Back to Journals » Breast Cancer: Targets and Therapy » Volume 12
Mirna21 Expression in the Breast Cancer Tumor Tissue is Independent of Neoadjuvant Chemotherapy
Authors Sales ACV , Gomes da Silva IIF , Leite MCB, Coutinho LL , Reis RBAC , Castoldi A , B. G. Martins D , Lima-Filho JL , Souto FO
Received 28 June 2020
Accepted for publication 12 September 2020
Published 8 October 2020 Volume 2020:12 Pages 141—151
DOI https://doi.org/10.2147/BCTT.S269519
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pranela Rameshwar
Alexandre Cesar Vieira Sales,1,2 Isaura Isabelle Fonseca Gomes da Silva,1 Matheus CB Leite,1 Leandro L Coutinho,1 Renata BAC Reis,1 Angela Castoldi,1 Danyelly BG Martins,1 José Luiz Lima-Filho,1 Fabricio Oliveira Souto1,2
1Keizo Asami Immunopathology Laboratory, LIKA/Federal University of Pernambuco – UFPE, Recife, PE, Brazil; 2Life Sciences Nucleus– NCV, Agreste Academic Center– CAA, Federal University of Pernambuco – UFPE, Caruaru, PE, Brazil
Correspondence: Alexandre Cesar Vieira Sales; Fabricio Oliveira Souto
Keizo Asami Immunopathology Laboratory, LIKA/Federal University of Pernambuco – UFPE, Av. Prof. Moraes Rego, 1235, Cidade Universitária, Recife, PE 52171-011, Brazil
+55 (81) 2126-8484
; +55 (81) 2126-8484
Email [email protected]; [email protected]
Introduction: MicroRNA-21 (miRNA-21) has been described as one of the most significantly upregulated miRNAs in human breast cancer. However, limited knowledge exists on miRNA-21 expression in breast cancer tissue after neoadjuvant chemotherapy (NAC).
Purpose: The aim of this study was to assess miRNA-21 expression in the tumor tissues of Brazilian patients with breast cancer who underwent NAC and its correlation with clinicopathological variables.
Patients and Methods: Utilizing qRT-PCR, miRNA-21 expression in tumor tissue was measured in a cohort of female patients with breast cancer who underwent NAC. The correlation of miRNA-21 expression with breast cancer molecular subtypes and other clinicopathological variables was also assessed.
Results: A total of 55 patients were included in the study, and 28 (50.9%) underwent NAC. miRNA-21 was upregulated in patients with breast cancer, regardless of previous exposure to chemotherapy, molecular subtypes, tumor-node-metastasis (TNM) staging and lymph node status of the axilla. miRNA-21 expression did not differ between patients with breast cancer who achieved a pathologic complete response after NAC and healthy controls.
Conclusion: miRNA-21 was upregulated in the tumor tissue of Brazilian patients with breast cancer regardless of NAC treatment, which reinforces its role as an “oncomiR” and a potential biomarker.
Keywords: miRNA-21, breast cancer, biomarker, tumor tissue, neoadjuvant chemotherapy
Introduction
With an annual estimate of 2.1 million new cases worldwide, breast cancer is the most common neoplasia among women and is the leading cause of cancer-related death in over 100 countries.1 Despite the significant progress in the comprehension of breast cancer molecular biology, mammography is still considered the gold standard method for screening and early diagnosis in asymptomatic patients,2 and the development of novel biomarkers for breast cancer diagnosis and disease outcome prediction is urgently needed.
