Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 15
Missense Variant rs28362680 in BTNL2 Reduces Risk of Coronary Heart Disease
Authors Zhuo J, Wu Y, Li W, Li Z, Ding Y , Jin T
Received 8 December 2021
Accepted for publication 4 March 2022
Published 6 May 2022 Volume 2022:15 Pages 449—464
DOI https://doi.org/10.2147/PGPM.S353085
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Jian Zhuo,1 Yingchun Wu,2 Wei Li,1 Zerong Li,1 Yipeng Ding,3 Tianbo Jin4,5
1Department of Emergency Service, People’s Hospital of Wanning, Wanning, Hainan, 571500, People’s Republic of China; 2Department of Intensive Care Unit, Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University, Haikou, 570311, Hainan, People’s Republic of China; 3Department of General Practice, Hainan General Hospital, Hainan affiliated Hospital of Hainan Medical University, Haikou, 570311, Hainan, People’s Republic of China; 4Key Laboratory of Resource Biology and Biotechnology in Western China, Ministry of Education, Northwest University, Xi‘an, Shaanxi, 710069, People’s Republic of China; 5Provincial Key Laboratory of Biotechnology of Shaanxi Province, Northwest University, Xi’an, People’s Republic of China
Correspondence: Yipeng Ding, Department of General Practice, Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University, No. 19, Xinhua Road, Xiuying District, Haikou, 570311, Hainan, People’s Republic of China, Tel +86-18976335858, Email [email protected] Tianbo Jin, Key Laboratory of Resource Biology and Biotechnology in Western China, Ministry of Education, Northwest University, #229 Taibai North Road, Xi’an, 710069, Shaanxi, People’s Republic of China, Tel/Fax +86-29-88895902, Email [email protected]
Background: The pathological basis of coronary heart disease (CHD) is atherosclerosis. BTNL2 can inhibit the activation of T cells. We aimed to explore the association between BTNL2 genetic variants and CHD risk in the southern Chinese Han population.
Methods: We recruited 1419 participants to perform an association analysis between missense variants in BTNL2 and CHD risk through SNPStats online software. Genotyping of all candidate SNPs were completed by the Agena MassARRAY. In addition, we used false-positive report probability analysis to detect whether the positive findings were noteworthy observations. We also used Haploview 4.2 software and SNPStats online software to conduct the haplotype analysis and analysis of linkage disequilibrium (LD). Finally, the interaction of SNP-SNP in CHD risk was evaluated by multi-factor dimensionality reduction (MDR).
Results: The results showed that BTNL2-rs35624343, -rs117896888, -rs41441651, -rs41417449, -rs28362680 and -rs2076523 were significantly associated with the CHD susceptibility. Especially for BTNL2-rs28362680, the allele A (OR = 0.68, p < 0.0001), genotype AA (OR = 0.40, p = 0.001) or GA (OR = 0.68, p < 0.0001) were associated with the reducing CHD risk. And -rs28362680 significantly reduced the CHD risk under all genetic models (dominant: OR = 0.64, p < 0.0001; recessive: OR = 0.47, p = 0.003; overdominant: OR = 0.73, p = 0.004; log-additive: OR = 0.66, p < 0.0001). And -rs28362680 was also closely associated with CHD risk reduction in all stratified analyses (age, gender, smoking, drinking, hypertension and diabetes). In addition, haplotype analysis showed that the “Crs117896888Crs41441651Trs41417449Ars28362680” (OR = 0.65, p < 0.0001) and “Grs117896888Trs41441651Crs41417449Ars28362680” (OR = 0.68, p = 0.013) may reduce CHD risk.
Conclusion: Missense variants (rs35624343, rs117896888, rs41441651, rs41417449, rs28362680, rs2076523) may be protective factors for the CHD risk. In particular, there were sufficient evidences that BTNL2-rs28362680 can protective CHD risk.
Keywords: coronary heart disease, BTNL2, missense variants, southern Chinese Han population
Introduction
Coronary heart disease (CHD), as a major cardiovascular disease, is the leading cause of death in the global population. The pathological basis of coronary heart disease is atherosclerosis (AS).1 CHD is a complex disease that involves multiple mechanisms, multiple cell types, and is affected by multiple environmental factors, including smoking,2 drinking,3 poor eating habits,4 lifestyle, etc.5 These life-related factors further promote the occurrence of traditional risk factors for coronary heart disease, including diabetes,6 dyslipidemia7 and hypertension.8,9 In addition, with the development of molecular Biology and the improvement and application of genetic testing technology, the influence of genetic factors on disease risk has been highly valued. More and more studies have proved that genetic factors play an important role in the occurrence and development of CHD.10 CHD has an important genetic basis and is considered equivalent to environmental factors. At present, some genes single nucleotide polymorphism (SNP) have been found to be associated with CHD susceptibility, such as XKR6,11 interleukin-6 receptor,12 T-cell immunoglobulin and mucin domain 4 gene,13 etc. However, it should be noted that the genetic mechanism of coronary heart disease is not fully clearly. The discovery of SNPs associated with the occurrence and development of diseases plays an important role in exploring the susceptibility and pathogenesis of diseases, making the prevention, diagnosis and treatment of diseases in the future full of hope.
BTNL2 belongs to the butyrophilin-like (BTNL) family. Studies have found that BTNL2 acts as a co-inhibitory molecule for T cell activation.14 In addition, the genetic polymorphism of BTNL2 is associated with the susceptibility of Kawasaki disease15 and dilated cardiomyopathy.16 Kawasaki disease is a child vasculitis with a higher risk of coronary heart disease in adulthood.17 In summary, BTNL2 is very likely to participate in the occurrence and development of coronary heart disease. But so far, there is no relevant report on the association between BTNL2 genetic polymorphism and coronary heart disease.
Therefore, this study intends to conduct a “case-control” study with the Han population in southern China as the research subject. The purpose is to explore the association between BTNL2 genetic polymorphism and CHD susceptibility. At the same time, we conducted a stratified analysis based on other potential risk factors for CHD (age, smoking, drinking, hypertension, diabetes, etc.) to assess the association between BTNL2 genetic polymorphisms and these influencing factors. This study will lay a foundation for the study on mechanism of BTNL2 in the occurrence and development of coronary heart disease. And it will provide new ideas for the individualized prevention and treatment of CHD.
