Back to Journals » International Journal of General Medicine » Volume 15
MMP2 Polymorphisms and Colorectal Cancer Susceptibility in a Chinese Han Population
Authors Liu X, Yang K, Li Z , Liu J
Received 29 April 2022
Accepted for publication 15 June 2022
Published 5 July 2022 Volume 2022:15 Pages 6009—6019
DOI https://doi.org/10.2147/IJGM.S364029
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xu Liu,1,* Kelaier Yang,2,* Zhangfu Li,3 Jikui Liu1
1Department of Hepato-Pancreato-Biliary Surgery, Peking University Shenzhen Hospital, Shenzhen, Guangdong, People’s Republic of China; 2Department of Endocrinology and Metabolism, Shenzhen University General Hospital, Shenzhen, Guangdong, People’s Republic of China; 3Department of Hepato-Pancreato-Biliary Surgery, Peking University Shenzhen Hospital, Shenzhen Peking University-The Hong Kong University of Science and Technology Medical Center, Shenzhen, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jikui Liu, Department of Hepato-Pancreato-Biliary Surgery, Peking University Shenzhen Hospital, #1120, Lianhua Road, Futian District, Shenzhen, Guangdong, 518036, People’s Republic of China, Tel/Fax +86-13841498565, Email [email protected]
Purpose: Colorectal cancer (CRC) is among the most common cancers worldwide and an important cause of cancer-related death. Inherited genetic variation plays a vital role in the occurrence and development of CRC. The aim of this study was to evaluate the association of single nucleotide polymorphisms (SNPs) in MMP2 with CRC risk.
Patients and Methods: Three candidates, MMP2 SNPs, rs1053605, rs243849, and rs14070, were selected and genotyped using the Agena MassARRAY RS1000 system, and their association with risk of CRC was evaluated in 663 CRC cases and 663 healthy controls by calculating odds ratio (OR) with 95% confidence interval (95% CI) values.
Results: The minor allele of rs243849 (T) was significantly less frequent in cases than controls (p = 0.021), and this SNP was associated with a decreased risk of CRC under co-dominant (p = 0.033), dominant (p = 0.021), and log-additive (p = 0.017) models, after adjusting for confounding factors. After stratification, rs243849 was found to be protective against CRC in patients who were non-smoking, consumed alcohol, and were ≥ 60 years old (p < 0.05). Conversely, rs1053605 was associated with disease occurrence in patients with CRC who consumed alcohol and were < 60 years old (p < 0.05). Furthermore, rs1053605 genotype was associated with an increased risk of colon cancer (p < 0.05), while that of rs243849 was associated with a decreased risk of rectal cancer (p < 0.05). The rs1053605-rs243849 CT haplotype exhibited a protective role in CRC risk, following adjustment for confounders (p = 0.014). The rs14070 SNP was not associated with CRC risk. Finally, the false discovery rate (FDR) method was used to validate the study results.
Conclusion: Overall, the MMP2 gene polymorphisms, rs243849 and rs1053605, may be useful for predicting CRC progression.
Keywords: single nucleotide, alleles, haplotypes, colorectal neoplasms
Introduction
Colorectal cancer (CRC) is a risk to human health that ranks third in terms of tumor incidence and is the second leading cause of cancer-related death worldwide; global cancer statistics estimated that more than 1.9 million new CRC cases, resulting in 935,000 deaths, occurred in 2020.1 Genetic variants, specific environmental factors, and biological processes are generally considered the main causes of CRC.2,3 The high heritability and frequent recurrence of CRC lead to considerable mortality and are associated with poor prognosis.4,5 Therefore, determining the pathological mechanisms underlying CRC and developing therapeutic agents to treat the disease are urgently required. Recent studies of gene polymorphisms have demonstrated that specific genetic variants have critical roles in CRC susceptibility;6,7 for example, polymorphisms of CABLES2,7 CRTC3,8 LDH2, and ADH1B6 are associated with CRC risk.
