Back to Journals » OncoTargets and Therapy » Volume 13
MiRNA505/NET1 Axis Acts as a CD8+ T-TIL Regulator in Non-Small Cell Lung Cancer
Authors Zhu P , Liu Z, Huang H, Zhong C, Zhou Y
Received 3 June 2020
Accepted for publication 27 August 2020
Published 1 October 2020 Volume 2020:13 Pages 9785—9795
DOI https://doi.org/10.2147/OTT.S265859
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Pengyuan Zhu,1 Zhenchuan Liu,2 Haitao Huang,3 Chongjun Zhong,3 Yongxin Zhou2
1Department of Thoracic and Cardiovascular Surgery, The Second Affiliated Hospital of Nantong University, School of Medicine, Nantong University, Nantong, Jiangsu 226001, People’s Republic of China; 2Department of Thoracic and Cardiovascular Surgery, Tongji Hospital, School of Medicine, Tongji University, Shanghai 200065, People’s Republic of China; 3Department of Thoracic and Cardiovascular Surgery, The Second Affiliated Hospital of Nantong University, Nantong, Jiangsu 226001, People’s Republic of China
Correspondence: Chongjun Zhong; Yongxin Zhou Tel +86-13906277618
; Tel +86-13681666828
Email [email protected]; [email protected]
Introduction: Lung adenocarcinoma (LUAD), which is the most important and common subtype of non-small cell lung cancer (NSCLC), is highly heterogeneous with a poor prognosis and poses great challenges to health worldwide. MicroRNAs (miRNAs) are regulators of gene expression with recognized roles in physiology and diseases, such as cancers, but little is known about their functional relevance to CD8+ T cell infiltration regulation in the tumor microenvironment (TME) of NSCLC patients, especially LUAD patients.
Methods: Bioinformatic analysis was used to analyze TCGA data. RT-PCT, Western blot, luciferase assay and immunohistochemistry were used to detect the expression levels and bindings of genes and miRNA. ELISA and cytotoxic assay were used to evaluate CD8+ T cell function.
Results: In this study, bioinformatic analysis unveiled the miR-505-3p/NET1 pair as a CD8+ T-tumor-infiltrating lymphocyte (TIL) regulator. Then, we confirmed the bioinformatic results with LUAD patient samples, and NET1 was shown to be a direct target of miR-505-3p in a luciferase assay. Functional experiments demonstrated that miR-505-3p enhanced CD8+ T-TIL function, while NET1 impaired CD8+ T-TIL function and partly reversed the effects of miR-505-3p. The observed effects might be exerted via the regulation of immunosuppressive receptors in T cells.
Discussion: Our study may provide novel insights into LUAD progression related to the TME mechanism and new possibilities for improving adoptive immunotherapy.
Keywords: lung adenocarcinoma, miR-505-3p, NET1, CD8+ T-TILs, TME
Introduction
Lung cancer is a malignant disease with high morbidity and mortality, and 80% of lung cancer cases are non-small cell lung cancer (NSCLC). It is estimated that the number of deaths from lung cancer will rise to 10,000,000 by 2030.1 NSCLC is the most common type of lung cancer and is further classified into adenocarcinoma (LUAD), squamous cell carcinoma, and large-cell carcinoma, and LUAD is the most important and common subtype, with an increasing incidence both globally and in China.2 Lung cancer is usually discovered in late stages; thus, despite improvements in therapeutic treatment and novel tumor biomarker detection, the prognosis of patients with NSCLC remains poor, with only a 1.0% 5-year survival rate for advanced patients.3,4 Currently, standard treatment of lung cancer is still surgical resection, with or without adjuvant chemotherapy. But with immunotherapy emerging, overall survival of lung cancer patients significantly increased accepting immunotherapy plus surgery.5,6 In the last few years, definitive proof for direct microRNA (miRNA) contributions to the initiation and progression of LUAD has been established. The oncogenetic basis of lung cancer is related primarily to changes in the profile of miRNAs. miRNAs are massively dysregulated during lung carcinogenesis and may be proposed as an early diagnostic tool in cancer to identify high-risk subjects and cancer in early stages by liquid biopsy.7,8
miRNAs are crucial, endogenous non-protein-coding RNAs and posttranscriptional regulators of multiple target genes in several cell types.9 By affecting finely tuned protein expression levels, miRNAs can provide quantitative control of gene output. The cooperative interactions of miRNAs with target mRNAs can influence various critical biological programs, such as cell senescence, metabolism, control of tumorigenesis and immune responses.10 Therefore, miRNA functions in tumor development are not limited to regulating cancer cell behavior. Despite the focus on genetic aberrations in tumor cells, it has been increasingly noticed that cancer cells exhibit a range of dependence on interactions with the nonmalignant cells and stromal matrix of the tumor microenvironment (TME).11
The TME contains a diversity of cells, including fibroblasts, immune cells, and endothelial cells. Recently, the cell-autonomous significance of macrophages, T cells, and endothelial cells and their putative implications on cancer have begun to be revealed.12–14 In the TME, CD8+ T cells are the major immune cells that eliminate cancer cells in a cell-cell contact-dependent manner. Tumor-infiltrating lymphocytes (TILs) include CD8+ T cells specific for tumor cells, which are activated by antigen recognition, proliferate and differentiate into cytotoxic T lymphocytes (CTLs).15,16 The molecular and cellular “crosstalk” exploited by NSCLC, especially LUAD, in the TME and thus contributing to the clinicopathological profile and the efficacy of immunotherapy, however, has not been comprehensively studied.
