-binding sequence and arginine-glycine-aspartic acid using a glycine-glycine-glycine linker. Binding to the Ti substrate and antimicrobial properties against streptococci were evaluated. Significant improvement in reduction of bacterial colonization onto the Ti surface was observed, with or without the presence of saliva or serum. The MC3T3-E1 cells grew well on the modified Ti surfaces compared with the control group.
Results: The data showed that the three peptide functional motifs maintained their respective functions, and that the antibiofilm mechanism of the chimeric peptide was via suppression of sspA and sspB gene expression.
Conclusion: These results indicated that the endogenous peptide fragments engineered on the Ti surface could provide an environmentally friendly approach for improving the biocompatibility of oral implants.
Keywords: multifunctional chimeric peptide, modification of titanium surface, human β-defensin-3, Ti-binding peptide-1, arginine-glycine-aspartic acid-containing peptides
Introduction
Dental implants have become an established and indispensable therapy for providing adequate support for the replacement of missing teeth.1,2 According to Geckili’s study, success rates of 76%–84% based on gender and age have been reported.3 Nevertheless, increasing evidence of the presence of peri-implantitis has been reported, representing one of the most frequent complications that can lead to the loss of the implant.4–6 Peri-implantitis is a progressive and irreversible disease of the hard and soft tissues surrounding the implant and is accompanied by bone resorption7 resulting from biofilm formation on the implant surface.8 The aim of this study, therefore, was to control the initial colonizers on the titanium (Ti) surface and thereby decrease or inhibit biofilm formation on the surface.
Oral biofilm formation is the initiating factor of a number of diseases. The microbial composition of peri-implantitis-associated biofilms is very similar to that observed in periodontitis.8 Biofilm formation normally involves three steps,9 the first of which requires the adhesion and attachment of initial colonizers to the surface. The importance of these early bacteria lies in the fact that they modify the environment for subsequent colonizers, such as Porphyromonas gingivalis.10 Streptococcus spp. are among the most important bacteria in the early stages of biofilm formation,11 which control the growth and attachment of streptococci on the implant surface and thereby disrupt subsequent pathogenic bacterial attachment, thus preventing bacterial biofilm formation on the surface.
Owing to the disadvantages of systematic administration of antibiotics, research has increasingly focused on the development of biomaterials that prevent bacterial adhesion or completely eliminate infection.12 To achieve this, various surface modification strategies have been developed that improve the antibacterial properties of biomaterials.13–16 One simple strategy that has few side effects is to engineer the implant surface by attaching antimicrobial peptides (AMPs), especially those of human origin. Human β-defensin-3 (hBD3) is an endogenous AMP with broad-spectrum activity against both gram-positive and gram-negative bacteria, and is not influenced by physiological NaCl concentration or the order of disulfide pairing.17–20 Recently, three fragments derived from hBD3 have shown cell- and skin-penetrating properties and exhibited anti-inflammatory activity.21 Investigation of whether these three sequences can exert antibacterial activity and inhibit biofilm formation on the surface of dental implants is of interest.
In this study, three fragments of hBD3 and arginine-glycine-aspartic acid (RGD)-containing peptides, which can induce osteoblast differentiation and mineral deposition,22 were bonded using glycine-glycine-glycine (GGG) to a chimeric peptide. Potential applications for this multifunctional peptide were explored. In addition, Ti-binding peptide-1 (TBP-1) was utilized to connect the chimeric peptide with the Ti surface.23 The purpose of this study was to investigate the application of the multifunctional chimeric peptide on implants for the prevention of biofilm formation.
Materials and methods
Peptide design and synthesis
The peptides were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The final chimeric sequences purity was>95% as determined by mass spectrometry. The information about the basic properties and the secondary structures of the sequences was obtained by means of the peptide property calculator (http://www.pepcalc.com/). Different concentrations of peptide solution were prepared with a sterile phosphate buffer (1 × PBS). The concentration of peptides used in this study was 0.5 MIC unless specially noted.
Preparation of Ti substrates
Ti substrates were prepared as pure Ti (99.998% purity) films with a thickness of 300 nm on silicon wafers by electron beam evaporation (Sharon Vacuum, Brockton, MA, USA). All Ti slices were ultrasonically rinsed with acetone and 70% (v/v) ethanol for 20 minutes, and then oven-dried at 80°C and sterilized at 120°C.
Preparation of modified Ti substrates
Ti substrates prepared according to the above method were placed into sterile PBS solution (2 mL) containing either three chimeric peptides (320 g of each) as the experimental group or no chimeric peptide for the control group. The resulting mixtures were cultured at room temperature for 2 hours, after which the Ti substrates were rinsed with sterile PBS and deionized water, and then dried under argon.
Circular dichroism (CD) spectroscopy
Samples (0.1 mg/mL) were placed in 1.00 mm cuvettes. CD measurements were obtained at 4°C or 37°C with a scanning speed of 60 nm/min by a CD spectrometer (J-810; BIO-LOGIC Corp., Seyssinet-Pariset, France), recorded from 350 to 190 nm with solvent subtraction. To obtain quantitative analysis, the acquired CD spectra of peptides were analyzed using a SELCON3 algorithm.