The matrix metalloproteinases (MMP) family of Zn2+ dependent endopeptidases has important roles in various biological processes by contributing to extracellular matrix remodeling, cell differentiation, and tissue repair.9 MMP2 maps to chromosome 16q12.2 and is a member of the MMP family whose protein product, type IV collagenase, can degrade various collagen substrates, including fibronectin, aggrecan, elastin, and nidogen, among others; this can contribute to tumor development by influencing cancer cell invasion and migration.10,11 Furthermore, MMP2 gene polymorphisms are associated with risk for multiple diseases. The rs2241146 single nucleotide polymorphism (SNP) in MMP2 is associated with an increased risk of steroid-induced osteonecrosis of the femoral head in the Chinese Han population,12 while another MMP2 SNP, rs243865, is not associated with susceptibility to breast cancer.13
Although there are many reports showing that CRC risk is influenced by genetic susceptibility, specific associated genes and details of mechanisms underlying the contributions of a major proportion of risk loci identified by polymorphism analysis remain largely unknown. Hence, we performed an association study to identify the putative susceptibility of SNPs in MMP2. Three SNPs (rs1053605, rs243849 and rs14070) were selected and associations between these polymorphisms and CRC risk explored. Our research provides additional information to support the limited evidence that MMP2 polymorphisms are associated with CRC risk in the Chinese Han population.
Materials and Methods
Study Population
In total, 1326 Chinese Han participants, comprising 663 patients with CRC and 663 unrelated healthy controls, matched for sex and age, were included in this study. Patients were diagnosed according to clinical and pathological data, and those with a history of other cancer, or hematologic disorders were excluded. The diagnosis criteria of CRC14 are based on clinical history, routine laboratory evaluation, and histopathological detection, and all cases were patients with positive colonoscopy results for malignancy. Results of CRC diagnosis and clinical information for patients with CRC (lymph node metastasis, and cancer stage and type) were collected from medical records and histopathology reports. Controls were recruited from the physical examination center and had no history of disease, including medical illnesses, family history of cancer, cardiovascular disease, hepatic disease, or pulmonary disease. Participant demographic data (age, sex, body mass index (BMI), alcohol consumption, and smoking habits) were collected using a standard clinical information questionnaire.
This study received ethical approval from the Peking University Shenzhen Hospital institutional review board and was conducted in accordance with the Helsinki declaration on human medical research. All participants were informed about the purpose of the study and signed an informed consent form.
DNA Extraction and SNP Genotyping
MMP2 candidate SNPs with minor allele frequency (MAF) >5% were selected based on the 1000 genome project database (Http://www.Internationalgenome.org/); three SNPs (rs1053650, rs243849, and rs14070) were finally identified for analysis in a case–control study.
Peripheral blood samples (5 mL) were collected from all subjects by a professional technician using a vacutainer and placed into tubes containing EDTA. Genomic DNA was isolated using a GoldMag whole blood genomic DNA purification kit (GoldMag Co. Ltd, Xi’an, China), according to the manufacturer’s protocol. DNA concentration and purity were evaluated using a Nano Drop One (ThermoFisher Scientific, Waltham, MA), and all samples met the quality requirements (OD 260/280 = 1.8–2.0).
SNP detection primers were designed using Agena Bioscience Assay Design Suite V2.0 software (https://agenacx.com/online-tools/) and synthesized by Sangon Biotechnology (Sangon Biotech Co. Ltd., Shanghai, China). SNPs were genotyped using an Agena MassARRAY RS1000 (Agena, San Diego, CA, USA), according to the standard recommended instructions. Agena Bioscience 4.0 software was used to analyze and manage data.