Materials and Methods
Sample Source
We recruited 710 patients with coronary heart disease and 709 healthy individuals for this case-control study. The 1419 participants were recruited in the same hospital (People’s Hospital of Wanning). We continuously recruited 710 CHD patients in the outpatient or inpatient department of cardiovascular. These patients were all newly diagnosed. Recruitment criteria: patients met the clear and diagnostic criteria of the World Health Organization (WHO) on coronary heart disease. Patients with coronary angiography indications were examined by coronary angiography, screening patients with coronary angiography at least one lumen diameter less than 50% or more. Exclusion criteria: patients with cardiomyopathy, heart valve disease, congenital heart disease or renal failure were excluded. During the same period, 709 healthy individuals were recruited in the health examination center as the control group in this study. Control group recruitment criteria: participants without family history of premature coronary heart disease, history of ischemic cardiomyopathy or history of chronic heart failure, and participants without hypertension, diabetes, hyperlipidemia, hyperuricemia and obesity will be enrolled in control group. We obtained the demographic information and basic epidemiological information of the participants by consulting medical records or professional questionnaire survey. After obtaining the informed consent of all participants, we collected their blood samples for further study. This study has been approved by the ethics committee of the Northwest University and People’s Hospital of Wanning before the beginning.
Selection of SNPs
First, we found that the physical position of the BTNL2 was on the Chromosome 6: 32393963–32407128 through the e!GRCh37 database (http://asia.ensembl.org/Homo_sapiens/Info/Index). Then, we use the online converter window (VCF to PED: http://grch37.ensembl.org/Homo_sapiens/Tools/VcftoPed) to download the related files of BTNL2 genetic variants. For the population, we choose CHB and CHS. Finally, after setting specific conditions, six candidate BTNL2 SNPs (rs35624343, rs117896888, rs41441651, rs41417449, rs28362680, rs2076523) were selected using Haploview software. These special conditions are as follows: Tagger r2 > 0.8, Min Genotype > 75%, MAF> 0.05 and HWE> 0.01.
DNA Extraction and Genotyping
In this study, we chose the kit (GoldMag Co. Ltd. Xi’an, China) to complete the extraction and purification of genomic DNA, and the specific experimental steps were carried out according to the instructions. According to the DNA sequence of BTNL2, we designed specific amplification primers and extension primers for candidate genetic loci (All primers can be found in Supplemental Table 1) by MassARRAY Assay Design software. Genotyping was performed by the MassARRAY ® -IPLEX SNP genotyping technology.
At the same time, we randomly selected 5% DNA samples for repeated experiments. The repetition rate of the results should reach >99% to ensure the reliability and repeatability of the experimental results.
Data Analysis
We used online software (HaploReg v4.1: https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) to predict the potential function of candidate SNPs, and the information related to SNPs was searched through dbSNP online database (https://www.ncbi.nlm.nih.gov/snp/). The statistical analysis of this study was mainly completed by SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). In this study, the association between CHD risk and candidate genetic polymorphism under multiple genetic models was completed by SNPStats online software (https://www.snpstats.net/start.htm?q=snpstats/start.htm). We mainly evaluated the impact of candidate SNPs on the risk of coronary heart disease through the Odds ratios (OR) and 95% confidence intervals (CI). In order to avoid the influence of confounding factors on the results, all the results were adjusted by the confounding factors, such as age, gender, smoking or drinking. We also used false-positive report probability (FPRP) analysis to detect whether all positive results are noteworthy at a prior probability level of 0.25 and FPRP threshold of 0.2. At the same time, the statistic power analysis will be completed. Haploview 4.2 software and SNPStats online software were used to perform the haplotype analysis of candidate SNPs and evaluation of linkage disequilibrium (LD). Finally, the interaction of candidate SNPs in IS risk was evaluated by multi-factor dimensionality reduction (MDR). The p < 0.05 indicated statistically significant.
Results
There were 1419 participants in the case group (710) and the control group (709) enrolled in this study. The average ages of the case and control groups were 62.58 ± 10.49 years and 62.23 ± 7.44 years (Table 1). The number of males and females in the case group were 481 (67.7%) and 229 (32.3%), and in the control group were 479 (67.6%) and 230 (32.4%), respectively. There was no statistical difference in gender and age between the participants in the two groups, which indicated that the two groups were matched in age and gender. The basic information of the participants can be found in Table 1.
 |
Table 1 Characteristics of Patients with CHD and Healthy Individuals |
Genotyping and Information About Candidate SNPs
Six BTNL2 candidate genetic loci (rs35624343 A/G, rs117896888 G/C, rs41441651 T/C, rs41417449 C/T, rs28362680 A/G, rs2076523 C/T) have been successfully genotyped. These genetic variants are all missense variants in BTNL2, and dbSNP online database shows that these missense variants may lead to amino acid changes (Table 2). After testing, the distribution of all candidate genetic loci is in accordance with Hardy-Weinberg equilibrium (HWE p > 5%). HaploReg online software was used to predict the potential functions of candidate genetic loci and found that the 6 SNPs may be regulated by a variety of factors. These factors include Promoter histone marks; Enhancer histone marks; DNAse; Proteins bound; Motifs changed; Selected eQTL hits, etc. (Table 2).
 |
Table 2 The Basic Information and HWE About the Selected SNPs of BTNL2 |
Association Analysis Between BTNL2 SNPS and CHD Risk (Overall Analysis)
The association analysis between candidate SNPs and the risk of CHD showed that BTNL2-rs35624343, -rs117896888, -rs41441651, -rs41417449 and -rs28362680 were significantly associated with the susceptibility of CHD among participants (Table 3). Specifically, compared with the GG genotype, the GA genotype of BTNL2-rs35624343 was a protective factor for CHD (GA vs GG, OR = 0.78, 95% CI = 0.62–0.99, p = 0.040). BTNL2-rs35624343 can significantly reduce the CHD risk under dominant (GA-AA vs GG, OR = 0.78, 95% CI = 0.62–0.98, p = 0.036), overdominant (GG-AA vs GA, OR = 0.79, 95% CI = 0.62–0.99, p = 0.045), log-additive genetic models (OR = 0.80, 95% CI = 0.65–0.99, p = 0.043). -rs117896888 can significantly reduce the CHD risk under allele (G vs C, OR = 0.74, 95% CI = 0.55–0.98, p = 0.036) and log-additive genetic models (OR = 0.73, 95% CI = 0.55–0.98, p = 0.035). -rs41441651 can significantly reduce the CHD risk under allele (T vs C, OR = 0.73, 95% CI = 0.54–0.97, p = 0.028), dominant (CT-TT vs CC, OR = 0.72, 95% CI = 0.53–0.98, p = 0.036) and log-additive genetic models (OR = 0.72, 95% CI = 0.54–0.97, p = 0.027). -rs41417449 can significantly reduce the CHD risk under allele (C vs T, OR = 0.73, 95% CI = 0.55–0.97, p = 0.031), dominant (CT-CC vs TT, OR = 0.73, 95% CI = 0.54–0.99, p = 0.040) and log-additive genetic models (OR = 0.73, 95% CI = 0.54–0.97, p = 0.030). Compared with wild allele G, allele A of -rs28362680 was a protective factor for the risk of CHD (A vs G, OR = 0.68, 95% CI = 0.57–0.81, p < 0.0001). Compared with the GG genotype, the AA (OR = 0.40, 95% CI = 0.23–0.67, p = 0.001) and GA (OR = 0.68, 95% CI = 0.54–0.84, p < 0.0001) genotypes were protective factors for CHD risk. In addition, -rs28362680 significantly reduced the risk of CHD among participants under all genetic models (dominant: OR = 0.64, 95% CI = 0.52–0.79, p < 0.0001; recessive: OR = 0.47, 95% CI = 0.28–0.78, p = 0.003; overdominant: OR = 0.73, 95% CI = 0.59–0.90, p = 0.004; log-additive: OR = 0.66, 95% CI = 0.55–0.79, p < 0.0001).
 |
Table 3 The Association Analysis Between Susceptibility of CHD and Single Nucleotide Polymorphism of BTNL2 |
We found no evidence that BTNL2-rs28362680 was associated with the CHD risk among the participants.
