Back to Journals » Cancer Management and Research » Volume 11
MNX1-AS1 is a functional oncogene that induces EMT and activates the AKT/mTOR pathway and MNX1 in breast cancer
Authors Cheng Y, Pan Y, Pan Y, Wang O
Received 18 September 2018
Accepted for publication 16 December 2018
Published 17 January 2019 Volume 2019:11 Pages 803—812
DOI https://doi.org/10.2147/CMAR.S188007
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Yue Cheng,1 Yiqi Pan,2 Yiyuan Pan,3 Ouchen Wang1
1Department of Thyroid and Breast Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, China; 2Wenzhou Medical University, Wenzhou, China; 3Department of Thyroid and Breast Surgery, Wenzhou People’s Hospital, Wenzhou, China
Purpose: lncRNAs have recently been identified as key regulators of basic biological processes as well as the pathogenesis of various diseases. Previous studies have shown that lncRNA MNX1-AS1 promotes cell migration and invasion in ovarian cancer; however, its role in regulating breast cancer-associated biological processes remains unclear.
Materials and methods: We obtained paired specimens of breast cancer tissues and adjacent normal tissues by modified radical mastectomy from 36 patients, in addition to four breast cancer cell lines (MDA-MB-231, MDA-MB-468, BT-549 and MCF-7). RNA was isolated from these tissues and cell lines and subsequently subjected to quantitative real-time polymerase chain reaction. This was followed by bisulfite deep sequencing. The cells were also transfected with siRNA against MNX1-AS1. The cells were then subject to cell proliferation, Transwell migration and invasion assays. Finally, Western blotting analysis was conducted to determine expression levels of MNX1, 5-cadherin, Snail and Slug.
Results: Our results show that MNX1-AS1 expression was significantly higher in breast cancer tissues than adjacent normal tissues. Moreover, knockdown/overexpression of MNX1-AS1 inhibits/promotes proliferation, migration and invasion of breast cancer cells. MNX1-AS1 and its natural sense transcript MNX1 are expressed synergistically in breast tumor tissues. Our results suggest that MNX1-AS1 is a functional oncogene that induces epithelial–mesenchymal transition, in addition to activating AKT/mTOR pathway and its natural sense transcript MNX1 in breast cancer cells.
Conclusion: Our data indicate that MNX1-AS1 can serve as a novel therapeutic target in breast cancer.
Keywords: MNX1-AS1, breast cancer, oncogene, MNX1, EMT, AKT/mTOR signaling pathway
Introduction
Breast cancer is the leading cause of cancer death in women. Currently, breast cancer therapy mainly relies on a combination of treatments, such as surgery, chemotherapy, radiotherapy, hormone therapy and molecular targeted therapy.1 In recent years, tumor gene therapy has become a new research hotspot in cancer treatment.2,3 Although a large number of studies on the pathogenesis of breast cancer exist, we still urgently need to clarify the new molecular mechanisms correlated with the development of breast cancer in order to better use gene therapy against breast cancer.
ncRNA accounts for 99% of the total RNA transcribed from human genome. Although they are abundant in cells, they are not valued for the lack of protein-coding ability. However, accumulating evidence has shed light on the critical role of ncRNA in cell functions and diseases.4 The role of lncRNAs in cancer has recently been focused on more. lncRNAs are longer than 200 nucleotides, which functions in diverse and complex mechanisms, strongly demonstrating the diversity of their biological roles. In fact, lncRNAs are involved in many biological and cellular pathways by interacting with proteins, chromatin, DNA as well as various RNA species.5,6 Several studies have demonstrated to be dysregulated expression of lncRNAs in breast cancer, which is strongly associated with oncogenes or tumor suppressor genes involved in carcinogenesis. Thus, lncRNAs might serve as potential markers for the diagnosis and prognosis in breast cancer.7,8 MNX1-AS1 is an lncRNA located on chromosome 7, a natural antisense transcript of MNX1, near the 5′ end of this protein-coding gene.9 Related studies have also shown that MNX1-AS1 is an oncogene in ovarian cancer and glioblastoma.10,11 Thus far, no relevant reports have described the relationship between MNX1-AS1 and breast cancer. In this study, we examined the expression of MNX1-AS1 in breast cancer tissues and matched adjacent normal breast tissues, and we investigated the function and underlying mechanism of MNX1-AS1 in breast cancer cell lines.