Statistical Analyses
All statistical analyses were performed using SPSS version 21.0 (SPSS, Chicago, USA). The chi-square test was used to analyze genotype distributions among cases and controls, and to assess Hardy-Weinberg equilibrium (HWE). Allele and genotype frequencies were compared between patients with CRC and controls using Pearson Chi-square test. Associations between polymorphisms in MMP2 and CRC risk were assessed by calculating odds ratios (ORs) with 95% confidence intervals (CIs) using logistic regression analysis adjusted for age, sex, smoking status, and alcohol consumption. Inheritance models (genotype, dominant, recessive, and additive) established using PLINK software version 1.9 were used to evaluate associations between polymorphisms and CRC risk. Pairwise linkage disequilibrium (LD), haplotype construction, and genetic associations of polymorphic loci were assessed using Haploview software (version 4.2). The false discovery rate (FDR) method was used to control for type I error due to multiple comparisons. Values of p < 0.05 were considered statistically significant.
Results
Characteristics of Study Subjects
A total of 663 patients (386 men and 277 women, 348 aged ≥60 years and 315 aged <60 years) with CRC and 663 controls (400 men and 263 women, 379 aged ≥60 years and 284 aged <60 years) were recruited to our study. The basic characteristics of all participants are presented in Table 1. There were no significant differences between CRC cases and controls in terms of sex (p = 0.467), age (p = 0.098), smoking (p = 0.321), or alcohol consumption (p = 0.912); however, BMI differed significantly between the case and control groups (p < 0.001). Among 663 CRC cases with available tumor type, 310 (46.8%) cases had colon cancer and 353 (53.2%) had rectal cancer. Information on tumor stage and lymph node metastasis in the case group was incomplete.
 |
Table 1 Characteristics of Cases and Controls |
Associations Between MMP2 Polymorphisms and CRC Risk
Three MMP2 SNPs (rs1053605, rs243849, and rs14070) were screened according to the criteria described above and successfully genotyped in all included samples. SNP data are presented in Table 2, and the genotype frequencies of all SNPs conformed to HWE in the control group (p > 0.05). Of the three SNPs, the minor allele of rs243849 (T) was significantly associated with decreased CRC risk under an allelic model (OR = 0.78, 95% CI = 0.63–0.96, p = 0.018), while no allelic associations were detected with either rs1053605 or rs14070. Moreover, there were no significant differences between patients and healthy controls on the application of the FDR test to correct for multiple comparisons (p−FDR > 0.05), implying that rs243849 was not associated with susceptibility to CRC in the allelic model.
 |
Table 2 Basic Characteristics of MMP2 Candidate SNPs and Their Relationship with CRC Risk Under the Allelic Model |
We next applied four genetic models (genotype, dominant, recessive, and additive) to further analyze the relationship between rs243849 and CRC risk (Table 3). The results indicated that individuals with the C/T genotype had a reduced risk of CRC compared to those with the CC genotype, under the genotype model (non-adjusted OR = 0.76, 95% CI = 0.60–0.97, p = 0.026; adjusted OR = 0.77, 96% CI = 0.60–0.98, p = 0.033). Under the dominant model, genotypes C/T and T/T were associated with reduced risk relative to the C/C genotype (non-adjusted OR = 0.75, 95% CI = 0.59–0.95, p = 0.017; adjusted OR = 0.76, 96% CI = 0.60–0.96, p = 0.022). Similarly, a 0.77-fold reduced risk was detected under the additive model (non-adjusted 95% CI = 0.62–0.95, p = 0.015; adjusted 95% CI = 0.62–0.96, p = 0.018). Furthermore, following the application of the FDR test, the significance of the protective role for rs243849 under the additive model was retained (p−FDR = 0.045).
 |
Table 3 Association Between rs243849 and CRC Risk Under Co-Dominant, Dominant, Recessive, and Log-Additive Models |
Correlation of MMP2 Polymorphisms with CRC Risk in Patients Stratified According to Age, Smoking, and Alcohol Consumption
Associations between these loci and CRC risk stratified by age, smoking, and alcohol consumption were also analyzed (Table 4). No associations between rs14070 and CRC risk were detected in data stratified according to these factors; however, rs1053605 was associated with increased risk of CRC in people <60 years old under the dominant (adjusted OR = 1.57, 95% CI = 1.054–2.33, p = 0.028) and additive (adjusted OR = 1.61, 95% CI = 1.10–2.38, p = 0.014) models, with the significance of the association under the additive model surviving FDR correction (p−FDR = 0.041). Among subjects ≥60 years old, rs243849 was associated with decreased CRC risk under the genotype (adjusted OR = 0.65, 95% CI = 0.47–0.91, p = 0.011), dominant (adjusted OR = 0.64, 95% CI = 0.46–0.89, p = 0.008), and additive (adjusted OR = 0.67, 95% CI = 0.50–0.91, p = 0.01) models, furthermore, all of these associations remained significant following application of the FDR test (p-FDR = 0.033, p-FDR = 0.024, and p-FDR = 0.029, respectively).