Raman spectroscopy
The samples (5 mg) were measured by a Raman spectrometer (Renishaw, Gloucestershire, UK) using 785 nm laser excitation (Coherent, Santa Clara, CA, USA) at an intensity of 2 mW and an exposure time of 10 seconds, aimed directly at the sample. The spectrum of the samples was performed at 300–1,800 cm−1 with a resolution of 16×16 μm.
Zeta potential analyses
The zeta potentials of the samples were determined using a ZetaPALS (BI-200SM; Brookhaven, Holtsville, NY, USA). Prior to the sample analysis, the instrument was calibrated with three pH 7 standards. Pressure was kept at 400 mbar and all analyses were performed at 37°C.
X-ray photoelectron spectroscopy (XPS) analysis
The chemical composition on the Ti surfaces was measured by XPS spectrometer (ESCALAB 250) equipped with a monochromatized Al Ka X-ray source (1486.6 eV photons). Take-off angle was at 45° with a power of 150 W by under vacuum (10−8 Torr or lower). The main C1s peak was set to a value of 284.80 eV as a standard to calibrate all other binding energies.
Antibacterial assays
The bacterial strains studied were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The strains used were Streptococcus oralis (ATCC No 9811), Streptococcus sanguinis (ATCC No 10556), and Streptococcus gordonii (ATCC No 10558). S. oralis was cultured aerobically on brain heart infusion (BHI) agar plate supplemented with 1% yeast extract for 24 hours at 37°C. S. gordonii and S. sanguinis were cultured separately in freshly prepared BHI agar plates supplemented with sterile defibrinated sheep blood (5%), hemin (1%), and menadione (0.1%) under anaerobic conditions of 80% N2, 10% H2, and 10% CO2 for 48 hours at 37°C.
Minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) measurements
The MIC of the target fragments was investigated against streptococci using the standard broth microdilution method of the National Committee for Clinical Laboratory Standards (NCCLS document M7-A3). The MIC was taken as the lowest concentration of peptides at which no growth of the bacteria was observed visually. After bacteria were cultured with various concentrations of peptide based on the MIC for 24 hours, 20 μL of each sample well was removed and spread on BHI agar plates. The concentration at which more than 99.9% of the cells were inhibited was considered the MBC.
Biofilm susceptibility assay
Biofilms of streptococci chosen for this study were incubated with peptides in 96-well microtiter plates at 37°C for 24 and 72 hours. The biofilms were fixed and stained with crystal violet (0.5% w/v), and the absorbance was measured with a microplate reader at 600 nm.
Confocal laser scanning microscopy (CLSM) examination of biofilm samples, with and without peptide treatment
We divided the Ti substrates into four treatment groups: 1) PBS (blank Ti), 2) peptide-modified Ti, 3) peptide-modified Ti with bovine calf serum (HyClone, Logan, UT, USA), and 4) peptide-modified Ti with saliva (from six volunteers) sterilized by syringe filter. We treated groups (3) and (4) with 20 μL of serum or saliva for 4 hours. For CLSM (IX71; Olympus, Tokyo, Japan), bacterial suspension (5×105 CFU/mL, 1 mL) was added to all wells and the 24-well plates were cultured for 24 and 36 hours. To recognize the peptides, 7-amino-4-methylcoumarin (AMC) was introduced for staining. The Ti specimens were dyed with AO/EB (1:1, 20 μL) to view live/dead cells simultaneously at room temperature.
Antibacterial efficacy
Streptococci cultures were diluted to 5×105 CFU/mL and cultured on the modified Ti surfaces. After a period of 24 or 72 hours, the Ti samples were treated by ultrasonic at 40 W for 5 minutes. The antibacterial rates were calculated with the following formula: R = (B - A)/B×100%, where A is the average number of viable bacteria on a modified Ti surface and B is the average number of viable bacteria on the untreated Ti surface.
Scanning electron microscopy (SEM) examination
Peptide-treated Ti samples were washed by PBS, and then S. gordonii suspension (5×105 CFU/mL) was added to every well. After culturing for 12 or 24 hours, samples were prefixed with phosphate-buffered 2.5% glutaraldehyde (BioChemika, Fluka), rinsed with PBS three times and observed by SEM (Jeol JSM-6310LV; JEOL Ltd., Tokyo, Japan).
Transmission electron microscopy (TEM) examination
After being treated with TBP-1-RGDS-hBD3-3, or without the peptide (control group), ultrathin slice samples for TEM were prepared using reported methods.24 After dehydration with ethanol, the samples were cut into ultrathin slices and stained with 2% uranyl acetate and 0.2% lead citrate before observing under electron microscopy (JEM-2100F; JEOL Ltd.).
Cytotoxicity test
MC3T3-E1 murine pre-osteoblasts were bought from the Chinese Academy of Sciences (Shanghai, China). They were cultured in DMEM with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 100 mg/mL streptomycin (Gibco) and 100 U/mL penicillin (Sigma). MC3T3-E1 cells were then planted on the Ti surfaces at 37°C. The cytotoxicity test was measured at different time points (1 day, 3 days, 5 days, or 7 days) by using Cell Counting Kit-8 (CCK8; Dojindo Laboratories, Kumamoto, Japan).