Association Analysis Between BTNL2 SNPS and CHD Risk (Stratified Analysis)
In order to further avoid the influence of other factors on the reliability of the association analysis results, we divided the participants according to different influencing factors and carried out a stratified analysis (age, gender, smoking, drinking, or CHD complicated with hypertension/diabetes).
Age: the results showed that BTNL2-rs28362680 was significantly associated with CHD risk in people ≤ 62 years old or > 62 years old (Supplemental Table 2). Among participants ≤ 62 years old, -rs28362680 significantly reduced the risk of CHD under all genetic models (Allele: OR = 0.66, 95% CI = 0.52–0.84, p = 0.001. Genotype (AA): OR = 0.66, 95% CI = 0.52–0.84, p = 0.001; Genotype (GA): OR = 0.67, 95% CI = 0.49–0.92, p = 0.012. Dominant: OR = 0.62, 95% CI = 0.46–0.84, p = 0.002. Recessive: OR = 0.37, 95% CI = 0.17–0.80, p = 0.008. Overdominant: OR = 0.73, 95% CI = 0.54–0.98, p = 0.037. Log-additive: OR = 0.63, 95% CI = 0.48–0.81, p < 0.0001). Among participants > 62 years old, -rs28362680 significantly reduced the risk of CHD under multiple genetic models (Allele: OR = 0.70, 95% CI = 0.55–0.90, p = 0.005. Genotype (AA): OR = 0.44, 95% CI = 0.20–0.93, p = 0.032; Genotype (GA): OR = 0.67, 95% CI = 0.48–0.93, p = 0.017. Dominant: OR = 0.66, 95% CI = 0.49–0.89, p = 0.007. Overdominant: OR = 0.71, 95% CI = 0.52–0.97, p = 0.033. Log-additive: OR = 0.70, 95% CI = 0.54–0.90, p = 0.006). In the age stratified analysis, the remaining 5 candidate SNPs were not associated with the CHD risk.
Gender: the results showed that BTNL2-rs28362680 was significantly associated with CHD susceptibility in both male and female participants (Supplemental Table 2). Among female participants, -rs28362680 significantly reduced the risk of CHD under all genetic models (Allele: OR = 0.57, 95% CI = 0.42–0.79, p = 0.001. Genotype (AA): OR = 0.31, 95% CI = 0.12–0.84, p = 0.020; Genotype (GA): OR = 0.56, 95% CI = 0.38–0.83, p = 0.004. Dominant: OR = 0.47, 95% CI = 0.32–0.70, p < 0.0001. Recessive: OR = 0.38, 95% CI = 0.14–1.01, p = 0.042. Overdominant: OR = 0.55, 95% CI = 0.36–0.82, p = 0.004. Log-additive: OR = 0.51, 95% CI = 0.36–0.73, p < 0.0001). Among male participants, -rs28362680 significantly reduced the risk of CHD under multiple genetic models (Allele: OR = 0.73, 95% CI = 0.59–0.90, p = 0.003. Genotype (AA): OR = 0.45, 95% CI = 0.24–0.85, p = 0.013; Genotype (GA): OR = 0.73, 95% CI = 0.56–0.96, p = 0.022. Dominant: OR = 0.69, 95% CI = 0.53–0.89, p = 0.004. Recessive: OR = 0.51, 95% CI = 0.27–0.95, p = 0.030. Log-additive: OR = 0.70, 95% CI = 0.56–0.87, p = 0.001).
In addition, allele A (A vs G: OR = 0.76, 95% CI = 0.59–0.97, p = 0.031) and genotype GA of BTNL2-rs35624343 (GA vs GG: OR = 0.74, 95% CI = 0.55–0.99, p = 0.040) were protective factors for CHD in male participants. BTNL2-rs35624343 also significantly reduced the risk of CHD under dominant (OR = 0.74, 95% CI = 0.56–0.98, p = 0.039) and log-additive genetic models (OR = 0.76, 95% CI = 0.58–0.98, p = 0.037).
Allele G (G vs C: OR = 0.71, 95% CI = 0.50–1.00, p = 0.049) and genotype CG of BTNL2-rs117896888 (CG vs CC: OR = 0.68, 95% CI = 0.47–0.98, p = 0.038) were protective factors for CHD in male participants. BTNL2-rs117896888 also significantly reduced the risk of CHD under dominant (OR = 0.67, 95% CI = 0.47–0.97, p = 0.032), overdominant (OR = 0.66, 95% CI = 0.46–0.96, p = 0.030) and log-additive genetic models (OR = 0.69, 95% CI = 0.49–0.99, p = 0.040).
Allele T (T vs C: OR = 0.69, 95% CI = 0.49–0.98, p = 0.035) and genotype CT of BTNL2-rs41441651 (CT vs CC: OR = 0.66, 95% CI = 0.46–0.95, p = 0.026) were protective factors for CHD in male participants. BTNL2-rs41441651 also significantly reduced the risk of CHD under dominant (OR = 0.65, 95% CI = 0.45–0.94, p = 0.022), overdominant (OR = 0.65, 95% CI = 0.45–0.94, p = 0.020) and log-additive genetic models (OR = 0.68, 95% CI = 0.48–0.96, p = 0.028).
Allele C (C vs T: OR = 0.70, 95% CI = 0.49–0.99, p = 0.040) and genotype CT of BTNL2-rs41417449 (CT vs TT: OR = 0.67, 95% CI = 0.46–0.96, p = 0.030) were protective factors for CHD in male participants. BTNL2-rs41417449 also significantly reduced the risk of CHD under dominant (OR = 0.66, 95% CI = 0.46–0.95, p = 0.026), overdominant (OR = 0.65, 95% CI = 0.45–0.95, p = 0.024) and log-additive genetic models (OR = 0.68, 95% CI = 0.48–0.97, p = 0.032).
We found no evidence that BTNL2-rs2076523 was associated with the CHD risk in age or gender stratified analysis.