 |
Table 4 Evaluation of Associations Between SNPs and CRC Risk Following Stratification for Age, Smoking, and Alcohol Consumption |
Further, the rs243849 variant was associated with protection against CRC risk among non-smokers under the genotype (adjusted OR = 0.63, 95% CI = 0.45–0.88, p = 0.006), dominant (adjusted OR = 0.62, 95% CI = 0.45–0.86, p = 0.004), and additive (adjusted OR = 0.65, 95% CI = 0.48–0.87, p = 0.004) models, with associations remaining significant following FDR correction (p-FDR = 0.018, p-FDR = 0.012, and p-FDR = 0.012, respectively).
Furthermore, the rs1053605 polymorphism was associated with increased CRC risk among patients who consumed alcohol under the genotype (adjusted OR = 1.50, 95% CI = 1.02–2.23, p = 0.041), dominant (adjusted OR = 1.52, 95% CI = 1.03–2.23, p = 0.031), and additive (adjusted OR = 1.47, 95% CI = 1.03–2.10, p = 0.032) models. Additionally, the differences remained significant under the dominant and additive models on application of the FDR test (p-FDR = 0.047 and p-FDR = 0.048, respectively).
Conversely, rs243849 was associated with reduced risk of CRC in patients who consumed alcohol under the genotype (adjusted OR = 0.62, 95% CI = 0.43–0.89, p = 0.009), dominant (adjusted OR = 0.61, 95% CI = 0.43–0.87, p = 0.006), and additive (adjusted OR = 0.63, 95% CI = 0.45–0.87, p = 0.006) models, and the results remained significant following FDR analysis under all three models (p-FDR = 0.028, p-FDR = 0.018, and p-FDR = 0.017, respectively).
Association Between MMP2 Variants and Tumor Risk Based on Tumor Type Stratification
Next, we stratified patients with CRC according to the type of cancer and analyzed the relationship between SNP variants and risk of colon and rectal cancers (Table 5). We found that the rs1053605 T allele was associated with a significantly higher likelihood of developing colon cancer relative to the C allele under the genotype (genotype “T/T” vs “C/C”, adjusted OR = 3.40, 95% CI = 1.1–10.52, p = 0.033) and recessive (genotype “T/T” vs “C/C-C/T”, adjusted OR = 3.53, 95% CI = 1.14–10.87, p = 0.028) models. In addition, the rs243849 polymorphism was significantly associated with reduced risk of rectal cancer under the genotype (adjusted OR = 0.73, 95% CI = 0.54–0.98, p = 0.039), dominant (adjusted OR = 0.74, 95% CI = 0.55–0.98, p = 0.036), and additive (adjusted OR = 0.77, 95% CI = 0.59–0.99, p = 0.049) models. Nevertheless, none of these associations survived FDR correction. The rs14070 variant was not associated with reduced tumor risk following stratification for tumor type.
 |
Table 5 Evaluation of Associations Between SNP Variants and Cancer Risk Based on Tumor Type Stratification |
LD and Haplotype Analysis
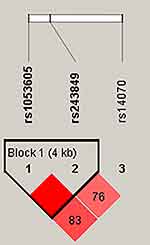
The results of pairwise LD analysis of the three MMP2 SNPs are presented in Figure 1. We detected a haplotype block with a strong LD between the rs1053605 and rs243849 SNPs. Further, the results of haplotype analysis indicated that the rs1053605-C/rs243849-T haplotype was associated with significantly decreased CRC risk (p = 0.013, chi-square test; OR = 0.76, 95% CI = 0.62–0.95, p = 0.014, logistic regression analysis with adjustment) (Table 6).