Quantitative RT-PCR analysis
The sspA and sspB gene expression was evaluated according to the 2(-DeltaDeltaC(T)) method.25 Total RNA was isolated using the Bacterial RNA Isolation Kit (Thermo Fisher Scientific). Reverse transcription PCR was performed using a PrimeScript RT Reagent Kit (Takara, Shiga, Japan) with gDNA Eraser. Quantitative RT-PCR was performed with STBR Green reactions (Stratagene). The products were measured on the Real-Time PCR system (iQ™5; Bio-Rad Laboratories Inc., Hercules, CA, USA). Gene-specific primers are listed in Table S1 and were developed by Primer3 (version 0.2).26
Statistical analysis
Each experiment was performed a minimum of three replicate times. The mean and SD values were calculated by a data analysis software (SPSS Inc., Chicago, IL, USA). ANOVA and independent sample t-tests were carried out to calculate the significance among groups, a P-value <0.05 was considered to be statistically significant.
Results
Physicochemical properties of peptides
To investigate the structure of the peptides, their theoretical physicochemical properties were first predicted using the peptide calculator. The data (Figure 1) revealed the basic parameters of these peptides, which indicated that all peptides were estimated to have satisfactory solubility. TBP-1-RGDS-hBD3-3 had more hydrophilic groups (Figure 1) and exhibited a higher net positive charge compared with the other peptides. The all three chimeric peptides had hydrophilic and hydrophobic segments.
  | Figure 1 (A–C) Molecular characteristics of the peptides (TBP-1-RGDS-hBD3-1, TBP-1-RGDS-hBD3-2, and TBP-1-RGDS-hBD3-3) showing amphipathic properties (top: hydrophilic; bottom: hydrophobic). |
Secondary peptide structure
More information of the secondary structure and properties of the peptides was obtained by CD, Raman spectroscopy, and Zeta potential measurements. The three peptides dissolved in PBS (0.1 mg/mL) had α-helical, β-strand, β-turn, and random coil structures at 37°C (Figure 2A and C) as shown in the CD spectra. Quantitative analysis indicated that the secondary structures of TBP-1-RGDS-hBD3-1 and TBP-1-RGDS-hBD3-2 contained similar secondary structure ratio. In TBP-1-RGDS-hBD3-3, the proportions of β-strands (24.1%) were higher and those of α-helices (20.7%) and β-turns (19%) were lower compared with those of the other two peptides (11%). Compared with the structural composition at 4°C (Table S2), in addition to TBP-1-RGDS-hBD3-2 structural changes, the other two segments were stable with no significant changes to the secondary structure observed at 37°C.
The results of Raman spectroscopy in the wave number range 300–1,800 cm−1 are summarized in Figure 2B and D. The bands at 1,240, 1,340, 1,430, and 1,666 cm−1 were attributed to a coil structure,27 α-helical conformation,28 CH2CH3 deformation,29 and β-strand structure,27 respectively. The Raman spectroscopy results showed that the three peptides shared α-helical and β-strand structures, which was consistent with the CD results. Based on the above results and previous studies, it was hypothesized that three chimeric peptides may exhibit antibacterial properties, which is worth further exploring.
The zeta potential of TBP-1-RGDS-hBD3-1, TBP-1-RGDS-hBD3-2, and TBP-1-RGDS-hBD3-3 was observed to be 5.58, 3.34, and 12.09 mV, respectively (Figure 2E). This was in agreement with the theoretical result obtained using the peptide calculator that these peptides were positively charged at pH 7.4. TBP-1-RGDS-hBD3-3 had the highest zeta potential, suggesting that it possesses good stability and greater potential for therapeutic applications.
Adsorption of chimeric peptides on the Ti surfaces
The binding elements’ compositions of the Ti surface were measured using XPS. Figure 3 shows the data of the surface elemental composition after PBS treatment (control) and peptide modification (experimental group). The chief components of C1s, O1s (531.18 eV), Na1s (1,071.83 eV), P2p (133.19 eV), Cl2p (198.69 eV), and Ti2p3 (458.30 eV) were confirmed in the XPS results shown in Figure 3. All binding energies were referenced to the C1s spectrum peak (284.80 eV) as an internal reference after calibrating peak positions. The peaks of Na1s, P2p, and Cl2p were primarily due to the presence of PBS. Compared with the results of the PBS-treated Ti samples (Figure 3, control), the appearance of the distinctive N1s and S2p peaks originated from peptides absorbed onto the Ti surface (Figure 3, TBP-1-RGDS-hBD3-1/2/3) appeared to verify that the peptides had successfully been immobilized on the surface of the Ti substrates. The quantitative XPS data also supported the results (Figure 3).