MicroRNAs (miRNAs) are small noncoding RNA molecules with a length of 19–23 nucleotides that regulate gene expression at the posttranscriptional level.3 Based on accumulated evidence, miRNAs play a crucial role in several physiological and pathological processes, such as metabolism, proliferation, differentiation and apoptosis. Their deregulation has been associated with a broad spectrum of diseases, such as Parkinson’s disease, Alzheimer’s dementia, obesity, diabetes, autoimmune disorders, cardiovascular diseases and cancer.4,5
With regard to cancer, miRNAs are differentially expressed between healthy and tumor cells. miRNAs can show different tissue-specific signatures among distinct types of cancer, and they can either promote (“oncomiRs”) or suppress tumor development and progression by influencing oncogenic processes such as initiation, promotion, malignant conversion, progression and metastasis.6 In addition, miRNAs can be detected in the intracellular environment and in several body fluids, such as blood, urine, saliva, cerebrospinal fluid, semen and breast milk; thus, they might represent a promising source of stable and noninvasive biomarkers for cancer diagnosis, prognosis and treatment.7,8
Several studies have been performed to identify the abnormal expression of circulating miRNAs for early breast cancer diagnosis. However, miRNAs have not been applied in clinical practice because of inconsistencies in the selection of specific miRNAs for diagnostic and prognostic purposes, methodological challenges, patient heterogeneity, and differences in genetic background, sex, age and metabolic status.9
miRNA-21 has been described as one of the most significantly upregulated miRNAs in human breast cancer and other neoplasms, such as gastric, colorectal, pancreatic, lung and ovarian carcinomas.10–12 In addition, its expression has been associated with tumor diagnosis and development as well as poor prognosis. Evidence suggests that miRNA-21 targets and inhibits tropomyosin 1 (TPM1), programmed cell death 4 (PDCD4) and phosphatase and tensin homolog (PTEN) as well as other tumor-related genes.13 Previous studies have shown that miRNA-21 could be measured stably in tumor tissues, formalin-fixed paraffin-embedded tissues and blood circulation.14
Various studies have consistently reported that miRNA-21 is upregulated in breast cancer tissues compared with normal breast tissues.14 However, some conflicting reports have been described in terms of its association with prognostic clinicopathological factors, such as breast cancer molecular subtypes, tumor size, tumor grade, tumor-node-metastasis (TNM) staging and lymph node involvement. Furthermore, there are scarce published data on miRNA-21 tumor expression patterns following neoadjuvant chemotherapy treatment (NAC) and in a Brazilian population that is characterized by miscegenation and a varied ethnic background.
The present study aimed to assess miRNA-21 expression in the tumor tissues of a cohort of female Brazilian patients with breast cancer who underwent NAC and its correlation with the clinicopathological characteristics of breast cancer.
Patients and Methods
Ethics Approval and Consent to Participate
Approval and written consent were obtained from all study participants for the use of their blood, healthy breast tissue and breast cancer tissue samples for research purposes. The study protocol was reviewed and approved by the Ethics Research Committee of Hospital do Cancer de Pernambuco (approval number: CAAE 75061717.3.0000.5205) and was carried out under the terms of the Helsinki Declaration.
Study Cohort and Clinical Sample Processing
The study included 55 patients with operable breast cancer at the Hospital do Cancer de Pernambuco from May 2018 to July 2018. Of the 55 patients, 28 were under NAC, and the choice of the most appropriate chemotherapy protocol was at the discretion of the attending physician. Patients with histologically confirmed invasive breast cancer and tumor-node-metastasis (TNM) clinical stages I to III (localized disease) were included in this study. Patients undergoing past cancer treatments, diagnosed with neurologic disorders and autoimmune disease were not included in this study. The clinicopathological and relevant demographic characteristics of the surgical patients were documented prospectively in the breast cancer database we maintained. Cancer staging was performed according to the 8th edition of the American Joint Committee on Cancer TNM criteria.15 The breast cancer molecular subtypes were defined according to the immunohistochemistry characteristics of estrogen, progesterone, HER-2 and Ki-67 receptor staining.18 Healthy breast tissue samples from nine women who had undergone plastic surgery for cosmetic purposes were collected to represent the control subjects. All tissue samples were obtained from surgical specimens, and fragments with a diameter of up to 0.5 cm were submerged in five volumes of RNAlaterTM (Qiagen, MD, USA) at room temperature for 2 hours (to allow the solution to thoroughly penetrate the tissue) and stored at −80°C until they were subjected to total RNA extraction.