Material and methods
Tissue sample collection
Paired specimens of breast cancer tissues and adjacent normal tissues were obtained by modified radical mastectomy from 36 patients at The First Affiliated Hospital of Wenzhou Medical University. None of the patients had undergone chemotherapy, radiotherapy or endocrine therapy before the surgery. After operation, all samples were snap-frozen in liquid nitrogen and preserved at –80°C. This research was complied with the Declaration of Helsinki and was approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University. Written informed consent was obtained from each patient before the surgery.
Cell culture
Four breast cancer cell lines (MDA-MB-231, MDA-MB-468, BT-549 and MCF-7) were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). MDA-MB-231 and MCF-7 cell lines were cultured in DMEM (Life Technologies) and incubated at 37°C with 5% CO2. The BT-549 cell lines were cultured in RPMI 1640 (Life Technologies) and incubated at 37°C with 5% CO2. The MDA-MB-468 cell line was cultured in L15 (Life Technologies) at 37°C without CO2. All media were supplemented with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA).
Total RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from clinical tissues and cultured breast cancer cells using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The cytoplasmic and nuclear RNA were isolated and purified by Cytoplasmic and Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA). For qRT-PCR, RNAs were reversely transcribed into cDNA using PrimeScript RT Master Mix (Takara, Cat. #RR036A). The reverse transcription was conducted at 37°C for 15 minutes, followed by 85°C for 5 seconds, according to the manufacturer’s protocols. Real-time PCR was performed with SYBR Premix Ex Taq II (Takara, Cat. #RR820A). The thermocycling conditions were as follows: 95°C for 5 minutes, followed by 50 cycles of denaturation at 95°C for 15 seconds and then 60°C for 30 seconds. Primer sequences were as follows: MNX1-AS1: forward AAGGTAGCCACCAAACAC and reverse AGACTCACGTAGCACTGT; MNX1: forward CGTTGCTGTAGGGGAAATGGT and reverse AGGAAGCGGAGAAACAGAAGG.
Bisulfite deep sequencing
Bisulfite-converted gDNA samples (500 ng) were obtained using EpiTect96 Bisulfite Kit (Qiagen NV, Venlo, the Netherlands) according to the manufacturer’s instructions. Bisulfite-converted DNA was amplified with the MNX1 unmethylation-specific primers. A 97-bp region of the MNX1 promoter was amplified. The PCR products were loaded on a 1.5% agarose gel for analysis and recovered for the library construction and next-generation sequencing (NGS) in the MiSeq platform (Illumina, San Diego, CA, USA). DNA methylation level of the candidate fragment was determined by analyzing the NGS data. Sequencing reads were mapped to the genome using the Bismark software (www.bioinformatics.babraham.ac.uk/projects/bismark/). MethylKIT R package software (http://www.bioconductor.org/packages/release/bioc/html/methylKit.html) was then used to obtain site-specific methylation information. PCR primer was available as shown in Table 1.
  | Table 1 PCR primers for bisulfite deep sequencing |
Transfection
MDA-MB-231 and MDA-MB-468 were transfected with siRNA (GenePharma Co, Shanghai, China) against MNX1-AS1 using LipoMax® (Thermo Fisher Scientific) in a six-well cell culture plate according to the manufacturer’s protocols. The siRNA sequences were as follows: 5′-GCUACGUGAGUCUUGCAAATT-3′ (sense) and 5′-UUUGCAAGACUCACGUAGCTT-3′ (antisense). Plasmid vectors, MNX1-AS1 vector and empty vector, were used (GenePharma Co). MDA-MB-231 and MDA-MB-468 were transfected with vectors to overexpress MNX1-AS1 using Lipofectamine® 3000 Reagent (Thermo Fisher Scientific) following the manufacturer’s instructions.
Cell proliferation
Cell proliferation was monitored everyday using Cell Counting Kit-8, according to the manufacturer’s instructions. Transfected cells (5,000) were seeded into 96-well plates and examined daily for 4 consecutive days. The absorbance was measured at 450 nm.
Transwell migration and invasion assays
To evaluate cell migration, 8×104 transfected cells in 200 µL serum-free medium were added to the upper chamber (Corning Costar Corp, Cambridge, MA, USA). The lower chamber was filled with 600 µL medium supplied with 10% FBS. For the cell invasion assay, 1.5×105 transfected cells in 200 µL serum-free medium were added to the upper chamber of BioCoat™ Matrigel Invasion Chamber (Corning, NY, USA), and the lower chamber was filled with 600 µL medium supplied with 20% FBS. After 24-hour incubation at 37°C, the membranes were fixed with pre-cooled methanol and stained with 0.5% crystal violet. PBS was used to wash and remove excess stain. Five randomly selected fields were counted and images were captured under a microscope.