 |
Table 6 Association of MMP2 Haplotypes and CRC Risk |
Discussion
In this study, we identified associations between rs1053605 and rs243849 in MMP2 and CRC susceptibility among a Chinese Han population. Our results indicate that the rs243849-T allele has a protective role in people ≥60-years-old, non-smokers, and drinkers, based on genotype, dominant, and additive models, with associations remaining significant following FDR correction. Conversely, the minor allele of rs1053605 was associated with an increased risk of CRC in patients ≥60-years-old and those who consumed alcohol, and the results remained significant following FDR analysis. Moreover, the rs1053605-C/rs243849-T haplotype was associated with decreased risk of CRC. Hence, the rs1053605 and rs243849 SNPs in MMP2 may be associated with CRC development.
CRC is a serious threat to human health and genetic factors play a vital role in the etiology of these tumors. Numerous reports have identified genetic polymorphisms significantly associated with CRC risk. For example, Gholamalizadeh et al performed a case–control study and found that the minor allele (A) of rs9939609 in the FTO gene was associated with significantly increased CRC risk in Iranian people.15 Semlali et al evaluated the TSLP rs10043985 and IL-7R rs1053496 SNPs and found that they could be used as markers for CRC development in the Saudi population, based on a case–control study.16 In addition, Li et al found that the ADCY9 polymorphism, rs2230742, was related to an increased risk of CRC in the Chinese Han population.17 Moreover, the rs12674822 variant of ANGPT2 was reported to be associated with CRC risk in Chinese subjects.18
SNPs in MMP2 have previously been found to be associated with CRC risk. The minor allele (T) of rs1053605 was found to have a protective role against CRC risk;19 however, we found no association between rs1053605 and the overall risk of CRC. Further, other MMP2 polymorphisms are reported to be related to CRC risk; rs243865 is significantly associated with decreased risk of CRC in the Kashmiri population,20 while this polymorphism was found to have no association with CRC risk in a meta-analysis.21 In addition, the rs2241145 variant in MMP2 was identified as associated with decreased overall survival of patients with CRC.22 To the best of our knowledge, this is the first study to explore the association between MMP2 rs243849 and CRC risk in the Chinese Han population. Future studies with large sample size are warranted to validate our findings.
MMPs are believed to play a vital role in tumor metastasis. Many studies have found that type IV collagenase, encoded by the MMP2 gene, is not only closely related to invasion and migration in tumor cells models23 but also correlated with tumor grade, invasion, and metastasis in human tumor biopsy tissues.24 Moreover, changes in MMP2 expression are related to the occurrence and development of CRC. For example, retinoic acid α promotes CRC carcinogenesis by increasing MMP2 transcription,25 and seven transmembrane domains containing 1 (ELTD1/ADGRL4) can promote CRC cell migration and invasion by activating MMP2 at the transcriptional level.11 Furthermore, TWIST1/2 can bind to the MMP2 promoter and induce its expression, resulting in increased epithelial-to-mesenchymal transition and invasion potential of CRC cells.26
In the present study, HaploReg v4.127 was used to investigate possible SNP functional effects. We found that both rs1053605 and rs243849 are synonymous variants, and that rs1053605 likely influences motif changes, GRASP QTL hits, and selected eQTL hits, while rs243849 likely affects enhancer histone marks, motif changes, GRASP QTL hits, and selected eQTL hits. Many studies have demonstrated that synonymous SNPs can confer susceptibility by impacting the splicing, regulation, and stability of mRNA,28,29 consequently influencing gene expression, and these SNPs may influence MMP2 expression via those mechanisms. Nevertheless, the biological processes underlying the effects of rs1053605 and rs243849 on CRC risk require further experimental exploration.