Tissue RNA Extraction and DNase Treatment
RNA extraction was performed using TRIzol® (Invitrogen, Life Technologies, UK) on both healthy breast (control group) and breast cancer tissues. Breast glandular and tumor tissues were carefully dissected free from surrounding fat tissue. One milliliter of TRIzol® was added per 50–100 mg of tissue. The tissue was then homogenized using a mechanical tissue homogenizer at 0°C. RNA was DNase treated with a commercially available kit (Ambion, UK) according to the manufacturer’s recommendations. After extraction, the RNA concentration and integrity were determined using a NanoDrop spectrophotometer (NanoDrop ND1000; Thermo Fisher Scientific, USA). The total RNA was stored at –80°C.
cDNA Synthesis and Gene Expression
Aliquots of 10 ng of eluted RNA samples were used to make cDNA using the TaqManTM MicroRNA Reverse Transcription Kit (Applied Biosystems, USA) and 5X RT Human TaqMan MicroRNA Assay primers (hsa-miR-21-5p: Assay ID_000397; RNU6B: Assay ID_001093) according to the manufacturer’s instructions. The reaction was performed in a Veriti thermal cycler (Applied Biosystems, USA) for 30 minutes at 16°C followed by 30 minutes at 42°C and an additional 5 minutes at 85°C.
Quantitative PCR was performed on a 7500 Real Time PCR system (Applied Biosystems, USA) with TaqMan™ Universal PCR Master Mix (Applied Biosystems, USA), 20X primers and nuclease-free water to a total volume of 10μL. The cycling process was performed as follows: 50°C for 2 minutes and incubation at 95°C for 10 minutes, followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. The threshold standard deviation (SD) for intra-assay and interassay replicates was 0.3.
All samples were analyzed in triplicate on the ABI 7500 platform (Applied Biosystems, USA). The cycle quantification (Cq) values were calculated with SDS 1.4 software (Applied Biosystems).
Data Processing and Statistical Analysis
The normalization process utilized the 2−ΔΔCt method.16 The Cq values from the target miRNA-21 were subtracted from the Cq values of the endogenous control RNU6B. A subsequent ΔΔCq value was calculated for the malignant tumors using the average of the control ΔCq values. The expression values are represented as the ΔΔCq value on a log2 scale. An absolute quantification method for miRNA-21 expression was also performed by utilizing the means of the Cq values from patients with breast cancer in comparison to those of the controls. Clinical and pathological characteristics between patients who were chemo-naïve and NAC were compared using the Mann–Whitney U-test for continuous variables and Pearson’s chi-squared test for categorical variables. The normality of the data was tested by the Shapiro–Wilk test. Variance homogeneity among groups was assessed with the Levene test. Numerical variables are presented as the mean ± standard error (SE), and categorical variables are shown as relative frequencies.
Group comparisons were performed utilizing the generalized linear model. We used the gamma model because of the nonparametric distribution of the data. When significance was reached, we conducted post hoc analysis for single comparisons and Bonferroni correction for multiple comparisons. To analyze the relationship between miRNA-21 and tumor size, we calculated Spearman’s rank correlation coefficient.
Numerical variables are presented as the mean ± SE, and categorical variables are shown as a relative frequency. For all the performed analyses, a value of p≤0.05 was considered statistically significant. The analysis procedures were accomplished using GraphPad Prism version 7.00 (GraphPad Software, La Jolla California, USA) and the Statistical Package for the Social Sciences software SPSS/PASW version 20 (IBM Corp., Armonk, NY).
Results
The clinical and pathological characteristics of the patients with breast cancer are summarized in Table 1. The average age in the control group was 37.8±2.4 years old.
 |
Table 1 Clinical and Pathological Characteristics of Patients with Breast Cancer |
Among the patients with breast cancer, those submitted to neoadjuvant chemotherapy were younger and had larger tumors at the initial diagnosis of the disease. The average age in the control group was 37.8±2.4 years old. Invasive carcinoma of nonspecific type was the most common histological type, and more than half of the patients were classified as the luminal molecular subtypes, followed by HER-2 and triple-negative subtypes. It is important to note that in terms of the breast cancer molecular subtypes, the HER-2 group included 10 patients with luminal hybrid tumors (positivity for estrogen and/or progesterone and HER-2 receptors in immunohistochemistry staining) and two patients with HER-2 overexpression.