Antibacterial assays
MIC and MBC
The MIC and MBC of the peptides against the planktonic streptococci were determined using the broth dilution technique to evaluate the antibacterial strength. As shown in Table 1, the MICs for TBP-1-RGDS-hBD3-3 indicated an average bacteriostatic concentration of 600, 850, and 850 μg/mL, respectively, against S. oralis, S. gordonii, and S. sanguinis, which was lower than that for TBP-1-RGDS-hBD3-1 and TBP-1-RGDS-hBD3-2 against these streptococci. MBC tests showed the same trend. According to these results, all the three chimeric peptides exhibited antibacterial activity, with TBP-1-RGDS-hBD3-3 exhibiting the greatest antibacterial properties among them.
Anti-biofilm test
The antimicrobial efficacy of peptides against streptococci biofilm states was investigated by anti-biofilm test. The three peptides were shown to be significantly effective (P<0.01) in disrupting the 24- and 72-hour biofilms (Figure 4). Among them, TBP-1-RGDS-hBD3-3 showed higher anti-biofilm activity (P<0.01). There was no significant difference (P>0.05) between the peptides’ activity in killing 24- or 72-hour biofilms.
Based on the results of the above comparison and analysis, it was confirmed that the antibacterial properties of TBP-1-RGDS-hBD3-3 merited further study and was therefore chosen as the most promising candidate to modify Ti surfaces in the subsequent experiments.
Antibacterial efficacy of TBP-1-RGDS-hBD3-3-modified Ti surfaces
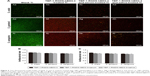
CLSM images (Figure 5A) show the total number of bacterial colonies using live/dead stain, attached to Ti slices modified with TBP-1-RGDS-hBD3-3 and cultured with 20 μL of serum or saliva for 4 or 24 hours. For the blank Ti substrate, green-stained cells (S. oralis, S. gordonii, and S. sanguinis) fully covered the substrate (Figure 5Aa, e, and i). For the substrates modified with TBP-1-RGDS-hBD3-3 (Figure 5Ab, f, and j) or modified with TBP-1-RGDS-hBD3-3 and treated with serum or saliva (Figure 5Ac, d, g, h, k, and l), the cells adhered to almost all of the substrate that were most red-stained. According to the CLSM images (Figure 5A), TBP-1-RGDS-hBD3-3 demonstrated high antibacterial efficacy against streptococci biofilm.
Antibacterial efficacy (R) against adherent bacteria on the TBP-1-RGDS-hBD3-3-modified Ti surfaces was further analyzed to verify the activity of surfaces treated alone or with serum or saliva. Figure 5B showed that the R value of samples toward S. gordonii and S. sanguinis was more than 80% and as high as 90% for S. oralis during the first 24 hours. Although resulting in a decreased R value, serum or saliva did not significantly affect the antibacterial efficacy of samples against colonizing bacteria compared with the Ti plates treated only with TBP-1-RGDS-hBD3-3. After 72 hours, the R value decreased gradually (Figure 5C), but there was no significant difference in the numbers of colonizing streptococci.
Effect of TBP-1-RGDS-hBD3-3 on bacterial membranes and biofilm structure
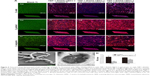
We used CLSM to investigate the inhibitory effect of TBP-1-RGDS-hBD3-3-modified Ti plates on streptococci biofilm formation. According to two- and three-dimensional biofilm thickness CLSM images (Figure 6A), we obtained a direct observation of the change in biofilm thickness, cell density, and adsorption conditions of the TBP-1-RGDS-hBD3-3 peptides on Ti plates after 36 hours. Consistent with the results obtained using AMC stain, TBP-1-RGDS-hBD3-3 peptides were adsorbed onto the Ti surfaces. As shown in Figure 6A, dead cells almost covered the surface of the modified Ti surface treated with TBP-1-RGDS-hBD3-3 alone or also with serum or saliva, compared to the bare Ti substrate that had green-stained S. oralis, S. gordonii, and S. sanguinis.
We performed SEM and TEM to directly observe the bacteria treated with TBP-1-RGDS-hBD3-3. The data (Figure 6B and C) showed that the membranes of S. gordonii cells treated with the chimeric peptide for 12 hours were damaged compared with the control group (Figure S1). This indicated that TBP-1-RGDS-hBD3-3 can disrupt bacterial membranes.
SspA and sspB from the antigen I/II family represent one of the major cell-surface adhesions and are expressed on the cell wall in S. gordonii to connect with receptors in saliva films or pellicles.30 We employed the real-time quantitative PCR to examine whether TBP-1-RGDS-hBD3-3 inhibited expressions of sspA and sspB and consequently affect S. gordonii biofilm formation on the modified Ti surfaces. The expression reduction of sspA and sspB as compared with the control group is shown in Figure 6D. The data show that the expression of these two genes reduced by 40% and 12%, respectively (P<0.05), suggesting that TBP-1-RGDS-hBD3-3 was inhibitory to expression of sspA and sspB in S. gordonii compared to the control (untreated bacteria).