Here, we report a facilitative role for miR-505-3p in anti-NSCLC immunity. We performed bioinformatic analysis using The Cancer Genome Atlas (TCGA) data to uncover prognosis-related miRNAs and CD8+ T-TIL infiltration-related mRNAs. Combining the results of miRNA target prediction programs, we selected the miRNA-mRNA pair miR-505-3p and neuroepithelial cell transforming 1 (NET1) as putative functional regulators of CD8+ T-TIL infiltration and cytotoxicity. Then, we confirmed the bioinformatic results in LUAD patient samples, and NET1 was shown to be a direct target of miR-505-3p in a luciferase assay. By coculture of CD8+ T-TILs and NSCLC cell lines, we demonstrated that NET1 in NSCLC cells influenced CD8+ T-TIL function and that its role might involve immunosuppressive receptor regulation. In this study, our results may provide novel insights into the NSCLC TME mechanism and adoptive immunotherapy.
Materials and Methods
Data Acquisition and Preprocessing
The RNA-Seq data for 510 samples and miRNA-Seq data for 508 samples were collected from the TCGA (http://cancergenome.nih.gov/) project, specifically the TCGA-LUAD cohort, via GDC Data Portal (https://portal.gdc.cancer.gov/) on August 13, 2019. For data preprocessing, we normalized the raw counts of RNA/miRNA expression data by TMM applied in edgeR and transformed them by voom implemented in limma. RNAs and miRNAs with a count per million reads mapped (cpm) > 1 in more than half of all samples were kept for subsequent analysis, while low-expression mRNAs and miRNAs were filtered out. Samples with incomplete clinical information were also screened out.
Human Samples
The study was approved by the Institutional Review Board of the Center for Medicine, Nantong University. Patients with LUAD undergoing surgical resection were enrolled in this study after providing informed consent, and samples were collected at The Second Affiliated Hospital of Nantong University. All studies were conducted in accordance with the Declaration of Helsinki. For PCR and Western blotting, human LUAD tissue specimens were quickly rinsed in cold PBS, rapidly frozen and stored in liquid nitrogen until protein or RNA extraction. For the CD8+ T-TIL functional assay, tumor tissue samples were dissected into approximately 2-mm3 fragments, plated in 24-well plate wells individually and digested using an enzyme mix including DNAse, collagenase, and hyaluronidase. Cells were then cultured with 6,000 IU/mL recombinant human IL-2 (rhIL-2) for 3 weeks until lymphocyte growth was evident and stimulated with 1 μg/mL PHA, 3000 IU/mL rhIL2 (Proleukin, Roche) and feeders for rapid expansion. CD8+ T-TILs were positively selected using CD8 MicroBeads (Miltenyi, Germany).
Cell Culture, Coculture and Transfection
Human normal bronchial epithelial cell line (BEAS-2B) and NSCLC cell lines (A549, H1975, H460, H1299, and PC9) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). BEAS-2B cells were cultured in complete medium of Lonza BEGM Kit (USA). NSCLC cell lines were cultured in RPMI-1640 medium (HyClone, USA) containing 10% fetal bovine serum (HyClone, USA) and 1% penicillin and streptomycin (HyClone, USA) in an atmosphere of 5% CO2 at 37°C. CD8+ T-TILs were separated from dissected fragments of tumor tissues and rapidly expanded with PHA 1 μg/mL, 3000 IU/mL rhIL2 (Proleukin, Roche) and feeders. CD8+ T-TILs were cultured in the complete medium described above supplemented with 1% L-glutamine, 25 mM HEPES, 0.1% β-mercaptoethanol. To achieve substantial expansion, cells were cultured in the presence of anti-CD3 and anti-CD28 antibodies bound to large particles (Miltenyi, Germany) (bead:T cell ratio = 1:1) overnight. Indirect coculture of tumor cells and CD8+ T cells was performed using Transwell chambers with 0.4-μm-pore membranes. MiR-505-3p mimics and inhibitors and the corresponding negative controls were designed and synthesized by Genepharma (China). A specific siRNA and recombinant plasmid for NET1 were generated and purchased from Ke Lei Biological Technology (China). Cell lines were transfected with Lipofectamine 2000 (Invitrogen, USA) following the manufacturer’s protocols.