Clinical stages II and III were documented in 83.6% of the patients, which means that tumors larger than 2 cm and/or there was involvement of regional lymph nodes. Among patients treated with neoadjuvant chemotherapy, a similar percentage of axillary negative lymph nodes was observed.
The clinical and pathological characteristics of the patients with breast cancer are summarized in Table 1.
Breast Cancer Tissues Presented Increased miRNA-21 Expression
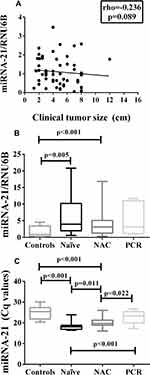
Comparative analysis of miRNA-21 expression between breast cancer and healthy breast tissues demonstrated upregulated levels of miRNA-21 among all patients with breast cancer included in the study, regardless of previous exposure to chemotherapy treatment (Figure 1A). In accordance with the relative expression, the means of the absolute Cq values also showed a significant difference between patients and controls (Figure 1B).
Tissue miRNA-21 Expression According to Breast Cancer Molecular Subtypes and Other Prognostic Factors
To clarify whether miRNA-21 expression is related to breast cancer molecular subtypes and prognostic factors, we analyzed miRNA-21 expression in tumor tissue samples according to different breast cancer molecular subtypes and observed upregulated levels of this biomarker for all breast cancer molecular subtypes. These results were more prominent for the HER-2-positive and luminal A groups in which its expression was 3.4 times higher than that in the control group. Although miRNA-21 expression in the triple-negative breast cancer group was 2.31 times higher than that in the control group, a significant difference was not reached. When the same analysis was performed considering the means of the Cq values for the breast cancer subgroups in comparison to those of the healthy controls, a significant difference in miRNA-21 expression was found for all the subtypes (Table 2).
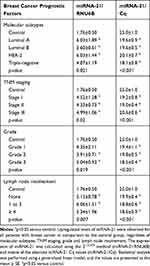 |
Table 2 miRNA-21 Expression According to Breast Cancer Prognostic Factors |
Analysis of miRNA-21 expression in relation to breast cancer TNM staging demonstrated upregulated levels of this molecule for all the groups in comparison to the control group. Such upregulated expression was more pronounced in the stage III group than in the stage II group when the means of the Cq values were considered (Table 2).
Regarding the relation between breast cancer histological grades and miRNA-21 tissue expression, a significant upregulation of the latter was perceived for all grades with significant results for grades 2 and 3. Patients with metastatic disease (stage IV) were not included in the study. When the same analysis was performed utilizing the means of the Cq values, a significant difference in miRNA-21 expression was found for all histological grades in comparison to the control group (Table 2). These findings suggest that the expression of miRNA-21 does not correlate with increased disease severity since there was no difference in its expression among the grades. A positive correlation was not found between tumor size and miRNA-21 expression in breast cancer tissues (Figure 2A). The lymph node analysis showed upregulated miRNA-21 expression in all the breast cancer groups, regardless of the status of the axilla. For both quantification methods, no intergroup difference was observed in terms of the quantity of positive lymph nodes (Table 2).
miRNA-21 Tissue Expression in Patients with Breast Cancer After Neoadjuvant Chemotherapy
To investigate whether NAC impacts miRNA-21 expression, we started a new set of analyses. The clinical and pathological characteristics of the 28 patients treated with NAC are described in Table 1. The average tumor size for the patients before treatment was 5.75±0.43 cm, and the pathological (in surgical specimens) size was 2.89±0.41 cm. Five out of 28 patients achieved pathologic complete response (PCR) in the breast and the axilla (Table 1).