Apoptotic sensitivity of osteoblasts on TBP-1-RGDS-hBD3-3-modified Ti
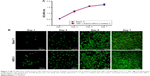
In order to evaluate the potential cytotoxicity of TBP-1-RGDS-hBD3-3, the MC3T3-E1 cells were chosen for the experiment. According to the data, the cells treated with TBP-1-RGDS-hBD3-3 at 640 μg/mL proliferated continuously during the entire incubation time (Figure 7). The mean proliferation rate of MC3T3-E1 cells was not significantly different from that of cells in the control sample (Figure 7A). For the sake of visually assessing the viability and proliferation of MC3T3-E1 cells, live/dead staining (AO/EB) was introduced to further confirm this finding. As shown in Figure 7B, live cells (green ones) could be seen on all substrates after 1–7 days of culture, and the number of dead cells (red ones) was low. These results clearly confirmed that TBP-1-RGDS-hBD3-3 exhibited no significant cytotoxicity toward MC3T3-E1 cells.
Discussion
In this study, three multifunctional peptides were developed for investigation of their suitability to improve the survival rate of dental implants. Ti substrates modified with these multifunctional peptides were demonstrated to have antimicrobial activity, thereby potentially enhancing biocompatibility for implant applications. Besides structure and stability, antimicrobial activity and the mechanism of action of the best-performing peptide were evaluated to elucidate the process of chimeric peptide modification on Ti implant surfaces.
Design of TBP-1-RGDS-hBD3-1/2/3 multifunctional peptides for modification of the surface of Ti
The multifunctional chimeric peptides comprised the major antimicrobial sequences from hBD3, a sequence that can promote osteogenesis (RGDS), a Ti-binding protein, and a linker (GGG). They were designed to prevent bacterial attachment on Ti surfaces. The purpose of this study was to achieve multifunctional modification using a single process.
Establishment of a biofilm occurs following microbial attachment, microcolony formation, biofilm maturation, and finally, propagation of infection. In order to avoid biofilm-associated infections, various strategies have been developed to improve the antimicrobial properties of biomaterial surfaces including loading with antibiotics,31 covalent attachment of AMPs,14–16 and polymer-based surface modification.32–35 Although these methods can inhibit bacterial attachment, the procedure is tedious and has a limited efficacy in practice. To overcome these limitations and combat the issues of antibiotic resistance and toxicity, a Ti-binding protein was used to connect the multifunctional chimeric peptides with the Ti surface. The main antimicrobial fragment was a short sequence from hBD3; this has a wide range of antibacterial properties as well as cell- and skin-penetrating properties and anti-inflammatory activity.21 In a recent study, its notable antibacterial performance was observed.36 After design of the main antimicrobial sequence, an RGDS sequence was introduced into the chimeric peptide to enhance osseointegration. The GGG linker was chosen to protect the principal function of the peptide from interference by the other chimeric peptide sequences. The multifunctional chimeric peptide was designed to investigate whether combining three functional sequences would achieve these various functions.
Structure and function analysis of the multifunctional chimeric peptides
The molecular structures of the chimeric peptides were significantly related to their function. Secondary structure determination by CD spectroscopy showed all three peptides as having molecular conformations (α-helical, β-strand) that exhibit antimicrobial activity (Figure 2A). Considering the proposed application, the peptides’ structures were determined at 37°C and 4°C: TBP-1-RGDS-hBD3-1 and TBP-1-RGDS-hBD3-3 retained stable structural composition at both temperatures, whereas TBP-1-RGDS-hBD3-2 did not. These results were consistent with those previously reported.37 Zeta potential measurements showed the chimeric peptides to have varying degrees of stability, with TBP-1-RGDS-hBD3-3 being the most stable (Figure 2E). Raman spectroscopy was used to further analyze the structures and predict their tertiary conformations. The bands at 1,340, 1,666, and 1,240 cm−1 were consistent with α-helical, β-strand, and coil structures, respectively, which agreed with the theoretical predictions using software and the CD spectra.
The structural similarity of the three chimeric peptides was shown by Raman spectroscopy, with the spectral bands of hBD3-1, hBD3-2, and hBD3-3 indicating that the hBD3 sequence preserved its 3D structure.37 TBP-1-RGDS-hBD3-3 possessed more positively charged residues (Arg, Lys), while all the peptides had both hydrophilic and hydrophobic domains, satisfying the characteristics of antibacterial peptides (Figure 1). The XPS results showed that TBP-1-RGDS-hBD3-1/2/3 were all successfully immobilized on the surface of Ti (Figure 3), which proves that the functional domain of TBP-1 was not influenced by the other chimeric peptide domains.