Protein Extraction and Western Blotting
To extract proteins, frozen tissue samples were pulverized with a mortar and pestle under liquid nitrogen conditions, and the cultured cells were rinsed 3 times with precooled PBS. Then, the cells were lysed with RIPA and PMSF at 4°C for 30 minutes and centrifuged at 14,000 x g at 4°C for 30 minutes. The precipitate was discarded, and the protein extracted in the supernatant was quantified with the BCA Protein Assay Kit (Thermo Fisher Scientific, USA). Proteins were separated by 8% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis, followed by transfer to polyvinylidene difluoride membranes (Millipore, USA). The membranes were blocked with 5% skim milk dissolved in Tris-buffered saline Tween 20 (TBST) for 1 h at room temperature. Subsequently, the membranes were rinsed with TBST 3 times and incubated with primary antibodies at 4°C overnight. The membranes were incubated with HRP-conjugated secondary antibodies, and immunoreactive bands were detected and analyzed. The primary antibodies used were anti-NET1 (Biorbyt, orb49116), anti-β-actin (Abcam, ab179467), anti-LAG3 (Abcam, ab40465), anti-GAPDH (Abcam, ab181602), anti-PD1 (Abcam, ab243644), anti-TIM3 (Abcam, ab47997), and anti-CD8 (Abcam, ab4055).
Total RNA Extraction and Real-Time PCR (RT-PCR)
Total RNA was isolated from primary and transfected cells with TRIzol reagent (Invitrogen, USA) following the manufacturer’s protocols. Total RNA was reverse transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan), while miRNAs were reverse transcribed individually with specific reverse transcription primers and the One Step miRNA cDNA Synthesis Kit (HaiGene Bio Inc., China). Quantitative real‐time polymerase chain reaction (qRT‐PCR) was performed with the SYBR Premix Ex Taq Reverse Transcription PCR Kit (Takara, Dalian, China) and carried out on an ABI QuantStudio 6 (USA). GAPDH was used as a housekeeping gene control for mRNA analysis. U6 RNA was used as an endogenous control for miRNA analysis. Each sample was tested in triplicate. Data were analyzed using the 2−ΔΔCt method. The primer used were as follows: NET1: 5’-GCGCTTGAATGCCTACAGAG-3’ (F) and 5’- ACATCAGGGTGCTCTTTTGG-3’ (R); PD1: 5’-CCAGGATGGTTCTTAGACTCCC-3’ (F) and 5’-TTTAGCACGAAGCTCTCCGAT-3’ (R); TIM3: 5’-AGACAGTGGGATCTACTGCTG-3’ (F) and 5’-CCTGGTGGTAAGCATCCTTGG-3’ (R); LAG3: 5’-GCGGGGACTTCTCGCTATG-3’ (F) and 5’-GGCTCTGAGAGATCCTGGGG-3’ (R); GAPDH: 5’‐TCAAGAAGGTGGTGAAGCAGG‐3’ (F) and 5’‐TCAAAGGTGGAGGAGTGGGT‐3’ (R); miR-505-3p: 5’‐TCTCCTGCGTCAACACTTGC‐3’ (F) and 5’‐CACTTCCTCAGCACTTGTTGGTAT‐3’ (R); U6: 5’‐CAGCACATATACTAAAATTGGAACG‐3’ (F) and 5’‐ACGAATTTGCGTGTCATCC‐3’ (R).
Luciferase Reporter Assay
Luciferase reporter plasmids containing h-NET1-3’UTR-wild type (wt) or h-NET1-3’UTR-mutant (mut) were purchased from and constructed by Hanbio (China). Wild-type or mutant vectors were cotransfected with miR-505-3p mimics or the appropriate negative control into NSCLC cells using Lipofectamine 2000 (Invitrogen, USA). The sequences are shown in Figure 1D. The cells were lysed 48 hours later, and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, USA) according to the manufacturer’s instructions.