A comparison of the miRNA-21 levels among the patients in the three groups, namely, the chemotherapy-naïve, NAC-treated and PCR groups, revealed significantly upregulated levels of this microRNA for the chemotherapy-naïve and NAC-treated groups in comparison to the control group. However, a considerable difference in miRNA-21 tissue expression was noted between the chemotherapy-naïve and NAC-treated groups when we compared the groups utilizing the means of the Cq values (Figure 2C and Supplementary Table 1). In this case, NAC patients presented higher Cq values than chemotherapy-naïve patients. Furthermore, in both comparison methods, no difference in miRNA-21 expression was observed between patients who achieved PCR and the controls (Figure 2B and C; Supplementary Table 1).
The analysis of miRNA-21 tissue expression among the luminal A, luminal B, HER-2 positive and triple-negative subtypes was focused on chemotherapy-naïve and NAC-treated patients. The results showed a significant difference in terms of miRNA-21 upregulation for the luminal A, luminal B and HER-2 subgroups (Table 3). However, when the same analysis was performed considering miRNA-21 as the means of the Cq values, a significant difference was observed for all the subgroups, especially for the luminal B and HER-2 subgroups in which a significant intergroup difference between the treated and chemotherapy-naïve groups was noticed.
 |
Table 3 Prognostic Factors and miRNA-21 Expression in Patients Treated with Neoadjuvant Chemotherapy |
A significant intergroup difference was found between stage III-treated and chemotherapy-naïve patients when the relative expression of miRNA-21 was analyzed according to TNM stage. In the former, miRNA-21 expression was 2.51 times higher in comparison to the latter. When the same analysis was performed by considering the means of the miRNA-21 Cq values, a significant intergroup difference was observed for clinical stages II and III (Table 3).
The analysis of miRNA-21 tissue expression within the three histological grades showed upregulated levels of this molecule in comparison to those in the control group detected for all the grades only when we analyzed the means of the miRNA-21 Cq values (Table 3).
The analysis of miRNA-21 expression for lymph node involvement between NAC and chemotherapy-naïve patients revealed upregulated levels of this biomarker for all breast cancer groups, regardless of exposure to previous chemotherapy. However, significant differences for all lymph node metastasis statuses were only demonstrated when we utilized the means of the miRNA-21 Cq values (Table 3).
Discussion
In this study, we analyzed the expression of miRNA-21 in the tumor tissues of 55 patients with breast cancer in comparison to that of healthy controls. We found that the expression of this biomarker was significantly higher in patients with breast cancer than in controls, regardless of their previous treatment with systemic chemotherapy. This finding is highly consistent with previous reports that identified miRNA-21 as one of the most commonly and prominently elevated miRNAs among breast cancer tumor tissues.14,17,18
In terms of the clinical and epidemiological aspects, the patients with breast cancer included in this study mirror the distribution of this disease in Brazil and in the rest of the world.19 Conversely, contrary to the trend in developed countries of diagnosing breast cancer in its initial stages, most patients included in this study received a disease diagnosis at a more advanced stage, which unfortunately reflects the specific practices of the public health system in the country.20
More than 90% of the cases were diagnosed with invasive carcinoma of a nonspecific type, which is consistent with the findings described in the literature.21 However, in terms of the molecular subtype distribution among patients with breast cancer, 28 (50.9%) luminal B cases were observed, which is higher than the proportion of 15–20% reported in the literature.22
Analysis of miRNA-21 expression between patients who underwent NAC and those who were chemotherapy naïve demonstrated upregulated levels of miRNA-21 expression for both groups in comparison to the control group. However, no significant difference was observed between the cancer groups. This finding is aligned with the report of Al-Khanbashi et al23 who assessed the expression of miRNA-21 in the tumor tissues of nine patients with breast cancer at different times of NAC and reported that the miRNA-21 levels did not show any significant alteration during different times of NAC.