Given the focus of this research on the prevention of biofilm formation, streptococci were chosen as the initial colonizers and the antimicrobial activity of multifunctional chimeric peptides was evaluated against S. oralis, S. gordonii, and S. sanguinis. The antibacterial and anti-biofilm activity of TBP-1-RGDS-hBD3-3 was much stronger than that of the other peptides (Table 1, Figure 4). Although there is no consensus on the exact mechanism by which hBD3 and its analogs operate, some trends are commonly agreed. It is widely acknowledged that positively charged peptides attach electrostatically to negatively charged microbial membranes. TBP-1-RGDS-hBD3-3 had a net charge of +8.9 (Figure 1) and a zeta potential of 12.09 mV (Figure 2E), which were significantly greater than those of the other peptides. β-strands can form an amphipathic symmetrical dimer structure through the β2 strand, resulting in the exposure of a greater positive surface charge after folding. Increasingly, evidence is emerging that amino acid composition and physicochemical properties have a greater impact on antimicrobial activity than specific peptide structure. Furthermore, the activity can depend more on the balance of electrostatic interactions in the initial steps and on hydrophobic interactions in the later steps during the binding-disruption process. A recent study has indicated that Arg42 and Arg43 play a key role in the broad antimicrobial activity and salt-resistant properties of hBD3 and may maintain its activity in the C-terminus.38 Therefore, positively charged residues and secondary structure may play a key role in the membrane disruption activity of TBP-1-RGDS-hBD3-3.
The antimicrobial activity of the TBP-1-RGDS-hBD3-3-modified Ti surface was further analyzed by CLSM, SEM, and TEM. According to the results, the surfaces modified by multifunctional peptides were found to significantly reduce bacterial adhesion when compared to adhesion on bare Ti (Figures 5 and 6A). 3D CLSM with peptides stained blue by AMC showed that the chimeric peptides were immobilized on the Ti surface, and the results were consistent with XPS (Figure 3). The R value of TBP-1-RGDS-hBD3-3 immobilized on the Ti surface was 80% against S. oralis, and 75%–80% for S. gordonii and S. sanguinis. While the R value decreased when treated with serum or saliva, the difference was not significant (Figure 5B and C). These results demonstrated the antimicrobial activity of the multifunctional chimeric peptides while immobilized on the surface of Ti, and also indirectly proved that the GGG linker prevented the active domains from interfering with each other as their function was retained. The antimicrobial efficacy of the modified Ti surface can be interpreted as arising from the antimicrobial activity of the hBD3-3 sequence, with the chimeric peptide competing for the bacterial attachment site.
The third function of the chimeric peptide via the RGDS moiety was presupposed to enhance the adhesion of osteoblasts and fibroblasts. According to the results of the cell proliferation test, the mean proliferation rates of MC3T3-E1 cells on the modified Ti surfaces were greater than that of cells on bare Ti at all time points, and the differences were significant (Figure 7). The chimeric peptide thus achieved the goal of protecting adherent cells from the effects of local apoptogens, by the RGDS sequence. In other words, the function of RGDS was preserved in the chimeric peptide. From another point of view, the number of binding sites on the Ti surface was limited. Because of the antibacterial effect of the chimeric peptide, the adhesion of the bacteria on the modified Ti surface was affected, which helped improve the effects of osseointegration.
The three chimeric peptides possessed the same Ti-binding protein and RGDS sequence with different antimicrobial domains. The GGG linker was connected to TBP-1 with a WG sequence and with GR and SG sequences connected to RGDS, while the GG sequence was connected to the hBD3 sequence. According to the present results, a possible explanation of the effectiveness of the GGG linker is that it formed a kink in the solid structure, which effectively separated the functional domain. These connection sequences provided the possibility for the design of multifunctional peptides.
Antimicrobial mechanism of the chimeric peptides
A consensus has not yet been reached regarding the exact mechanisms of the antibacterial action of hBD3. It has been assumed that hBD3 damages bacterial membranes via the carpet model39 due to its highly cationic nature and because it would not easily form a 3D amphiphilic structure. Conversely, it is more likely to dimerize and/or oligomerize through the β-2 strand in solution.39,40 Another study has shown that the dimers or oligomers on the surface of bacterial membranes make anionic lipid (POPG) clusters and form two types of regions in the membrane, which results in membrane disruption.41 However, dimerization is not an essential requirement for the activity of hBD3 against bacteria. This study showed that TBP-1-RGDS-hBD3-3 inhibited biofilm formation by interrupting the integrity of bacterial membranes (Figure 6B and C). However, the suspected mechanisms do not fully explain this phenomenon. Inferring from the present data, it is easy to tell that cell integrity has been destroyed, but this is difficult to capture using SEM. One reason is that the experimental conditions do not fully reflect the actual situation in vivo. In addition, other antimicrobial/anti-biofilm mechanisms of the chimeric peptide should be considered; for example, the mechanism of biofilm resistance by the attachment of proteins sspA and sspB in S. gordonii was explored. The results showed that TBP-1-RGDS-hBD3-3 may inhibit adhesion and impact biofilm formation of S. gordonii by downregulating the expression of sspA and sspB genes (Figure 6D). However, it is still not clear whether TBP-1-RGDS-hBD3-3 regulates the genes through other protein(s) or by direct impaction.