Immunohistochemistry (IHC) and Scoring
IHC was performed on formalin-fixed, paraffin-embedded (FFPE) tissue sections. Each FFPE sample was sliced (3 µm) onto a charged glass slide and heated at 60°C for 30 minutes. The slides were deparaffinized in three consecutive xylene baths for 3 minutes each, rehydrated with a graded series of ethanol with 5-minute incubations, and then washed in distilled water for 5 minutes. Intrinsic peroxidase activity was blocked with 3% H2O2 (Zhongshan Jinqiao Biotechnology Co., China) at 37°C for 15 minutes. The slides were then rinsed three times with PBS. To block nonspecific antibody binding, the sections were incubated with goat serum in a humidified chamber at 37°C for 60 minutes. The slides were washed with PBS and then incubated with either an anti-NET1 or anti-CD8 primary antibody overnight at 4°C. After the incubation, the sections were washed with PBS and then incubated with a goat anti-rabbit monoclonal secondary antibody (Zhongshan Jinqiao Biotechnology Co., China) at 37°C for 30 minutes. Finally, the specimens were stained with DAB and counterstained with hematoxylin (Beyotime Institute of Biotechnology, China).
For each patient’s specimen, slides were scanned at low magnification (10x magnification) first and 10 non-overlapping selected fields at high magnification (400x magnification) were assessed. At least two investigators trained in lung cancer blindly assessed the immunohistochemical staining and a final consensus was achieved. For scoring NET1 expression, staining intensity was categorized into 0 (no expression), 1 (weak), 2 (moderate) and 3 (high); and the percentage of NET1-positive cells was scored as 1 (positive cells ≤25%), 2 (25%< positive cells ≤50%), 3 (50% < positive cells ≤75%) and 4 (positive tumor cells >75%). The evaluation of histochemical score (H-score) was achieved by multiplying the staining intensity and the percentage of positive cells, ranging from 0 to 300. The mean of H-score was considered as the cut-off point and the samples were classified as low (H-score <180) or high (H-score ≥180) NET1 expression.17–19 For scoring CD8+ T-TILs, any cell with CD8 staining was counted as CD8+ T-TIL.17 CD8+ T-TILs were counted manually in each high-power field and scored as follows: 0 (none), 1 (1–2 CD8+ T-TILs), 2 (3–19 CD8+ T-TILs), 3 (≥20 CD8+ T-TILs).20 NET1 and CD8 expression was analyzed with nonparametric tests.
ELISA
CD8+ T cells were cocultured with A549 or H1975 cells (effector:target (E:T) ratio = 30:1), and the cell culture supernatant was collected and centrifuged to remove the cell precipitate. According to the detection ranges of IFN-γ, TNF-α, and IL-2 ELISA kits (Elabscience, China), the supernatant was diluted at the proper ratio. A standard working solution was used to generate a standard curve to determine the concentrations in the samples. IFN-γ, TNF-α, and IL-2 concentrations were measured by ELISA according to the manufacturer’s protocol. Optical density (OD) values were immediately measured with a microplate reader at a wavelength of 450 nm.
Cytotoxic Assay
The CytoTox 96 Nonradioactive Cytotoxicity Assay (Promega, USA) was used for the cytotoxicity assay. The NSCLC cell line A549 or H1975 was cocultured with CD8+ T cells at an E:T ratio of 3:1, 10:1 or 30:1. LDH analysis was performed according to the manufacturer’s protocol. Controls for spontaneous LDH release by effector or target cells, as well as maximum target cell release, were prepared. The percentage of cell lysis was calculated as follows: % Cytotoxicity=[(experimental–effector spontaneous−target spontaneous)/(target maximum−target spontaneous)]×100%.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software. All data are presented as the mean ± SD of at least three independent experiments. Differences between nonparametric data were analyzed by the Wilcoxon rank-sum test. A p value < 0.05 was considered significant. Data were available when required.
Results
NET1 Was Correlated with CD8+ T-TILs and a Potential Target of miR-505-3p
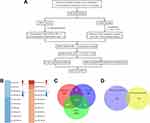
A flowchart of the bioinformatic analysis performed in this study is shown in Figure 2A. To explore crucial miRNAs involved in LUAD, survival analysis was performed using Cox regression analysis. Prognosis-predicting miRNAs with a p value < 0.05 were detected and sorted by hazard ratios (HRs). The top 10 poor prognosis- and favorable prognosis-related miRNAs are listed in Figure 2B. Among the top 5 favorable miRNAs, miR-505-3p had the highest relative abundance. Using the miRNA target prediction software PITA, microT and miRanda, we identified 114 predicted miRNA targets of miR-505-3p (Figure 2C). CIBERSORT, an in silico flow cytometry tool, is an emerging method for characterizing fractional representations of immune cell types in tumor tissue samples from gene expression profiles.21 We used the LM22 signature gene matrix, which includes 7 T cell types, 2 B cell types, plasma cells, NK cells, and myeloid lineage subsets. The infiltrating fractions of immune cell types were estimated in previously collected and preprocessed mRNA-Seq data using CIBERSORT. Immune cell infiltration-related mRNAs were identified with a CIBERSORT p value < 0.05. Genes with a negative correlation with the CD8+ T-TIL fraction had a great chance to be expressed in LUAD tumor cells, which provided the possibility that these genes could be targeted in the clinic. For coexpression analysis, we simultaneously analyzed the sequencing data profiles of miRNAs associated with a favorable prognosis and mRNAs negatively correlated with the CD8+ T-TIL fraction. For miR-505-3p, 149 potential miRNA-mRNA functional pairs were selected with r < 0 and p < 0.01. Four final predicted immune functional targets were identified in the intersection between the results of the coexpression analysis and those of bioinformatic programs (EPN2, NET1, SF1, and MAML3) (Figure 2D). Among the targets, NET1 had the most significant r and p values, which implied the presence of the miR-505-3p and NET1 functional pair in LUAD.