Smoking: the results showed that BTNL2-rs28362680 was significantly associated with CHD susceptibility among smokers or non-smokers (Supplemental Table 3). Among smoking participants, -rs28362680 significantly reduced the risk of CHD under all genetic models (Allele: OR =0.65, 95% CI = 0.51–0.82, p < 0.0001. Genotype (AA): OR = 0.37, 95% CI = 0.18–0.80, p = 0.011; Genotype (GA): OR = 0.59, 95% CI = 0.43–0.81, p = 0.001. Dominant: OR = 0.56, 95% CI = 0.41–0.77, p < 0.0001. Recessive: OR = 0.47, 95% CI = 0.22–0.99, p = 0.040. Overdominant: OR = 0.64, 95% CI = 0.47–0.87, p = 0.005. Log-additive: OR = 0.60, 95% CI = 0.46–0.78, p < 0.0001). Among non-smoking participants, -rs28362680 significantly reduced the risk of CHD under multiple genetic models (Allele: OR = 0.71, 95% CI = 0.56–0.91, p = 0.007. Genotype (AA): OR = 0.40, 95% CI = 0.19–0.85, p = 0.018. Dominant: OR = 0.70, 95% CI = 0.52–0.95, p = 0.020. Recessive: OR = 0.46, 95% CI = 0.22–0.96, p = 0.033. Log-additive: OR = 0.70, 95% CI = 0.54–0.91, p = 0.006).
Compared with wild allele G, allele A of BTNL2-rs35624343 was significantly associated with reducing CHD risk among smokers (A vs G, OR = 0.70, 95% CI = 0.52–0.94, p = 0.019). BTNL2-rs35624343 also significantly reduced the risk of CHD under log-additive genetic model (OR = 0.71, 95% CI = 0.52–0.97, p = 0.032) among smoking participants.
Compared with wild allele C, allele G of BTNL2-rs117896888 was significantly associated with reducing CHD risk among smokers (G vs C, OR = 0.64, 95% CI = 0.42–0.95, p = 0.027). BTNL2-rs117896888 also significantly reduced the risk of CHD under dominant (OR = 0.64, 95% CI = 0.42–0.99, p = 0.043) and log-additive genetic model (OR = 0.65, 95% CI = 0.43–0.97, p = 0.035) among smoking participants.
Compared with wild allele C, allele T of BTNL2-rs41441651 was significantly associated with reducing CHD risk among smokers (T vs C, OR = 0.63, 95% CI = 0.42–0.94, p = 0.023). Genotype (CT) of BTNL2-rs35624343 was significantly associated with reducing CHD risk among smokers (CT vs CC, OR = 0.64, 95% CI = 0.41–0.98, p = 0.041). BTNL2-rs35624343 also significantly reduced the risk of CHD under dominant (OR = 0.63, 95% CI = 0.41–0.97, p = 0.036) and log-additive genetic model (OR = 0.64, 95% CI = 0.42–0.96, p = 0.030) among smoking participants.
Compared with wild allele T, allele C of BTNL2-rs41417449 was significantly associated with reducing CHD risk among smokers (C vs T, OR = 0.63, 95% CI = 0.42–0.95, p = 0.026). Genotype (CT) of BTNL2-rs41417449 was significantly associated with reducing CHD risk among smokers (CT vs TT, OR = 0.64, 95% CI = 0.42–1.00, p = 0.048). BTNL2-rs41417449 also significantly reduced the risk of CHD under dominant (OR = 0.64, 95% CI = 0.41–0.99, p = 0.042) and log-additive genetic model (OR = 0.64, 95% CI = 0.43–0.97, p = 0.034) among smoking participants.
Compared with wild allele T, allele C of BTNL2-rs2076523 was significantly associated with reducing CHD risk among smokers (C vs T, OR = 0.78, 95% CI = 0.63–0.96, p = 0.020). Genotype (CC) of BTNL2-rs2076523 was significantly associated with reducing CHD risk among smokers (CC vs TT, OR = 0.58, 95% CI = 0.37–0.91, p = 0.017). BTNL2-rs2076523 also significantly reduced the risk of CHD under dominant (OR = 0.72, 95% CI = 0.52–0.99, p = 0.041), recessive (OR = 0.66, 95% CI = 0.44–0.99, p = 0.045) and log-additive genetic model (OR = 0.76, 95% CI = 0.61–0.95, p = 0.013) among smoking participants.
This study found that with the exception of BTNL2-rs28362680, the remaining five candidate SNPs were not associated with the risk of CHD in non-smoking participants.
Drinking: the results showed that BTNL2-rs28362680 was significantly associated with CHD susceptibility among drinking or non-drinking participants (Supplemental Table 3). Among drinking participants, -rs28362680 significantly reduced the risk of CHD under multiple genetic models (Allele: OR =0.65, 95% CI = 0.51–0.83, p = 0.001. Genotype (AA): OR = 0.29, 95% CI = 0.13–0.67, p = 0.004; Genotype (GA): OR = 0.68, 95% CI = 0.50–0.94, p = 0.018. Dominant: OR = 0.64, 95% CI = 0.47–0.87, p = 0.004. Recessive: OR = 0.33, 95% CI = 0.15–0.76, p = 0.006. Log-additive: OR = 0.63, 95% CI = 0.48–0.82, p = 0.001). Among non-drinking participants, -rs28362680 significantly reduced the risk of CHD under multiple genetic models (Allele: OR = 0.71, 95% CI = 0.56–0.90, p = 0.005. Genotype (GA): OR = 0.66, 95% CI = 0.48–0.89, p = 0.007. Dominant: OR = 0.64, 95% CI = 0.47–0.86, p = 0.003. Overdominant: OR = 0.69, 95% CI = 0.51–0.93, p = 0.015. Log-additive: OR = 0.68, 95% CI = 0.53–0.88, p = 0.003).
We found no evidence that the five remaining candidate SNPs were associated with the CHD risk in drinking stratified analysis.
CHD complicated with hypertension: the results showed that BTNL2-rs28362680 was significantly associated with the risk of CHD patients complicated with or without hypertension (Supplemental Table 4). Among CHD patients complicated with hypertension, -rs28362680 significantly associated with the CHD risk under all genetic models (Allele: OR =0.69, 95% CI = 0.56–0.84, p < 0.0001. Genotype (AA): OR = 0.35, 95% CI = 0.18–0.68, p = 0.002; Genotype (GA): OR = 0.72, 95% CI = 0.56–0.92, p = 0.010. Dominant: OR = 0.67, 95% CI = 0.53–0.86, p = 0.001. Recessive: OR = 0.40, 95% CI = 0.21–0.77, p = 0.003. Overdominant: OR = 0.78, 95% CI = 0.61–0.99, p = 0.044. Log-additive: OR = 0.67, 95% CI = 0.55–0.83, p < 0.0001). Among CHD patients complicated without hypertension, -rs28362680 significantly associated with the risk of CHD under multiple genetic models (Allele: OR = 0.66, 95% CI = 0.52–0.85, p = 0.001. Genotype (AA): OR = 0.47, 95% CI = 0.23–0.96, p = 0.038; Genotype (GA): OR = 0.62, 95% CI = 0.46–0.84, p = 0.002. Dominant: OR = 0.60, 95% CI = 0.45–0.80, p = 0.001. Overdominant: OR = 0.66, 95% CI = 0.49–0.89, p = 0.005. Log-additive: OR = 0.64, 95% CI = 0.50–0.83, p < 0.0001).