In this study, there was no further determination of the antimicrobial activity from the fragments of hBD3 in the secondary structure because of the lack of Raman database. Oral biofilm formation is a complex process; data from other colonizers from different time stages of biofilm formation are lacking, especially from the later colonizer-pathogenic bacteria. Therefore, a more comprehensive evaluation of the effects of chimeric peptides on biofilm formation is needed, including the effects on promotion of osteoblast differentiation and the mechanism of action. In particular, the mechanism of gene regulation of chimeric peptides requires exploration as to whether it is a single cause or a multi-factorial system. In contrast to previous studies, it was found that the chimeric peptide sequence needs to be short with multifunctional domains; this should be the approach in further studies.
Conclusion
This study provides a simple method for the modification of Ti surfaces to enable antimicrobial function by engineering of chimeric peptides. The chimeric peptide sequence consisted of Ti-binding, antimicrobial, and RGDS motifs. The multifunctional peptide relied on a Ti-binding protein that allowed the functional domains to perform their required tasks. The efficacy of TBP-1-RGDS-hBD3-3 was evaluated both in solution and on the surface of Ti against initial colonization by biofilm-forming bacteria. The results indicated that the chimeric peptide had good Ti-binding and antimicrobial activity on the surface.
In view of the chimeric peptide designation, multifunctional chimeric peptides wherein different effective domains are connected provide a promising approach. Further studies are necessary to consider the complete mechanism of antimicrobial activity of chimeric peptides that comprise a variety of protein sequences.
Acknowledgments
The authors thank Dr RuiXue for her assistance with Raman spectroscopy. The work was supported by the National Natural Science Foundation of China (Grant No 81501798 and Grant No 81501994) and by the China Postdoctoral Science Foundation (Grant No 2016M601276).
Disclosure
The authors report no conflicts of interest in this work.
References
Albrektsson T, Zarb G, Worthington P, Eriksson AR. The long-term efficacy of currently used dental implants: a review and proposed criteria of success. Int J Oral Maxillofac Implants. 1986;1(1):11–25. | ||
Esposito M, Hirsch J, Lekholm U, Thomsen P. Differential diagnosis and treatment strategies for biologic complications and failing oral implants: a review of the literature. Int J Oral Maxillofac Implants. 1999;14(4):473–490. | ||
Geckili O, Bilhan H, Geckili E, Cilingir A, Mumcu E, Bural C. Evaluation of possible prognostic factors for the success, survival, and failure of dental implants. Implant Dent. 2014;23(1):44–50. | ||
Smeets R, Henningsen A, Jung O, Heiland M, Hammächer C, Stein JM, Definition SJM. Definition, etiology, prevention and treatment of peri-implantitis – a review. Head Face Med. 2014;10:34. | ||
Elemek E, Almas K. Etiology Peri-Implantitis: diagnosis and treatment: an update. N Y State Dent J. 2014;80(1):26–32. | ||
Mccrea SJ. Advanced peri-implantitis cases with radical surgical treatment. J Periodontal Implant Sci. 2014;44(1):39–47. | ||
Wilson V. An insight into peri-implantitis: a systematic literature review. Prim Dent J. 2013;2(2):69–73. | ||
Belibasakis GN. Microbiological and immuno-pathological aspects of peri-implant diseases. Arch Oral Biol. 2014;59(1):66–72. | ||
Veerachamy S, Yarlagadda T, Manivasagam G, Yarlagadda PK. Bacterial adherence and biofilm formation on medical implants: a review. Proc Inst Mech Eng H. 2014;228(10):1083–1099. | ||
Li J, Helmerhorst EJ, Leone CW, et al. Identification of early microbial colonizers in human dental biofilm. J Appl Microbiol. 2004;97(6):1311–1318. | ||
Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8(7):471–480. | ||
Campoccia D, Montanaro L, Arciola CR. The significance of infection related to orthopedic devices and issues of antibiotic resistance. Biomaterials. 2006;27(11):2331–2339. | ||
Ma M, Kazemzadeh-Narbat M, Hui Y, et al. Local delivery of antimicrobial peptides using self-organized TiO2 nanotube arrays for peri-implant infections. J Biomed Mater Res A. 2012;100(2):278–285. | ||
Kazemzadeh-Narbat M, Lai BF, Ding C, Kizhakkedathu JN, Hancock RE, Wang R. Multilayered coating on titanium for controlled release of antimicrobial peptides for the prevention of implant-associated infections. Biomaterials. 2013;34(24):5969–5977. | ||
Costa F, Carvalho IF, Montelaro RC, Gomes P, Martins MC. Covalent immobilization of antimicrobial peptides (AMPs) onto biomaterial surfaces. Acta Biomater. 2011;7(4):1431–1440. | ||
Tan XW, Goh TW, Saraswathi P, et al. Effectiveness of antimicrobial peptide immobilization for preventing perioperative cornea implant-associated bacterial infection. Antimicrob Agents Chemother. 2014;58(9):5229–5238. | ||