NET1 Correlated with miR-505-3p and CD8+ T-TILs in LUAD Specimens
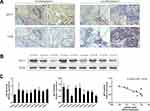
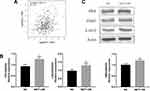
To confirm the bioinformatic prediction, 60 paraffin-embedded LUAD samples collected at diagnosis were immunohistochemically stained with anti-NET1 and anti-CD8 antibodies. In these patient samples, high expression of NET1 was observed with relatively few CD8-positive cells, while NET1 expression was low when more CD8+ T-TILs were present. Accordingly, the results demonstrated a negative correlation between NET1 expression and the CD8+ T-TIL fraction (Wilcoxon test, p <0.05), and representative figures are shown (Figure 1A). We then examined miR-505-3p and NET1 expression in 9 LUAD samples by Western blotting and RT-PCR, and tumor tissue samples exhibited higher NET1 but lower miR-505-3p expression levels or vice versa (Figure 1B-C). Furthermore, NET1 mRNA levels in LUAD samples obtained from 9 patients were significantly negatively correlated with miR-505-3p expression levels using a linear regression model (Figure 1C). The inverse correlation between miR-505-3p and NET1 expression in LUAD tumor cells implicated miR-505-3p as a regulator of NET1.
NET1 Was a Direct Target of miR-505-3p
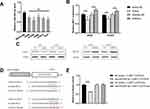
To evaluate the level of miR-505-3p in NSCLC cell lines, total RNA was harvested from a normal human bronchial epithelial cell line (BEAS-2B) and 5 human NSCLC cell lines (A549, H1975, H460, H1299, and PC9), and miR-505-3p expression levels were determined by semiquantitative RT-PCR (Figure 3A). Compared with BEAS-2B, NSCLC cell lines presented downregulated miR-505-3p, but no significant difference was observed among the 5 NSCLC cell lines. We chose the commonly used cell lines A549 and H1975 for subsequent experiments. We further examined NET1 expression levels in miR-505-3p mimic- and inhibitor-transfected cells and in the corresponding negative controls. NET1 levels were decreased with miR-505-3p mimic transfection but increased with miR-505-3p inhibitor transfection (Figure 3B-C). A luciferase reporter assay was then performed to confirm the predicted binding sites of miR-505-3p in the 3ʹ-untranslated region (3ʹ-UTR) of NET1. We cloned wild-type and mutant 3ʹ-UTRs into a luciferase reporter vector. The predicted target sites and relevant mutated sequences are shown in Figure 3D. Luciferase activity was considerably decreased with overexpression of miR-505-3p compared with negative control transfection in cells with the wild-type 3ʹ-UTR, while the luciferase activity of the NET1-mutant 3ʹ-UTR vector remained unchanged (Figure 3E). The results demonstrate that NET1 is a direct target of miR-505-3p.
miR-505-3p Facilitated Immune Function by Regulating NET1 Expression
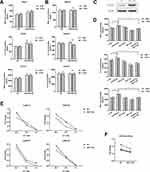
Primary CD8+ T cells were cocultured with NSCLC cells followed by anti-CD3/CD28 Dynabeads activation, and IFN-γ, TNF-α, and IL-2 production was used to measure effects, either direct or indirect, on T cell function. Interestingly, miR-505-3p significantly improved cytokine production, but miR-505-3p inhibition suppressed T cell function in cocultures allowing direct contact between CD8+ T cells and NSCLC cells (Figure 4A). However, although miR-505-3p expression was up- or downregulated in NSCLC cells, no significant differences in CD8+ T cell responses were observed in groups where tumor cells were segregated by cell-impermeable membranes (Figure 4B). The results implied that miR-505-3p alteration in NSCLC cells influenced the immune response via cell-surface molecules. A NET1 overexpression plasmid (NET1-OE) was transfected into A549 cells, and NET1 overexpression was confirmed by Western blotting (Figure 4C). We further assessed whether the CD8+ T cell regulation process was mediated by targeting NET1. For CD8+ T cell cytokine production analysis, CD8+ T cells were directly cocultured with A549 or H1975 cells transfected with either the NET1 overexpression plasmid or miR-505-3p or the corresponding scramble plasmid. The promotive effects on CD8+ T cells exerted by miR-505-3p were partly reversed by NET1 overexpression (Figure 4D). We collected CD8+ T cells from 4 LUAD patients, and cell killing assays were performed using NET1-OE and negative control NSCLC cells. When the highest lysis values were considered (E:T ratio = 30:1), a significant reduction in cytotoxicity was observed in the NET1-OE cells (Figure 4E-F).