Genotype (CC) of BTNL2-rs2076523 was significantly associated with CHD risk among CHD patients complicated without hypertension (CT vs TT, OR = 0.71, 95% CI = 0.52–0.97, p = 0.032). BTNL2-rs2076523 were also significantly associated with CHD risk under overdominant genetic model (OR = 0.75, 95% CI = 0.56–1.00, p = 0.048) among CHD patients complicated without hypertension.
We found no evidence that the remaining candidate SNPs were associated with the CHD risk in stratified analysis (CHD complicated with hypertension).
CHD complicated with diabetes: the results showed that BTNL2-rs28362680 was significantly associated with the risk of CHD patients complicated with or without diabetes (Supplemental Table 4). Among CHD patients complicated with diabetes, -rs28362680 significantly associated with the CHD risk under multiple genetic models (Allele: OR =0.72, 95% CI = 0.55–0.94, p = 0.014. Genotype (AA): OR = 0.35, 95% CI = 0.13–0.89, p = 0.028. Dominant: OR = 0.72, 95% CI = 0.52–0.99, p = 0.042. Recessive: OR = 0.38, 95% CI = 0.15–0.98, p = 0.024. Log-additive: OR = 0.70, 95% CI = 0.53–0.93, p = 0.012). Among CHD patients complicated without diabetes, -rs28362680 significantly associated with the CHD risk under all genetic models (Allele: OR =0.66, 95% CI = 0.55–0.80, p < 0.0001. Genotype (AA): OR = 0.42, 95% CI = 0.23–0.75, p = 0.003; Genotype (GA): OR = 0.64, 95% CI = 0.51–0.82, p < 0.0001. Dominant: OR = 0.61, 95% CI = 0.48–0.77, p < 0.0001. Recessive: OR = 0.50, 95% CI = 0.28–0.89, p = 0.014. Overdominant: OR = 0.68, 95% CI = 0.54–0.87, p = 0.002. Log-additive: OR = 0.64, 95% CI = 0.53–0.78, p < 0.0001).
Compared with wild allele G, allele A of BTNL2-rs35624343 was significantly associated with the risk of CHD patients complicated without diabetes (A vs G, OR = 0.77, 95% CI = 0.61–0.97, p = 0.027). Genotype (GA) of BTNL2-rs35624343 was significantly associated with the risk of CHD patients complicated without diabetes (GA vs GG, OR = 0.74, 95% CI = 0.57–0.97, p = 0.026). BTNL2-rs35624343 was also significantly associated with the risk of CHD under dominant (OR = 0.74, 95% CI = 0.57–0.96, p = 0.020), Overdominant (OR = 0.75, 95% CI = 0.58–0.97, p = 0.030) and log-additive genetic models (OR = 0.76, 95% CI = 0.60–0.96, p = 0.022) among CHD patients complicated without diabetes.
Compared with wild allele C, allele G of BTNL2-rs117896888 was significantly associated with the risk of CHD patients complicated without diabetes (G vs C, OR = 0.65, 95% CI = 0.47–0.91, p = 0.010). Genotype (CG) of BTNL2-rs117896888 was significantly associated with the risk of CHD patients complicated without diabetes (CG vs CC, OR = 0.67, 95% CI = 0.47–0.95, p = 0.024). BTNL2-rs117896888 was also significantly associated with the risk of CHD under dominant (OR = 0.65, 95% CI = 0.46–0.92, p = 0.013), Overdominant (OR = 0.67, 95% CI = 0.47–0.95, p = 0.024) and log-additive genetic models (OR = 0.65, 95% CI = 0.47–0.90, p = 0.009) among CHD patients complicated without diabetes.
Compared with wild allele C, allele T of BTNL2-rs41441651 was significantly associated with the risk of CHD patients complicated without diabetes (T vs C, OR = 0.64, 95% CI = 0.46–0.89, p = 0.008). Genotype (CT) of BTNL2-rs41441651 was significantly associated with the risk of CHD patients complicated without diabetes (CT vs CC, OR = 0.66, 95% CI = 0.46–0.93, p = 0.018). BTNL2-rs41441651 was also significantly associated with the risk of CHD under dominant (OR = 0.64, 95% CI = 0.45–0.90, p = 0.010), Overdominant (OR = 0.66, 95% CI = 0.47–0.94, p = 0.018) and log-additive genetic models (OR = 0.64, 95% CI = 0.46–0.89, p = 0.007) among CHD patients complicated without diabetes.
Compared with wild allele T, allele C of BTNL2-rs41417449 was significantly associated with the risk of CHD patients complicated without diabetes (C vs T, OR = 0.65, 95% CI = 0.46–0.90, p = 0.009). Genotype (CT) of BTNL2-rs41417449 was significantly associated with the risk of CHD patients complicated without diabetes (CT vs TT, OR = 0.66, 95% CI = 0.47–0.94, p = 0.020). BTNL2-rs41417449 was also significantly associated with the risk of CHD under dominant (OR = 0.64, 95% CI = 0.45–0.91, p = 0.011), Overdominant (OR = 0.66, 95% CI = 0.47–0.94, p = 0.020) and log-additive genetic models (OR = 0.64, 95% CI = 0.46–0.90, p = 0.008) among CHD patients complicated without diabetes.
Compared with wild allele T, allele C of BTNL2-rs2076523 was significantly associated with the risk of CHD patients complicated without diabetes (C vs T, OR = 0.82, 95% CI = 0.69–0.96, p = 0.017). Genotype CT (CT vs TT, OR = 0.69, 95% CI = 0.48–0.99, p = 0.045) and CC (CC vs TT, OR = 0.74, 95% CI = 0.58–0.95, p = 0.020) of BTNL2-rs2076523 was significantly associated with the risk of CHD patients complicated without diabetes. BTNL2-rs2076523 was also significantly associated with the risk of CHD under dominant (OR = 0.73, 95% CI = 0.58–0.93, p = 0.010) and log-additive genetic models (OR = 0.81, 95% CI = 0.68–0.96, p = 0.014) among CHD patients complicated without diabetes.
This study found that with the exception of BTNL2-rs28362680, the remaining five candidate SNPs were not associated with the risk of CHD among CHD patients complicated with diabetes.