However, when we performed the analysis of miRNA-21 expression by utilizing the experimental quantification method, it was observed that the Cq values increased significantly in the group of patients who underwent NAC in comparison to those patients who remained chemotherapy naïve, indicating a possible restoration of miRNA-21 levels as a consequence of the response to systemic treatment. This finding must be further explored in future studies.
In addition, when we analyzed the expression of miRNA-21 in the tumor tissue of patients with breast cancer who underwent NAC and achieved PCR, no difference was observed in comparison to that in the control group, which reinforces a possible restoration of this biomarker following an optimal response to chemotherapy. The analysis of residual breast cancer tumors after NAC may be useful for identifying new biomarkers to assess the response to systemic treatment and predict clinical outcomes. Previous studies reported on the roles of the expression of certain miRNAs, such as miRNA-18a, miRNA-7 and miRNA-340, in the tumor tissue of patients who achieved PCR in predicting survival outcomes and response to chemotherapy.24–27
The association between miRNA-21 expression and clinicopathological features in our study demonstrated an upregulation of this biomarker for clinical stages II and III, axillary lymph node metastasis and all breast cancer subtypes regardless of previous treatment with NAC. Conversely, there were no significant correlations of miRNA-21 expression with tumor size or tumor histological grade, which is in accordance with the findings of previous reports.28,29 Nevertheless, it is important to highlight that miRNA-21 was significantly upregulated for all histological grades when we performed the analysis considering the means of the Cq absolute values.
These aforementioned findings are inconsistent with previous data described in the literature in which upregulated miRNA-21 levels correlated with more advanced TNM staging and lymph node metastasis.30,31 Our results indicate that the levels of miRNA-21 are upregulated in all patients with breast cancer, regardless of TNM staging, lymph node status and molecular subtypes, which can be attributed to the ethnogenetic characteristics of the sample, sample size and miRNA normalization method. These findings warrant further investigation in future studies.
Our results revealed that the expression of miRNA-21 was similarly upregulated in all breast cancer molecular subtypes but did not reach a significant difference for the triple-negative breast cancer (TNBC) subgroup, which could be attributed to the limited sample size of this study or even to the choice of the normalization method. Dong et al32 reported upregulated levels of miRNA-21 in 72 TNBC specimens. Savad et al33 found that the expression of miRNA-21 was similar in different tumor subtypes and did not appear to discriminate between different breast tumor subtypes, reinforcing our results. However, an insignificant association between miRNA-21 expression and breast cancer subtypes has also been reported.13,34,35
In this study, a relative quantification method was utilized to compare miRNA-21 tissue expression between patients with breast cancer and healthy controls. However, a comparison of these two groups was also performed by analyzing the mean of the absolute miRNA-21 Cq values. Due to the analysis of miRNA-21 as a single investigational biomarker in this study, it was not possible to utilize the global mean normalization method.36–39 As a result, we performed an experimental normalization method by comparing the means of the absolute miRNA-21 Cq values between breast cancer tissue samples and healthy control tissue samples. As previously described, significant differences were found between those groups, most of the time reflecting the same results obtained when we utilized the relative quantification method: among patients with breast cancer, lower miRNA-21 Cq values in comparison to those of healthy controls denoted more copies of this molecule in tumor tissue, and this finding must be further evaluated.
Conclusion
Our study reinforced the role of miRNA-21 as an “oncomiR” with upregulated levels in the tumor tissues of a cohort of female Brazilian patients with breast cancer, regardless of their previous exposure to systemic chemotherapy. In addition, we demonstrated that miRNA-21 expression did not differ between patients with breast cancer who achieved a pathologic complete response and healthy controls, indicating a possible restoration of this biomarker in response to NAC as an effect of tumor destruction and surrounding tissue regeneration.
The correlation of miRNA-21 expression with clinicopathological variables was positive, with upregulated levels of this biomarker for all breast cancer molecular subtypes, regardless of previous NAC, TNM staging and lymph node metastasis. Tumor size and histological grade did not demonstrate any correlation with miRNA-21 expression, suggesting that miRNA-21 could be used as a diagnostic marker for lymph node metastasis.