Western blot analysis
Protein expression was detected by Western blot analysis. Total transfected cells were collected and lysed using the lysis buffer (RIPA) supplemented with protease inhibitors. MNX1, 5-cadherin, Snail and Slug were the detecting antibodies. GAPDH and β-actin were selected as the housekeeping genes. All these antibodies were purchased from Abcam.
Statistical analysis
The data were collected from three independent experiments and are presented as mean ± SD. An unpaired Student’s t-test was used to calculate the difference with GraphPad software version 5.0. P<0.05 was considered statistically significant.
Results
MNX1-AS1 is a bona fide lncRNA and a natural antisense transcript of MNX1
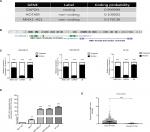
We used Coding Potential Calculator 2 (CPC 2.0) to predict the protein-coding probability of MNX1-AS1 to prove that it is a bona fide lncRNA. CPC 2.0 has an overall accuracy of 0.961, sensitivity of 0.952 and specificity of 0.970.12 The result showed that MNX1-AS1 had a low coding probability (Figure 1A). MNX1-AS1 is a 992-bp RNA transcribed from chromosome 7, the opposite strand of MNX1 (7q36.6), consisting of two exons (Figure 1B). To detect the subcellular localization of MNX1-AS1, we fractionated MDA-MB-231 and MDA-MB-468 cells into nuclear and cytoplasmic fractions. MNX1-AS1 molecules were found to be localized more in the nucleus than in the cytoplasm (Figure 1C).
Expression of MNX1-AS1 was increased in breast cancer tissues and cell lines
To explore the role of MNX1-AS1 in breast cancer, we collected 34 pairs of cancerous tissues along with adjacent normal tissues post-surgery. qRT-PCR results revealed that the expression of lncRNA MNX1-AS1 in cancer tissues was significantly increased compared to that in adjacent normal tissues. In addition, we examined MNX1-AS1 expression in four human breast cancer cell lines and in the normal breast epithelial cell line MCF10A. We found higher MNX1-AS1 expression levels in the breast cancer cell lines than in the normal breast epithelial cell line (Figure 1D). To validate MNX1-AS1 more robustly, we downloaded RNA sequencing data specific for breast cancer from TCGA. This database contains 837 breast cancer tissues and 105 noncancer tissues. The results consistently showed that the expression of MNX1-AS1 was significantly increased in breast cancer tissues (Figure 1E).
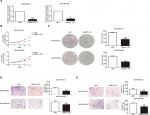
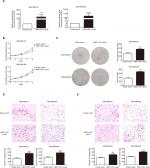
MNX1-AS1 was correlated with cell proliferation, migration and invasion
We used siRNA to knock down MNX1-AS1 and used plasmid vectors to overexpress MNX1-AS1 in MDA-MB-231 and MDA-MB-468 cells. qRT-PCR analysis showed that MNX1-AS1 was effectively downregulated by targeted siRNAs (Figure 2A, P<0.01). To determine the effect of MNX1-AS1 on cell growth, we performed CCK-8 and colony formation experiments. Results showed that compared with the control group, the proliferation rate and colony number of MNX1-AS1-knockdown cells were significantly decreased (Figure 2B, C). Further, we performed migration and invasion tests to examine the role of MNX1-AS1 in cell deformability. The results showed that MNX1-AS1 knockdown significantly inhibited the migration and invasion in MDA-MB-231 and MDA-MB-468 cells (Figure 2D, E). We used plasmid vectors to overexpress MNX1-AS1 in MDA-MB-231 and MDA-MB-468 cells. qRT-PCR analysis showed that MNX1-AS1 was highly upregulated using plasmid vectors (Figure 3A, P<0.01). To determine the effect of MNX1-AS1 on cell growth, we performed CCK-8 and colony formation experiments. Results showed that compared with the control group, the proliferation rate and colony number of cells overexpressing MNX1-AS1 were markedly increased (Figure 3B, C). Further, we performed migration and invasion tests to examine the role of MNX1-AS1 in cell deformability. The results showed that MNX1-AS1 overexpression promoted cell migration and invasion when compared with the control group (Figure 3D, E).
MNX1-AS1 was correlated with MNX1 expression
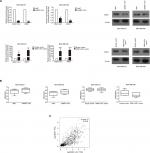
We performed PCR and Western blot techniques to demonstrate the relationship between MNX1-AS1 and MNX1 expression. The results showed that MNX1-AS1 knockdown significantly decreased MNX1 expression at the mRNA and protein levels. In contrast, MNX1-AS1 overexpression increased MNX1 expression at the mRNA and protein levels (Figure 4A). In addition, knockdown or overexpression of MNX1-AS1 did not cause any alterations in the methylation of the MNX1 promoter (Figure 4B). Furthermore, it was confirmed by the TCGA database that MNX1-AS1 was expressed coordinately with MNX1 in breast tumor tissues (Figure 4C).