FPRP Analysis
The results of FPRP analysis showed that a very few positive results may be accidental, and most of them were noteworthy findings at the prior probability level of 0.25 and FPRP threshold of 0.2 (Supplemental Table 5). And the statistical power value of noteworthy positive results in this study reached more than 90%.
Specifically, under certain genetic models, BTNL2-rs28362680 was potentially associated with the CHD risk among women (genotype AA: prior probability = 0.269; recessive: prior probability = 0.351), > 62 years old (genotype AA: prior probability = 0.204), smoking (recessive: prior probability = 0.245), non-smoking participants (recessive: prior probability = 0.219), as well as CHD patients complicated with diabetes (genotype AA: prior probability = 0.266; recessive: prior probability = 0.323). The above positive results may not be worth noting. The remaining positive findings were all noteworthy in this study.
Haplotype Analysis
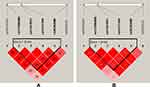
The results are shown in Figure 1, color represents the degree of linkage imbalance, and the redder the color, the stronger the linkage disequilibrium. We can see that the four candidate BTNL2 SNPs (rs117896888, rs41441651, rs41417449, rs28362680) can composed one linkage disequilibrium block. And the results of haplotype analysis showed that the haplotype “Crs117896888Crs41441651Trs41417449Ars28362680” (OR = 0.65, CI = 0.53–0.80, p < 0.0001) and “Grs117896888Trs41441651Crs41417449Ars28362680” (OR = 0.68, CI = 0.51–0.92, p = 0.013) all reduced the susceptibility of CHD among southern Chinese Han population (Table 4).
 |
Table 4 Haplotype Analysis of Candidate BTNL2 Genetic Polymorphisms with CHD Risk |
MDR Analysis
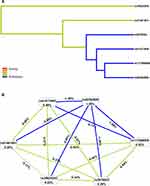
The MDR results showed that the single locus model (rs28362680) has the highest test accuracy (Table 5). As is shown in Figure 2A, the dendrogram has described the interaction between the six candidate SNPs. The color of the lines in the dendrogram represents the level of redundancy or synergy. The closer to the red, the stronger the synergy, and the closer to the blue, the stronger the redundancy. In addition, information gain (IG) is used to evaluate attribute interactions. As shown in Figure 2B, the IG value of rs28362680 is the highest. Therefore, we chose the unit locus model composed of rs28362680 as the best model for predicting CHD risk, with a test accuracy of 0.558 and a perfect CVC = 10/10.
 |
Table 5 BTNL2 SNP–SNP Interaction Models Analyzed by the MDR Method |
Discussion
In this study, we conducted an association study between BTNL2 genetic polymorphism and CHD susceptibility in southern Chinese Han population among 1419 participants. Participants were grouped according to the common CHD risk factors, and the association between BTNL2 genetic polymorphism and potential CHD risk factors was evaluated. Our results showed a strong association between BTNL2-rs28362680 and CHD susceptibility. Compared with the allele G and genotype GG, participants with allele A and genotype AA, GA of BTNL2-rs28362680 had lower risk of CHD (OR < 1). More importantly, the BTNL2-rs28362680 genetic polymorphism was significantly associated with a decreased risk of CHD in both overall and stratified analyses. Our study suggest that the allele A of BTNL2-rs28362680 can reduce the incidence of CHD in southern Chinese Han population.
It is worth noting that the results of all stratified analyses showed that the BTNL2-rs28362680 was significantly associated with the CHD susceptibility, which was not affected by other potential risk factors. That is to say, gender (male or female), age (≤62 years oldor >62 years old), smoking (smoking or non-smoking), drinking (drinking or non-drinking), hypertension (CHD complicated with hypertension or not), diabetes (CHD complicated with diabetes or not) cannot change the fact that BTNL2-rs28362680 was significantly associated with CHD susceptibility among participants. However, we all known that the occurrence and development of CHD will be affected by a variety of external factors.18,19 Hajek et al have reported that the genetic risk score of CHD can predict the risk of cardiovascular disease in men but not women.20 Many studies have shown that age and gender are the main determinants of CHD risk.21,22 Male,23,24 smokers,25 and older people26 are at higher risk of CHD. In addition, compared with non-diabetic patients, diabetes is the main risk factor for coronary artery disease (CAD).27 Hypertension induces endothelial dysfunction and exacerbates the process of atherosclerosis.28 And hypertension is also a risk factor for CHD.18 However, the results of our study showed that the A allele of BTNL2-rs28362680 was significantly associated with reduction of the CHD risk in all subgroup analyses. And this protection was not affected by other risk factors. So far, only one study has reported that BTNL2-rs28362680 was a new risk site for Crohn’s disease (CD) in Koreans.29 This study is the first to find that BTNL2-rs28362680 is a protective factor of CHD in Southern Chinese Han population. Based on the FPRP analysis and large sample size of this study, our new findings are very noteworthy.
In addition to finding that BTNL2-rs28362680 was significantly associated with participants’ CHD susceptibility, we also found that BTNL2-rs35624343, -rs117896888, -rs41441651 and -rs41417449 were associated with CHD susceptibility in the overall analysis and multiple stratified analyses. In this study, the above four candidate SNPs were significantly associated CHD risk reduction in the overall analysis, and similar findings were found in stratified analysis (male, smoking, and CHD patients without diabetes). A large-scale genome-wide association study showed that CHD and diabetes share a common genetic background.30 Studies have reported that BTNL2 genetic polymorphism is associated with the susceptibility of diabetes in Caucasians.31 Combined with the results of our study, we speculate that BTNL2-rs35624343, -rs117896888, -rs41441651 and -rs41417449 may reduce the risk of diabetes and thereby reduce the risk of CHD. Secondly, male and smoking have been reported as risk factors for CHD. Based on the results of this study, we speculate that these four candidate SNPs are protective genetic factors for CHD in southern Chinese Han population, and this protective effect will not be affected by potential risk factors for CHD. Our study was the first to find out the evidence that BTNL2-rs35624343, -rs117896888, -rs41441651 and -rs41417449 were significantly associated with CHD susceptibility. In this study, we identified some genetic variants that protect the risk of coronary heart disease in Southern Chinese Han population, and these SNPs are reported for the first time.
In previous studies on the association between BTNL2 and coronary artery, Hsueh et al found that BTNL2 gene polymorphism may be a genetic marker for the susceptibility of Taiwanese children to Kawasaki disease and the risk of coronary complications.32 Kawasaki disease is a childhood vasculitis that particularly affects the coronary arteries, and the risk of ischemic heart disease increases in the future.32 Previous studies have suggested that BTNL2 is potentially associated with the occurrence and development of coronary heart disease and this speculation has also been confirmed in the results of this study. Subsequently, Lu et al conducted a genome-wide association study on patients with coronary heart disease and found that the C6orf10-BTNL2 gene was a susceptibility gene for CHD in the Southern Chinese Han population, and the mutation of the BTNL2-rs9268402 locus can lead to the occurrence of coronary heart disease.33 In previous studies, whether it is Kawasaki disease or coronary heart disease, BTNL2 genetic polymorphism is the susceptibility genetic locus of the disease (OR> 1). However, the candidate BTNL2 genetic locus was associated with a reduced risk of CHD in this study. We speculate that this may be due to the differences of genetic background and geographical of the study subjects.