Immunosuppressive Receptor Expression Was Upregulated in CD8+ T Cells in the Context of Reduced Expression of NET1 in NSCLC Cells
Considering the different results observed in the direct and indirect coculture experiments, correlation analysis between NET1 and PDL1 was performed with LUAD data using GEPIA2 (http://gepia2.cancer-pku.cn). NET1 had a positive correlation with PDL1 (Figure 5A). We then isolated CD8+ T cells from 4 patients and cocultured them with NET1-OE- or corresponding negative control-transfected A549 cells (E:T ratio = 30:1). PD-1, LAG3, and TIM-3 expression levels were examined by RT-PCR (Figure 5B) and Western blotting (Figure 5C). The results indicated that NET1 might change tumor cell functions and impair CD8+ T cell infiltration and functions by upregulating immune checkpoint molecule expression.
Discussion
Accumulating evidence indicates that miRNAs contribute to cancer pathogenesis. As miRNAs may serve as tumor suppressors or oncogenes, dysregulation of miRNAs is associated with cancer initiation and progression.22,23 In the current study, miR-505-3p was selected as a favorable prognosis-related miRNA. Moreover, both bioinformatic analysis and a luciferase assay confirmed that NET1 is a direct target of miR-505-3p. In NSCLC cell lines, enforced miR-505-3p expression enhanced the function of cocultured CD8+ T cells, while overexpression of NET1 played a suppressive role in cocultured CD8+ T cells. In addition, after coculture with NET1-OE NSCLC cells, PD1, TIM3 and LAG3 expression was upregulated in CD8+ T cells. These data suggested that miR-505-3p might serve as a novel biomarker or therapeutic target in NSCLC.
Over the past few decades, hundreds of functional miRNAs have been identified and revealed to be involved in biological processes and diseases.24 miRNA dysregulation has been demonstrated to affect cancer proliferation, angiogenesis, metastasis, and drug resistance development through interactions among malignant cells, nonmalignant stromal cells, and noncellular components in the TME.25 In the TME, dysregulated miRNAs, such as miR-199a-3p, can suppress tumor growth, migration, invasion and angiogenesis;26 they can also influence the inflammatory TME and epithelial-mesenchymal transition.27 In addition, miRNAs interact with the TME via tumor-derived exosomes. miR-23a expression was found to be significantly upregulated in exosomes from lung cancer cells under hypoxic conditions and to increase angiogenesis and vascular permeability.28 Furthermore, miRNAs regulate immune cells in multiple ways. In LUAD, miR-31 expressed in myeloid dendritic cells results in the expression of tumor-supporting factors and increased invasiveness of NSCLC cells with an impact on clinical outcomes29 while miR-141 reduces the recruitment of regulatory T cells (Tregs) to malignant pleural effusion sites in NSCLC patients.30 Among the various types of immune cells, T-TILs are one of the most important types utilized for adoptive immunotherapy. CD8+ T cells are the major cells executing cytotoxic functions, encouraging us to explore ways of enhancing their antitumor function.
NET1 was previously identified as an inhibitor of ERK1/2 and PI3K/Akt1 in the PyMT mouse model and was shown to have upregulated expression in NSCLC as a biomarker and regulate cancer cell stemness in cancer-associated fibroblasts.31–33 In our study, we first identified NET1 as a suppressor of CD8+ T cells with its upstream regulator miR-505-3p, which was confirmed by a luciferase assay and functional experiments with CD8+ T cells. Accordingly, in our bioinformatic analysis, miR-505-3p was a potential prognostic marker for prolonged survival and facilitated CD8+ T cell functions. The inverse association between miR-505-3p and NET1 and the negative correlation between NET1 expression and CD8+ T cell infiltration were both verified in LUAD patient specimens. Remarkably, the expression of the immunosuppressive molecules PD1, TIM3 and LAG3 was upregulated in CD8+ T cells cocultured with NET1-OE NSCLC cells, implying the probable mechanisms underlying the immunoregulatory function of NET1.