We also tested an experimental normalization method by comparing the means of the absolute miRNA-21 Cq values in patients with breast cancer with those of the controls. Most of the time, the results reflected those obtained with the relative quantification method and need to be further explored.
Abbreviations
PCR, pathological complete response; NAC, neoadjuvant chemotherapy; Naïve, chemotherapy naïve; NST, invasive carcinoma of no special type; ILC, invasive lobular carcinoma; HER-2, HER-2 overexpression; TNM, tumor-node-metastasis breast cancer staging.
Author Contributions
All authors made a significant contribution to the reported work, whether in the conception, study design, execution, data acquisition, analysis and interpretation, or in all of these areas. In addition, the authors collaborated with the writing, critical review of the article and gave the final approval of the version to be published. After reviewing this study, all authors agreed with the journal to which the article was submitted; and agree to be responsible for all aspects of the work.
Funding
We would like to thank the Department of Mastology from Hospital do Câncer de Pernambuco – HCP, especially the surgeons who helped us in collecting the tissue samples, and all the patients and volunteers who participated in the study. This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq (Grant: 470702/2014-1). AC was supported by Fundação de Amparo a Ciência e Tecnologia do Estado de Pernambuco-FACEPE (Grant: BFP-0102-1.11/19). We also appreciate the financial support from the Coordination for the Improvement of Higher Education Personnel - CAPES for this study.
Disclosure
The authors report no conflicts of interest in this work. The authors also report no financial interest with this study.
References
1. Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–1953. doi:10.1002/ijc.31937
2. Nelson HD, Zakher B, Cantor A, et al. Risk factors for breast cancer for women aged 40 to 49 years: a systematic review and meta-analysis. Ann Intern Med. 2012;156(9):635–648. doi:10.7326/0003-4819-156-9-201205010-00006
3. Vishnoi A, Rani S. MiRNA Biogenesis and Regulation of Diseases: An Overview. InMicroRNA Profiling. Humana Press; 2017:1–10.
4. Shah PP, Hutchinson LE, Kakar SS. Emerging role of microRNAs in diagnosis and treatment of various diseases including ovarian cancer. J Ovarian Res. 2009;2(1):11. doi:10.1186/1757-2215-2-11
5. Chen JQ, Papp G, Szodoray P, et al. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016;15(12):1171–1180. doi:10.1016/j.autrev.2016.09.003
6. Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer. 2015;113(4):569. doi:10.1038/bjc.2015.253
7. Cortez MA, Bueso-Ramos C, Ferdin J, et al. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467.
8. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857. doi:10.1038/nrc1997
9. Grimaldi AM, Incoronato M. Clinical translatability of “identified” circulating miRNAs for diagnosing breast cancer: overview and update. Cancers. 2019;11(7):901. doi:10.3390/cancers11070901
10. Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi:10.1073/pnas.0510565103
11. Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120(5):1046–1054.
12. Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14(9):2690–2695. doi:10.1158/1078-0432.CCR-07-1731
13. Asaga S, Kuo C, Nguyen T, et al. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91. doi:10.1373/clinchem.2010.151845
14. Chen J, Wang X. MicroRNA-21 in breast cancer: diagnostic and prognostic potential. Clin Transl Oncol. 2014;16(3):225–233. doi:10.1007/s12094-013-1132-z
15. Giuliano AE, Edge SB, Hortobagyi GN. Eighth edition of the AJCC cancer staging manual: breast cancer. Ann Surg Oncol. 2018;25(7):1783–1785. doi:10.1245/s10434-018-6486-6
16. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408.
17. Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi:10.1158/0008-5472.CAN-05-1783
18. Si ML, Zhu S, Wu H, et al. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799. doi:10.1038/sj.onc.1210083
19. Ban KA, Godellas CV. Epidemiology of breast cancer. Surg Oncol Clin N Am. 2014;23(3):409–422. doi:10.1016/j.soc.2014.03.011
20. dos-Santos-Silva I, De Stavola BL, Junior NL, et al. Ethnoracial and social trends in breast cancer staging at diagnosis in Brazil, 2001–14: a case only analysis. Lancet Glob Health. 2019;7(6):e784–97. doi:10.1016/S2214-109X(19)30151-2
21. Li CI, Uribe DJ, Daling JR. Clinical characteristics of different histologic types of breast cancer. Br J Cancer. 2005;93(9):1046. doi:10.1038/sj.bjc.6602787
22. Kumar N, Patni P, Agarwal A, et al. Prevalence of molecular subtypes of invasive breast cancer: a retrospective study. Med J Armed Forces India. 2015;71(3):254–258. doi:10.1016/j.mjafi.2015.04.006
23. Al-Khanbashi M, Caramuta S, Alajmi AM, et al. Tissue and serum mirna profile in locally advanced breast cancer (labc) in response to neo-adjuvant chemotherapy (nac) treatment. PLoS One. 2016;11(4):e0152032. doi:10.1371/journal.pone.0152032
24. García-Vazquez R, Ruiz-García E, Meneses Garcia A, et al. A microRNA signature associated with pathological complete response to novel neoadjuvant therapy regimen in triple-negative breast cancer. Tumor Biol. 2017;39(6):1010428317702899. doi:10.1177/1010428317702899
25. Luengo-Gil G, García-Martínez E, Chaves-Benito A, et al. Clinical and biological impact of miR-18a expression in breast cancer after neoadjuvant chemotherapy. Cell Oncol. 2019;21:1–8.
26. Raychaudhuri M, Bronger H, Buchner T, et al. MicroRNAs miR-7 and miR-340 predict response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2017;162(3):511–521. doi:10.1007/s10549-017-4132-9
27. Zheng Y, Li S, Boohaker RJ, et al. A microRNA expression signature in taxane-anthracycline-based neoadjuvant chemotherapy response. J Cancer. 2015;6(7):671.
28. Huang GL, Zhang XH, Guo GL, et al. Clinical significance of miR-21 expression in breast cancer: SYBR-green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep. 2009;21(3):673–679.
29. Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14(11):2348–2360.
30. Ozgun A, Karagoz B, Bilgi O, et al. MicroRNA-21 as an indicator of aggressive phenotype in breast câncer. Onkologie. 2013;36(3):115–118. doi:10.1159/000348678
31. Wang F, Zheng Z, Guo J, et al. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119(3):586–593.
32. Dong G, Liang X, Wang D, et al. High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol. 2014;31(7):57. doi:10.1007/s12032-014-0057-x
33. Savad S, Mehdipour P, Miryounesi M, et al. Expression analysis of MiR-21, MiR-205, and MiR-342 in breast cancer in Iran. Asian Pac J Cancer Prev. 2012;13(3):873–877. doi:10.7314/APJCP.2012.13.3.873
34. Sempere LF, Christensen M, Silahtaroglu A, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67(24):11612–11620. doi:10.1158/0008-5472.CAN-07-5019
35. Si H, Sun X, Chen Y, et al. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139(2):223–229. doi:10.1007/s00432-012-1315-y
36. Schwarzenbach H, Da Silva AM, Calin G, et al. Data normalization strategies for microRNA quantification. Clin Chem. 2015;61(11):1333–1342.
37. Kok MG, Halliani A, Moerland PD, et al. Normalization panels for the reliable quantification of circulating microRNAs by RT-qPCR. FASEB J. 2015;29(9):3853–3862. doi:10.1096/fj.15-271312
38. Kang K, Peng X, Luo J, et al. Identification of circulating miRNA biomarkers based on global quantitative real-time PCR profiling. J Anim Sci Biotechnol. 2012;3(1):4. doi:10.1186/2049-1891-3-4
39. Arabkari V, Clancy E, Dwyer RM, et al. Relative and absolute expression analysis of micrornas associated with luminal a breast cancer–a comparison. Pathol Oncol Res. 2019;6:1–2.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.