The pathological basis of coronary heart disease is atherosclerosis, which is a chronic inflammatory disease. CHD is characterized by the formation of arterial plaques composed mainly of lipids, calcium, and inflammatory cells.34 Atherosclerotic plaques are rich in immune cells, which can dominate and influence the inflammatory response.35 T cells are crucial to the development of atherosclerotic plaques in the development of CHD.36 Helper T cell 1 (Th1)/Th17-mediated pro-inflammatory response can aggravate atherosclerosis, while regulatory T cells (T reg) play a protective role against atherosclerosis by limiting inflammation and balancing plaque formation.37 BTNL2 can inhibit the activation of T cells and reduce the secretion of pro-inflammatory cytokines, leading to immune dysfunction.38 In chronic inflammation, CD4 T cells can secrete large amounts of Th1 cytokines, TNF-α and IFN-γ,39 which are effective atherogenic cytokines.40 Sinisalo et al found that the level of BTNL2 mRNA were higher in coronary artery samples from subjects with acute coronary artery disease. At the same time, the study also found that inhibition of BTNL2 significantly enhanced the proliferation of CD4(+)FOXP3(+) T reg cells.41 Considering the results of previous studies and our study, we analyzed the reasons why these missense variants in BTNL2 in this study are associated with the reduction of CHD risk among participants in this study. We speculate that these missense variants (rs35624343 A/G, rs117896888 G/C, rs41441651 T/C, rs41417449 C/T, rs28362680 A/G, rs2076523 C/T) in BTNL2 may play a protective role in atherosclerosis by regulating BTNL2 mRNA expression and participating in a certain inflammatory pathway, thereby limiting inflammatory response and balancing plaque formation. However, the specific molecular mechanism needs to be further studied.
Our study provides data supplement for the genetic loci associated with CHD susceptibility in Han population from southern China. Although the mechanism of these missense variants in coronary heart disease is still unclear, the study provides new ideas for the risk assessment and clinical individualized prevention and treatment of CHD in Han population from southern China. In addition, we found these missense variations in BTNL2 could cause changes in amino acid sequences through dbSNP database. Changes in amino acid sequence can lead to changes in protein structure, which is directly related to its function.42 Combined with the results of our study, we speculated that these missense variants caused changes in amino acid sequence, which resulted in structural changes of BTNL2 protein, thus affecting its biological functions. Accordingly, we proposed a hypothesis that these missense variants are protective factors for CHD in Chinese Han population, which may be achieved by affecting the amino acid sequence of BTNL2 and then changing its protein structure and function. But the above is just speculation, and further mechanistic studies are needed to confirm it. In any case, this study has laid a solid theoretical foundation for the mechanism of BTNL2 in the occurrence and development of CHD.
However, we must face the fact that there are certain limitations in this study. In order to ensure the reliability and repeatability of the results, a large sample size verification study is necessary. Secondly, studies on the mechanism of BTNL2 genetic polymorphism in CHD development need to be further determined. Nevertheless, to the best of our knowledge, this study is the first to investigate the association between BTNL2 genetic polymorphism and CHD susceptibility in China and to find positive results. This study will lay a reliable theoretical foundation for the study of the mechanism of BTNL2 in the occurrence and development of CHD, and provide new ideas for the risk assessment and clinical individualized prevention and treatment of CHD in Chinese Han population.
Conclusion
In summary, this study showed that missense variants (rs35624343, rs117896888, rs41441651, rs41417449, rs28362680, rs2076523) in BTNL2 were associated with coronary heart disease susceptibility. In particular, there is sufficient evidence that BTNL2-rs28362680 was associated with a reduced risk of coronary heart disease in the southern Chinese Han population. This study has laid a reliable theoretical foundation for the study of the mechanism of BTNL2 in the occurrence and development of coronary heart disease.
Data Sharing Statement
The datasets used and analyzed in the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
This study was conducted under the standard approved by the ethics committee of Northwest University and People’s Hospital of Wanning. And conformed to the ethical principles for medical research involving humans of the World Medical Association Declaration of Helsinki. All participants signed informed consent forms before participating in this study.
Consent to Publication
All authors agreed to publish the manuscript.
Acknowledgments
We thank all authors for their contributions and support.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study did not receive any funding.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. doi:10.1371/journal.pmed.0030442
2. Chen S, Kawasaki Y, Hu H, et al. Smoking cessation, weight gain, and the trajectory of estimated risk of coronary heart disease: 8-year follow-up from a prospective cohort study. Nicotine Tob Res. 2021;23(1):85–91. doi:10.1093/ntr/ntz165
3. Liu YT, Tantoh DM, Wang L, Nfor ON, Hsu SY, Ho CC. Interaction between coffee drinking and TRIB1 rs17321515 single nucleotide polymorphism on coronary heart disease in a Taiwanese population. Nutrients. 2020;12(5):1301.
4. DiNicolantonio JJ, Lucan SC, O’Keefe JH. The evidence for saturated fat and for sugar related to coronary heart disease. Prog Cardiovasc Dis. 2016;58(5):464–472. doi:10.1016/j.pcad.2015.11.006
5. Bai MF, Wang X. Risk factors associated with coronary heart disease in women: a systematic review. Herz. 2020;45(Suppl 1):52–57. doi:10.1007/s00059-019-4835-2
6. Yang J, Zhou Y, Zhang T. Fasting blood glucose and HbA(1c) correlate with severity of coronary artery disease in elective PCI patients With HbA(1c) 5.7% to 6.4. Angiology. 2020;71(2):167–174. doi:10.1177/0003319719887655
7. Wang Y, Xu D. Effects of aerobic exercise on lipids and lipoproteins. Lipids Health Dis. 2017;16(1):132. doi:10.1186/s12944-017-0515-5
8. Challa HJ, Ameer MA, Uppaluri KR. DASH Diet To Stop Hypertension. In: StatPearls. Treasure Island (FL): StatPearls Publishing; 2021. Copyright © 2021, StatPearls Publishing LLC..