In summary, combining bioinformatic and functional analyses, our study demonstrated that the miR-505-3p/NET1 axis influenced primary CD8+ T cell functions, including cytokine secretion and cytotoxicity. This study provides a new viewpoint for understanding the roles of miRNA and CD8+ T cell functions and a potential immunoregulatory mechanism in NSCLC. Additional pathogenesis and therapeutic studies concentrated on miRNAs are warranted.
Conclusion
We demonstrated that miR-505-3p was a favorable prognosis-related miRNA. Moreover, in bioinformatic analysis and a luciferase assay, NET1 acts as a direct target of miR-505-3p. In NSCLC cell lines, enforced miR-505-3p expression enhanced the function of cocultured CD8+ T cells, while overexpression of NET1 partially reversed this effect and played a suppressive role. In addition, after coculture with NET1-OE NSCLC cells, PD1, TIM3 and LAG3 expression was upregulated in CD8+ T cells. These data implied miR-505-3p as a novel biomarker and therapeutic target in NSCLC.
Abbreviations
LUAD, Lung adenocarcinoma; TME, tumor microenvironment; TIL, tumor-infiltrating lymphocyte; NSCLC, non-small cell lung cancer; miRNA, microRNA; CTLs, cytotoxic T lymphocytes; TCGA, The Cancer Genome Atlas; NET1, neuroepithelial cell transforming 1.
Acknowledgments
This study was supported by Natural Science Foundation of China (Grant No.: 81670463), China postdoctoral science foundation (Grant No.: 2018M632352), The Science and Technology Support Program of Nantong (Grant No.: JC2018099 to S. Ding), The Fund Project of Nantong University (Grant No.: 2019JY003 to S. Ding), Jiangsu Youth Medical Key Talents (Grant No.: QNRC2016405 to H. Huang), Jiangsu Commission of Health Support Program (Grant No.: H2017056 to C. Zhong).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work. All authors gave their consent for publication.
References
1. Aggarwal A, Lewison G, Idir S, et al. The State of Lung Cancer Research: A Global Analysis. J Thorac Oncol. 2016;11(7):1040–1050. doi:10.1016/j.jtho.2016.03.010
2. Han B, Sun Y, Yang D, et al. USP22 promotes development of lung adenocarcinoma through ubiquitination and immunosuppression. Aging. 2020;12.
3. Jin R, Wang X, Zang R, et al. Desmoglein-2 Modulates Tumor Progression and Osimertinib Drug Resistance through the EGFR/Src/PAK1 Pathway in Lung Adenocarcinoma. Cancer Lett. 2020;483:46–58. doi:10.1016/j.canlet.2020.04.001
4. Raghav L, Chang YH, Hsu YC, et al. Landscape of Mitochondria Genome and Clinical Outcomes in Stage 1 Lung Adenocarcinoma. Cancers. 2020;12(3):3. doi:10.3390/cancers12030755
5. Zhu J, Li R, Tiselius E, et al. Immunotherapy (excluding checkpoint inhibitors) for stage I to III non-small cell lung cancer treated with surgery or radiotherapy with curative intent. Cochrane Database Sys Rev. 2017;12(12):Cd011300.
6. Roudi R, Mohammadi SR, Roudbary M, Mohsenzadegan M. Lung cancer and β-glucans: review of potential therapeutic applications. Invest New Drugs. 2017;35(4):509–517. doi:10.1007/s10637-017-0449-9
7. Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 2016;118(1):3243–3250. doi:10.1096/fj.09-135251
8. Izzotti A, Carozzo S, Pulliero A, Zhabayeva D, Ravetti JL, Bersimbaev R. Extracellular MicroRNA in liquid biopsy: applicability in cancer diagnosis and prevention. Am J Cancer Res. 2016;6(7):1461–1493.
9. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
10. Meltzer PS. Cancer genomics: small RNAs with big impacts. Nature. 2005;435(7043):745–746. doi:10.1038/435745a
11. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
12. Piao L, Canguo Z, Wenjie L, Xiaoli C, Wenli S, Li L. Lipopolysaccharides-stimulated macrophage products enhance Withaferin A-induced apoptosis via activation of caspases and inhibition of NF-kappaB pathway in human cancer cells. Mol Immunol. 2017;81:92–101. doi:10.1016/j.molimm.2016.10.010
13. Chamorro-Jorganes A, Lee MY, Araldi E, et al. VEGF-Induced Expression of miR-17-92 Cluster in Endothelial Cells Is Mediated by ERK/ELK1 Activation and Regulates Angiogenesis. Circ Res. 2016;118(1):38–47. doi:10.1161/CIRCRESAHA.115.307408
14. Jiang S, Li C, Olive V, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118(20):5487–5497. doi:10.1182/blood-2011-05-355644
15. Jensen MC. IMMUNOLOGY. Synthetic immunobiology boosts the IQ of T cells. Science. 2015;350(6260):514–515. doi:10.1126/science.aad5289
16. Jiang X, Xu J, Liu M, et al. Adoptive CD8(+) T cell therapy against cancer: challenges and opportunities. Cancer Lett. 2019;462:23–32. doi:10.1016/j.canlet.2019.07.017
17. Foroozan M, Roudi R, Abolhasani M, Gheytanchi E, Mehrazma M. Clinical significance of endothelial cell marker CD34 and mast cell marker CD117 in prostate adenocarcinoma. Pathol Res Pract. 2017;213(6):612–618. doi:10.1016/j.prp.2017.04.027
18. Sadeghi A, Roudi R, Mirzaei A, Zare Mirzaei A, Madjd Z, Abolhasani M. CD44 epithelial isoform inversely associates with invasive characteristics of colorectal cancer. Biomark Med. 2019;13(6):419–426. doi:10.2217/bmm-2018-0337
19. Kalantari E, Abolhasani M, Roudi R, et al. Co-expression of TLR-9 and MMP-13 is associated with the degree of tumour differentiation in prostate cancer. Int J Exp Pathol. 2019;100(2):123–132. doi:10.1111/iep.12314
20. Goode EL, Block MS, Kalli KR, et al. Dose-Response Association of CD8+ Tumor-Infiltrating Lymphocytes and Survival Time in High-Grade Serous Ovarian Cancer. JAMA Oncology. 2017;3(12):e173290. doi:10.1001/jamaoncol.2017.3290
21. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
22. He GY, Hu JL, Zhou L, et al. The FOXD3/miR-214/MED19 axis suppresses tumour growth and metastasis in human colorectal cancer. Br J Cancer. 2016;115(11):1367–1378. doi:10.1038/bjc.2016.362
23. Zhang P, Tang WM, Zhang H, et al. MiR-646 inhibited cell proliferation and EMT-induced metastasis by targeting FOXK1 in gastric cancer. Br J Cancer. 2017;117(4):525–534. doi:10.1038/bjc.2017.181
24. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: background, Mechanisms, and Therapeutic Approaches. Physiol Rev. 2016;96(4):1297–1325. doi:10.1152/physrev.00041.2015
25. Li J, Guan J, Long X, Wang Y, Xiang X. mir-1-mediated paracrine effect of cancer-associated fibroblasts on lung cancer cell proliferation and chemoresistance. Oncol Rep. 2016;35(6):3523–3531. doi:10.3892/or.2016.4714
26. Ghosh A, Dasgupta D, Ghosh A, et al. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017;8(3):e2706. doi:10.1038/cddis.2017.123
27. Heinrich EL, Walser TC, Krysan K, et al. The inflammatory tumor microenvironment, epithelial mesenchymal transition and lung carcinogenesis. Cancer Microenviron. 2012;5(1):5–18. doi:10.1007/s12307-011-0089-0
28. Hsu YL, Hung JY, Chang WA, et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36(34):4929–4942. doi:10.1038/onc.2017.105
29. Pyfferoen L, Brabants E, Everaert C, et al. The transcriptome of lung tumor-infiltrating dendritic cells reveals a tumor-supporting phenotype and a microRNA signature with negative impact on clinical outcome. Oncoimmunology. 2017;6(1):e1253655. doi:10.1080/2162402X.2016.1253655
30. Lv M, Xu Y, Tang R, et al. miR141-CXCL1-CXCR2 signaling-induced Treg recruitment regulates metastases and survival of non-small cell lung cancer. Mol Cancer Ther. 2014;13(12):3152–3162. doi:10.1158/1535-7163.MCT-14-0448
31. Fang L, Zhu J, Ma Y, Hong C, Xiao S, Jin L. Neuroepithelial transforming gene 1 functions as a potential prognostic marker for patients with non-small cell lung cancer. Mol Med Rep. 2015;12(5):7439–7446. doi:10.3892/mmr.2015.4385
32. Sung PJ, Rama N, Imbach J, et al. Cancer-Associated Fibroblasts Produce Netrin-1 to Control Cancer Cell Plasticity. Cancer Res. 2019;79(14):3651–3661. doi:10.1158/0008-5472.CAN-18-2952
33. Zuo Y, Ulu A, Chang JT, Frost JA. Contributions of the RhoA guanine nucleotide exchange factor Net1 to polyoma middle T antigen-mediated mammary gland tumorigenesis and metastasis. Breast Cancer Res. 2018;20(1):41. doi:10.1186/s13058-018-0966-2
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.