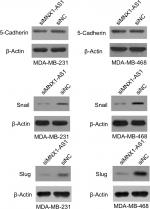
MNX1-AS1 could promote the expression of epithelial–mesenchymal transition (EMT)-related proteins in breast cancer cells
We found that MNX1-AS1 promoted breast cancer cell proliferation, migration and invasion; however, the underlying mechanism remained unclear. Therefore, we subsequently explored the relationship between MNX1-AS1 and EMT-related proteins. We found that MNX1-AS1 knockdown inhibited EMT in breast cancer cells (Figure 5). Thus, we suspect that MNX1-AS1 might affect the proliferation, migration and invasion of breast cancer cells by regulating EMT.
MNX1-AS1 regulated the AKT/mTOR signaling pathway in breast cancer
The AKT/mTOR signaling pathway regulates breast cancer cell proliferation and metastasis. To further investigate the possible mechanism of action of MNX1-AS1 in breast cancer, we used Western blot to detect the expression level of key molecules involved in the AKT/mTOR signaling pathway. The results showed that p-AKT expression in cells transfected with the MNX1-AS1 vector was significantly increased; moreover, its downstream target genes, such as CDK4, BCL2, cyclinD1 and c-MYC, were markedly upregulated (Figure 6).
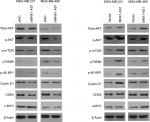  | Figure 6 The effect of MNX1-AS1 knockdown/overexpression on its downstream targets in AKT/mTOR signaling pathway was assessed by Western blot. |
Discussion
It is widely accepted that lncRNAs are key players in gene regulation processes and are associated with carcinogenesis. Therefore, lncRNAs may help us build a novel cancer diagnosis and treatment platform. Previous studies have shown that the lncRNA MNX1-AS1 plays an important role in proliferation and invasion in ovarian cancer and glioblastoma.10,11 However, the functional role and the detailed mechanism of MNX1-AS1 in the development of breast cancer has not been studied yet. Our study is the first to demonstrate that MNX1-AS1 was significantly upregulated in breast cancer tissues and cancer cell lines. To study the function of MNX1-AS1 in breast cancer, we either knocked down or overexpressed MNX1-AS1 in MDA-MB-231 and MDA-MB-468 cell lines. MNX1-AS1 knockdown significantly inhibited cell proliferation, migration and invasion, while its overexpression markedly promoted these cancer cell characteristics.
EMT plays an important role in invasion and metastasis in tumor development.13–16 Therefore, elucidating the role of MNX1-AS1 in EMT could be essential for understanding the latter’s regulatory mechanism. Our results showed that MNX1-AS1 knockdown decreased the expressions of 5-cadherin, snail and slug, suggesting that MNX1-AS1 regulates EMT in breast cancer. In addition, it is possible that MNX1-AS1 does not regulate the expression of MNX1 by regulating MNX1 promoter methylation.
We further clarified the downstream signaling pathways by which MNX1-AS1 might exert its tumor-promoting function in breast cancer and found that MNX1-AS1 upregulated the AKT signaling pathway. The AKT pathway plays a key role in cancer development and progression by modulating cell proliferation, apoptosis and angiogenesis.17–20 Our gain- and loss-of-function analysis of MNX1-AS revealed that it promoted/suppressed AKT activation by upregulating/downregulating phospho-AKT expression. Thus, the tumor-promoting function of MNX1-AS1 in breast cancer cells might be explained, at least in part, by the upregulated AKT signaling pathway.
mTOR is known to be a direct target of the AKT oncogenic signal.21,22 It drives tumorigenesis by alleviating 4E-BP1-mediated inhibition of eukaryotic translation initiation factor 4E and activating p70S6K to oncogene translation, both of which associate with mRNA and regulate mRNA translation initiation and extension. Therefore, we were next interested in whether MNX1-AS1 affects the activation of mTOR and its substrates p70S6K and 4E-BP1. Our results suggested that MNX1-AS1 could promote phosphorylation of p70S6K and 4E-BP1. In addition, many well-known downstream targets regulated by the translation of p70S6K and 4E-BP1, such as the key pro-growth proteins cyclinD1 and cyclin-dependent kinase 4, and the oncogenic transcription factor c-MYC, were upregulated by MNX1-AS1. CyclinD1, CDK4 and c-MYC contribute to tumor growth by promoting cell cycle progression.23,24 Therefore, MNX1-AS1 may promote the proliferation of breast cancer cells via the AKT/mTOR signaling pathway.