Krishnakumari V, Singh S, Nagaraj R. Antibacterial activities of synthetic peptides corresponding to the carboxy-terminal region of human beta-defensins 1–3. Peptides. 2006;27(11):2607–2613. | ||
Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276(8):5707–5713. | ||
Hoover DM, Wu Z, Tucker K, Lu W, Lubkowski J. Antimicrobial characterization of human beta-defensin 3 derivatives. Antimicrob Agents Chemother. 2003;47(9):2804–2809. | ||
Klüver E, Schulz-Maronde S, Scheid S, Meyer B, Forssmann WG, Adermann K. Structure-activity relation of human beta-defensin 3: influence of disulfide bonds and cysteine substitution on antimicrobial activity and cytotoxicity. Biochemistry. 2005;44(28):9804–9816. | ||
Lee JY, Suh JS, Kim JM, et al. Identification of a cell-penetrating peptide domain from human beta-defensin 3 and characterization of its anti-inflammatory activity. Int J Nanomedicine. 2015;10:5423–5434. | ||
Secchi AG, Grigoriou V, Shapiro IM, et al. RGDS peptides immobilized on titanium alloy stimulate bone cell attachment, differentiation and confer resistance to apoptosis. J Biomed Mater Res A. 2007;83(3):577–584. | ||
Sano K, Sasaki H, Shiba K. Specificity and biomineralization activities of Ti-binding peptide-1 (TBP-1). Langmuir. 2005;21(7):3090–3095. | ||
Mei L, Lu Z, Zhang W, et al. Bioconjugated nanoparticles for attachment and penetration into pathogenic bacteria. Biomaterials. 2013;34(38):10328–10337. | ||
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. | ||
Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. | ||
Akhtar W, Edwards HG. Fourier-transform Raman spectroscopy of mammalian and avian keratotic biopolymers. Spectrochim Acta A Mol Biomol Spectrosc. 1997;53A(1):81–90. | ||
Mensch C, Barron LD, Johannessen C. Ramachandran mapping of peptide conformation using a large database of computed Raman and Raman optical activity spectra. Phys Chem Chem Phys. 2016;18(46):31757–31768. | ||
Edwards HG, Hunt DE, Sibley MG. FT-Raman spectroscopic study of keratotic materials: horn, hoof and tortoiseshell. Spectrochim Acta A Mol Biomol Spectrosc. 1998;54A(5):745–757. | ||
Demuth DR, Duan Y, Brooks W, Holmes AR, Mcnab R, Jenkinson HF. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol Microbiol. 1996;20(2):403–413. | ||
Popat KC, Eltgroth M, Latempa TJ, Grimes CA, Desai TA. Decreased Staphylococcus epidermis adhesion and increased osteoblast functionality on antibiotic-loaded titania nanotubes. Biomaterials. 2007;28(32):4880–4888. | ||
Shi J, Liu Y, Wang Y, Zhang J, Zhao S, Yang G. Biological and immunotoxicity evaluation of antimicrobial peptide-loaded coatings using a layer-by-layer process on titanium. Sci Rep. 2015;5:16336. | ||
Muszanska AK, Rochford ET, Gruszka A, et al. Antiadhesive polymer brush coating functionalized with antimicrobial and RGD peptides to reduce biofilm formation and enhance tissue integration. Biomacromolecules. 2014;15(6):2019–2026. | ||
Zhang L, Ning C, Zhou T, et al. Polymeric nanoarchitectures on Ti-based implants for antibacterial applications. ACS Appl Mater Interfaces. 2014;6(20):17323–17345. | ||
Zhao L, Wang H, Huo K, et al. Antibacterial nano-structured titania coating incorporated with silver nanoparticles. Biomaterials. 2011;32(24):5706–5716. | ||
Geng H, Yuan Y, Adayi A, et al. Engineered chimeric peptides with antimicrobial and titanium-binding functions to inhibit biofilm formation on Ti implants. Mater Sci Eng C Mater Biol Appl. 2018;82:141–154. | ||
Zhang X, Adayi A, Geng H, et al. Antimicrobial Effects of Peptides from Human Beta-Defensin-3 on Planktonic and Biofilm States of Streptococci. Int J Pept Res Ther. 2017;53A(1):1–9. | ||
Sakagami-Yasui Y, Shirafuji Y, Yamasaki O, et al. Two arginine residues in the COOH-terminal of human β-defensin-3 constitute an essential motif for antimicrobial activity and IL-6 production. Exp Dermatol. 2017;26(11):1026–1032. | ||
Dhople V, Krukemeyer A, Ramamoorthy A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochim Biophys Acta. 2006;1758(9):1499–1512. | ||
Wu Z, Hoover DM, Yang D, et al. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc Natl Acad Sci U S A. 2003;100(15):8880–8885. | ||
Zhao X, Yu H, Yang L, Li Q, Huang X. Simulating the antimicrobial mechanism of human β-defensin-3 with coarse-grained molecular dynamics. J Biomol Struct Dyn. 2015;33(11):2522–2529. |
Supplementary materials
  | Table S1 Oligonucleotide primers used in this study |
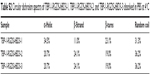  | Table S2 Circular dichroism spectra of TBP-1-RGDS-hBD3-1, TBP-1-RGDS-hBD3-2, and TBP-1-RGDS-hBD3-3 dissolved in PBS at 4°C |
  | Figure S1 SEM images of Streptococcus gordonii biofilms for 24 hours on untreated Ti surface. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.