Although several positive associations were observed in this study, some limitations should be considered. First, the sample size was relatively small, with only 663 cases and 663 controls enrolled. Further, all participants were recruited from the same hospital; hence, inherent selection bias cannot be excluded and our findings cannot be extrapolated to other ethnic groups. Moreover, comprehensive clinical informations, such as tumor stage and lymph node metastasis data, etc., were incomplete, and are very important for analysis of associations between polymorphisms and clinical clinicopathological characteristics. Given these limitations, further studies with larger sample size and more comprehensive clinical information will be required to confirm our findings. Despite these limitations, our findings provide additional information on the limited evidence supporting the role of MMP2 polymorphisms in CRC risk in the Chinese Han population.
Conclusion
In summary, this study provides data on the impact of the rs1053605 and rs243849 polymorphisms in MMP2 on CRC susceptibility in a Chinese Han population, and our findings suggest that MMP2 variants are potential genetic markers of CRC risk.
Acknowledgments
This work was supported by the Sanming Project of Medicine in Shenzhen (No. SZSM201612021) and the Science and Technology Development Fund Project of Shenzhen (JCYJ20190809100217290).
Disclosure
The authors declare that they have no conflicts of interest related to this work.
References
1. Sung H, Ferlay J, Siegel RL. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi:10.3322/caac.21660
2. Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients. 2019;11:164. doi:10.3390/nu11010164
3. Li J, Ma X, Chakravarti D, Shalapour S, DePinho RA. Genetic and biological hallmarks of colorectal cancer. Genes Dev. 2021;35:787–820. doi:10.1101/gad.348226.120
4. Bhullar DS, Barriuso J, Mullamitha S, Saunders MP, O’Dwyer ST, Aziz O. Biomarker concordance between primary colorectal cancer and its metastases. EBioMedicine. 2019;40:363–374. doi:10.1016/j.ebiom.2019.01.050
5. Skrede OJ, De Raedt S, Kleppe A, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350–360. doi:10.1016/S0140-6736(19)32998-8
6. Choi CK, Shin MH, Cho SH, et al. Association between ALDH2 and ADH1B polymorphisms and the risk for colorectal cancer in Koreans. Cancer Res Treat. 2021;53:754–762. doi:10.4143/crt.2020.478
7. Guo X, Lin W, Wen W, et al. Identifying novel susceptibility genes for colorectal cancer risk from a transcriptome-wide association study of 125,478 subjects. Gastroenterology. 2021;160:1164–1178.e1166. doi:10.1053/j.gastro.2020.08.062
8. He Y, Timofeeva M, Zhang X. Colorectal cancer risk variants rs10161980 and rs7495132 are associated with cancer survival outcome by a recessive mode of inheritance. Int J Cancer. 2021;148:2774–2778. doi:10.1002/ijc.33465
9. Pittayapruek P, Meephansan J, Prapapan O, Komine M, Ohtsuki M. Role of matrix metalloproteinases in photoaging and photocarcinogenesis. Int J Mol Sci. 2016;17:868–887. doi:10.3390/ijms17060868
10. Han Y, Ma L, Zhao L, Feng W, Zheng X. Rosmarinic inhibits cell proliferation, invasion and migration via up-regulating miR-506 and suppressing MMP2/16 expression in pancreatic cancer. Biomed Pharmacother. 2019;115:108878–108886. doi:10.1016/j.biopha.2019.108878
11. Sun J, Zhang Z, Chen J, Xue M, Pan X. ELTD1 promotes invasion and metastasis by activating MMP2 in colorectal cancer. Int J Biol Sci. 2021;17:3048–3058. doi:10.7150/ijbs.62293
12. Tian Y, An F, Wang J, et al. MMP2 and MMP10 polymorphisms are related to steroid-induced osteonecrosis of the femoral head among Chinese Han population. Biomed Res Int. 2019;2019:8298193. doi:10.1155/2019/8298193