MNX1-AS1 is a natural antisense transcript of MNX1; antisense transcripts are common in cells and most transcribed regions have antisense transcripts. However, the mechanism by which the majority of antisense lncRNAs regulate the comparable sense transcripts remains unknown. MNX1 is reported to be an oncogene in the rare malignancy infantile acute myeloid leukemia, hepatocellular cancer and prostate cancer.25–27 Here, we demonstrated that MNX1-AS1 is co-upregulated with its sense transcript MNX1 in tumor tissues. Knockdown of MNX1-AS1 significantly decreased MNX1 mRNA and protein levels, while its overexpression increased these levels.
Conclusion
In conclusion, this is the first study to demonstrate upregulated MNX1-AS1 expression in breast cancer tissues and cell lines. The results revealed that MNX1-AS1 is a functional oncogene by virtue of inducing EMT and activating the AKT/mTOR pathway and its natural sense transcript MNX1 in breast cancer cells. Therefore, MNX1-AS1 can be used as a novel therapeutic target in breast cancer.
Author contributions
OW conceived and designed the study; YC performed the majority of experiments and wrote the manuscript; YQP contributed to the sample collection; and YYP contributed to data analysis. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Ribeiro Pereira ACP, Koifman RJ, Bergmann A. Incidence and risk factors of lymphedema after breast cancer treatment: 10 years of follow-up. Breast. 2017;36:67–73. | ||
Shuvalov O, Petukhov A, Daks A, et al. Current genome editing tools in gene therapy: new approaches to treat cancer. Curr Gene Ther. 2015;15(5):511–529. | ||
Zhao XB, Ren GS. LncRNA taurine-upregulated gene 1 promotes cell proliferation by inhibiting microRNA-9 in MCF-7 cells. J Breast Cancer. 2016;19(4):349–357. | ||
Kim T, Croce CM. Long noncoding RNAs: undeciphered cellular codes encrypting keys of colorectal cancer pathogenesis. Cancer Lett. 2018;417:89–95. | ||
Schmitt AM, Chang HY. Long noncoding RNAs: at the intersection of cancer and chromatin biology. Cold Spring Harb Perspect Med. 2017;7(7):a026492. | ||
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. | ||
Ke H, Zhao L, Feng X, et al. NEAT1 is required for survival of breast cancer cells through FUS and miR-548. Gene Regul Syst Bio. 2016;10(Suppl 1):11–17. | ||
Xing Z, Park PK, Lin C, Yang L. LncRNA BCAR4 wires up signaling transduction in breast cancer. RNA Biol. 2015;12(7):681–689. | ||
Goyal A, Myacheva K, Groß M, Klingenberg M, Duran Arqué B, Diederichs S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017;45(3):e12. | ||
Lv Y, Li H, Li F, Liu P, Zhao X. Long noncoding RNA MNX1-AS1 knockdown inhibits cell proliferation and migration in ovarian cancer. Cancer Biother Radiopharm. 2017;32(3):91–99. | ||
Gao Y, Xu Y, Wang J, Yang X, Wen L, Feng J. LncRNA MNX1-AS1 promotes glioblastoma progression through inhibition of miR-4443. Oncol Res. Epub 2018 Apr 20. | ||
Kang YJ, Yang DC, Kong L, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–W16. | ||
Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21–45. | ||
Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–494. | ||
Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35(4):645–654. | ||
Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. | ||
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. | ||
Franke TF. PI3K/Akt: getting it right matters. Oncogene. 2008;27(50):6473–6488. | ||
Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122(Pt 20):3589–3594. | ||
Mamane Y, Petroulakis E, Lebacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25(48):6416–6422. | ||
Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. | ||
Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med (Berl). 2011;89(3):221–228. | ||
Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11(8):558–572. | ||
Cicenas J, Valius M. The CDK inhibitors in cancer research and therapy. J Cancer Res Clin Oncol. 2011;137(10):1409–1418. | ||
Wang Y, Wang J, Zhang L, et al. RGS12 is a novel tumor-suppressor gene in African American prostate cancer that represses AKT and MNX1 expression. Cancer Res. 2017;77(16):4247–4257. | ||
Das M. MNX1: a novel prostate cancer oncogene. Lancet Oncol. 2016;17(12):e521. | ||
Zhang L, Wang J, Wang Y, et al. MNX1 is oncogenically upregulated in African-American prostate cancer. Cancer Res. 2016;76(21):6290–6298. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.