9. Dugani SB, Moorthy MV, Li C, et al. Association of lipid, inflammatory, and metabolic biomarkers with age at onset for incident coronary heart disease in women. JAMA cardiol. 2021;6(4):437–447. doi:10.1001/jamacardio.2020.7073
10. Aragam KG, Natarajan P. Polygenic scores to assess atherosclerotic cardiovascular disease risk: clinical perspectives and basic implications. Circ Res. 2020;126(9):1159–1177. doi:10.1161/CIRCRESAHA.120.315928
11. Li KG, Yin RX, Huang F, Chen W-X, Wu J-Z, Cao X-L. XKR6 rs7014968 SNP increases serum total cholesterol levels and the risk of coronary heart disease and ischemic stroke. Clin Appl Thromb Hemost. 2020;26:1076029620902844. doi:10.1177/1076029620902844
12. Swerdlow DI, Holmes MV, Kuchenbaecker KB, et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a Mendelian randomisation analysis. Lancet. 2012;379(9822):1214–1224.
13. Khounphinith E, Yin RX, Cao XL, Huang F, Wu JZ, Li H. TIMD4 rs6882076 SNP is associated with decreased levels of triglycerides and the risk of coronary heart disease and ischemic stroke. Int J Med Sci. 2019;16(6):864–871. doi:10.7150/ijms.31729
14. Nagano T, Mitchell JA, Sanz LA, et al. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–1720. doi:10.1126/science.1163802
15. Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi:10.1038/nature10398
16. Cheng L, Zhao R, Jin Z, Ren K, Deng C, Yu S. Association of genetic polymorphisms on BTNL2 with susceptibility to and prognosis of dilated cardiomyopathy in a Chinese population. Int J Clin Exp Pathol. 2015;8(9):10488–10499.
17. Brown CJ, Ballabio A, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi:10.1038/349038a0
18. Wang L, Ai D, Zhang N. Exercise benefits coronary heart disease. Adv Exp Med Biol. 2017;1000:3–7.
19. Shao C, Wang J, Tian J, Tang YD. Coronary artery disease: from mechanism to clinical practice. Adv Exp Med Biol. 2020;1177:1–36.
20. Hajek C, Guo X, Yao J, et al. Coronary heart disease genetic risk score predicts cardiovascular disease risk in men, not women. Circ Genom Precis Med. 2018;11(10):e002324. doi:10.1161/CIRCGEN.118.002324
21. Khera AV, Chaffin M, Aragam KG. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi:10.1038/s41588-018-0183-z
22. Inouye M, Abraham G, Nelson CP, et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J Am Coll Cardiol. 2018;72(16):1883–1893. doi:10.1016/j.jacc.2018.07.079
23. Barrett-Connor E. Gender differences and disparities in all-cause and coronary heart disease mortality: epidemiological aspects. Best Pract Res Clin Endocrinol Metab. 2013;27(4):481–500. doi:10.1016/j.beem.2013.05.013
24. Barrett-Connor E. Sex differences in coronary heart disease. Why are women so superior? The 1995 Ancel Keys lecture. Circulation. 1997;95(1):252–264. doi:10.1161/01.CIR.95.1.252
25. Kantaria M, Buleishvili M, Kipiani NV, Ormotsadze G, Sanikidze T. Risk-factors of coronary ARTERY Disease. Georgian Med News. 2020;2020(299):78–82.
26. Damluji AA, Chung SE, Xue QL, et al. Physical frailty phenotype and the development of geriatric syndromes in older adults with coronary heart disease. Am J Med. 2021;134(5):662–71.e1. doi:10.1016/j.amjmed.2020.09.057
27. Naito R, Miyauchi K. Coronary artery disease and type 2 diabetes mellitus. Int Heart J. 2017;58(4):475–480. doi:10.1536/ihj.17-191
28. Escobar E. Hypertension and coronary heart disease. J Hum Hypertens. 2002;16(Suppl 1):S61–3. doi:10.1038/sj.jhh.1001345
29. Hong SN, Park C, Park SJ, et al. Deep resequencing of 131 Crohn’s disease associated genes in pooled DNA confirmed three reported variants and identified eight novel variants. Gut. 2016;65(5):788–796. doi:10.1136/gutjnl-2014-308617
30. Strawbridge RJ, van Zuydam NR. Shared genetic contribution of type 2 diabetes and cardiovascular disease: implications for prognosis and treatment. Curr Diab Rep. 2018;18(8):59. doi:10.1007/s11892-018-1021-5
31. Orozco G, Eerligh P, Sánchez E, et al. Analysis of a functional BTNL2 polymorphism in type 1 diabetes, rheumatoid arthritis, and systemic lupus erythematosus. Hum Immunol. 2005;66(12):1235–1241. doi:10.1016/j.humimm.2006.02.003
32. Hsueh KC, Lin YJ, Chang JS, Wan L, Tsai FJ. BTNL2 gene polymorphisms may be associated with susceptibility to Kawasaki disease and formation of coronary artery lesions in Taiwanese children. Eur J Pediatr. 2010;169(6):713–719. doi:10.1007/s00431-009-1099-5
33. Lu X, Wang L, Chen S, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44(8):890–894. doi:10.1038/ng.2337
34. Li H, Sun K, Zhao R, et al. Inflammatory biomarkers of coronary heart disease. Front Biosci. 2018;10(1):185–196. doi:10.2741/s508
35. Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685–1695. doi:10.1056/NEJMra043430
36. Ketelhuth DF, Hansson GK. Cellular immunity, low-density lipoprotein and atherosclerosis: break of tolerance in the artery wall. Thromb Haemost. 2011;106(5):779–786. doi:10.1160/TH11-05-0321
37. Huang L, Zheng Y, Yuan X, et al. Decreased frequencies and impaired functions of the CD31(+) subpopulation in T(reg) cells associated with decreased FoxP3 expression and enhanced T(reg) cell defects in patients with coronary heart disease. Clin Exp Immunol. 2017;187(3):441–454. doi:10.1111/cei.12897
38. Afrache H, Gouret P, Ainouche S, Pontarotti P, Olive D. The butyrophilin (BTN) gene family: from milk fat to the regulation of the immune response. Immunogenetics. 2012;64(11):781–794. doi:10.1007/s00251-012-0619-z
39. Vallejo AN, Weyand CM, Goronzy JJ. T-cell senescence: a culprit of immune abnormalities in chronic inflammation and persistent infection. Trends Mol Med. 2004;10(3):119–124. doi:10.1016/j.molmed.2004.01.002
40. Kleemann R, Zadelaar S, Kooistra T. Cytokines and atherosclerosis: a comprehensive review of studies in mice. Cardiovasc Res. 2008;79(3):360–376. doi:10.1093/cvr/cvn120
41. Sinisalo J, Vlachopoulou E, Marchesani M, et al. Novel 6p21.3 risk haplotype predisposes to acute coronary syndrome. Circ Cardiovasc Genet. 2016;9(1):55–63. doi:10.1161/CIRCGENETICS.115.001226
42. Tian K, Zhao X, Wan X, Yau SS. Amino acid torsion angles enable prediction of protein fold classification. Sci Rep. 2020;10(1):21773. doi:10.1038/s41598-020-78465-1
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.