13. Ou YX, Bi R. Meta-analysis on the relationship between the SNP of MMP-2-1306 C>T and susceptibility to breast cancer. Eur Rev Med Pharmacol Sci. 2020;24:1264–1270. doi:10.26355/eurrev_202002_20181
14. Qaseem A, Crandall CJ, Mustafa RA, et al. Screening for colorectal cancer in asymptomatic average-risk adults: a guidance statement from the American College of Physicians. Ann Intern Med. 2019;171:643–654. doi:10.7326/M19-0642
15. Gholamalizadeh M, Akbari ME, Doaei S, et al. The association of fat-mass-and obesity-associated gene polymorphism (rs9939609) with colorectal cancer: a case-control study. Front Oncol. 2021;11:732515. doi:10.3389/fonc.2021.732515
16. Semlali A, Almutairi MH, Alamri A, Reddy Parine N. Expression and polymorphism of TSLP/TSLP receptors as potential diagnostic markers of colorectal cancer progression. Genes. 2021;12:1386. doi:10.3390/genes12091386
17. Li H, Liu Y, Liu J, et al. Assessment of ADCY9 polymorphisms and colorectal cancer risk in the Chinese Han population. J Gene Med. 2021;23:e3298. doi:10.1002/jgm.3298
18. Du Z, Tang CH, Li LJ, et al. Angiopoietin-2 gene polymorphisms are biomarkers for the development and progression of colorectal cancer in Han Chinese. Int J Med Sci. 2020;17:97–102. doi:10.7150/ijms.37675
19. Wang N, Zhou S, Fang XC, et al. MMP-2, −3 and TIMP-2, −3 polymorphisms in colorectal cancer in a Chinese Han population: a case-control study. Gene. 2020;730:144320. doi:10.1016/j.gene.2019.144320
20. Banday MZ, Sameer AS, Mir AH, Mokhdomi TA, Chowdri NA, Haq E. Matrix metalloproteinase (MMP) −2, −7 and −9 promoter polymorphisms in colorectal cancer in ethnic Kashmiri population - A case-control study and a mini review. Gene. 2016;589:81–89. doi:10.1016/j.gene.2016.05.028
21. McColgan P, Sharma P. Polymorphisms of matrix metalloproteinases 1, 2, 3 and 9 and susceptibility to lung, breast and colorectal cancer in over 30,000 subjects. Int J Cancer. 2009;125:1473–1478. doi:10.1002/ijc.24441
22. Scherer D, Deutelmoser H, Balavarca Y, Toth R. Polymorphisms in the angiogenesis-related genes EFNB2, MMP2 and JAG1 are associated with survival of colorectal cancer patients. Int J Mol Sci. 2020;21:5359. doi:10.3390/ijms21155395
23. Liu D, Kang H, Gao M, Jin L, Zhang F, Chen D. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol. 2020;14:1365–1380. doi:10.1002/1878-0261.12637
24. Tryggvason K, Höyhtyä M, Pyke C. Type IV collagenases in invasive tumors. Breast Cancer Res Treat. 1993;24:209–218. doi:10.1007/BF01833261
25. Huang GL, Chen QX, Ma JJ, Sui SY, Wang YN, Shen DY. Retinoic acid receptor α facilitates human colorectal cancer progression via Akt and MMP2 signaling. Onco Targets Ther. 2019;12:3087–3098. doi:10.2147/OTT.S200261
26. Lu K, Dong J, Fan W. Twist1/2 activates MMP2 expression via binding to its promoter in colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22:8210–8219. doi:10.26355/eurrev_201812_16514
27. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–881. doi:10.1093/nar/gkv1340
28. Karambataki M, Malousi A, Kouidou S. Risk-associated coding synonymous SNPs in type 2 diabetes and neurodegenerative diseases: genetic silence and the underrated association with splicing regulation and epigenetics. Mutat Res. 2014;770:85–93. doi:10.1016/j.mrfmmm.2014.09.005
29. Hussain SK, Madeleine MM, Johnson LG, et al. Nucleotide variation in IL-10 and IL-12 and their receptors and cervical and vulvar cancer risk: a hybrid case-parent triad and case-control study. Int J Cancer. 2013;133:201–213. doi:10.1002/ijc.28000
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.