Back to Journals » OncoTargets and Therapy » Volume 10
miRNA expression profiling in formalin-fixed paraffin-embedded endometriosis and ovarian cancer samples
Authors Braicu OL, Budisan L, Buiga R , Jurj A , Achimas-Cadariu P, Pop LA, Braicu C , Irimie A, Berindan-Neagoe I
Received 15 March 2017
Accepted for publication 31 May 2017
Published 28 August 2017 Volume 2017:10 Pages 4225—4238
DOI https://doi.org/10.2147/OTT.S137107
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr William C. Cho
Ovidiu-Leonard Braicu,1 Liviuta Budisan,2 Rares Buiga,2,3 Ancuta Jurj,2 Patriciu Achimas-Cadariu,1,4 Laura Ancuta Pop,2 Cornelia Braicu,2 Alexandru Irimie,1,4 Ioana Berindan-Neagoe2,5,6
1Department of Surgery, Iuliu Hatieganu University of Medicine and Pharmacy, Cluj-Napoca, 2Research Center for Functional Genomics, Biomedicine and Translational Medicine, Iuliu Hatieganu University of Medicine and Pharmacy, 3Pathology Department, The Oncology Institute Prof. Dr. Ion Chiricuta, Cluj-Napoca, 4Department of Surgical Oncology, The Oncology Institute Prof. Dr. Ion Chiricuta, Cluj-Napoca, 5MEDFUTURE-Research Center for Advanced Medicine, University of Medicine and Pharmacy Iuliu-Hatieganu, Cluj-Napoca, 6Department of Functional Genomics, Proteomics and Experimental Pathology, The Oncology Institute Prof. Dr. Ion Chiricuta, Cluj-Napoca, Romania
Abstract: Endometriosis is an inflammatory pathology associated with a negative effect on life quality. Recently, this pathology was connected to ovarian cancer, in particular with endometrioid ovarian cancer. microRNAs (miRNAs) are a class of RNA transcripts ~19–22 nucleotides in length, the altered miRNA pattern being connected to pathological status. miRNAs are highly stable transcripts, and these can be assessed from formalin-fixed paraffin-embedded (FFPE) samples leading to the identification of miRNAs that could be developed as diagnostic and prognostic biomarkers, in particular those involved in malignant transformation. The aim of our study was to evaluate miRNA expression pattern in FFPE samples from endometriosis and ovarian cancer patients using PCR-array technology and also to compare the differential expression pattern in ovarian cancer versus endometriosis. For the PCR-array study, we have used nine macrodissected FFPE samples from endometriosis tissue, eight samples of ovarian cancers and five normal ovarian tissues. Quantitative real-time PCR (qRT-PCR) was used for data validation in a new patient cohort of 17 normal samples, 33 endometriosis samples and 28 ovarian cancer macrodissected FFPE samples. Considering 1.5-fold expression difference as a cut-off level and a P-value <0.05, we have identified four miRNAs being overexpressed in endometrial tissue, while in ovarian cancer 15 were differentially expressed (nine overexpressed and six downregulated). The expression level was confirmed by qRT-PCR for miR-93, miR-141, miR-155, miR-429, miR-200c, miR-205 and miR-492. Using the interpretative program Ingenuity Pathway Analysis revealed several deregulated pathways due to abnormal miRNA expression in endometriosis and ovarian cancer, which in turn is responsible for pathogenesis; this differential expression of miRNAs can be exploited as a therapeutic target. A higher number of altered miRNAs were detected in endometriosis versus ovarian cancer tissue, most of them being linked with epithelial-to-mesenchymal transition.
Keywords: endometriosis, ovarian cancer, miRNA, formalin-fixed paraffin-embedded samples
Introduction
Endometriosis is presented as a benign gynecological condition that affects ~5%–10% of premenopausal women and is characterized by the presence of ectopic endometrial implants in pelvic area; however, these implants can also be extended to the abdomen and even to the central nervous system. In some cases, these implants can undergo malignant transformation and can lead to a condition called endometriosis-associated ovarian cancer (EAOC).1 This progression is now supported by epidemiological studies and some molecular data; the development of cancer from endometriosis is a slowly progressing process that is difficult to be revealed by clinical evidences.1,2 A recent study affirms that ~2.5% of women diagnosed with ovarian endometriosis undergo malignant conversion, but this percentage can be seriously underestimated due to unspecific molecular processes that intervene in the transit from benign to malignant status.3 It is consequently anticipated to be a multifaceted process, based on the characteristic features of the disease, where coding and noncoding transcripts play an important role.4
The molecular characteristics of this carcinogenic event are still poorly understood, although there are a number of genes and molecular signaling pathways that are currently associated with the possible transition from endometriosis to ovarian cancer being regulated by an altered pattern of key for tumor suppressor genes or oncogenes,5,6 microsatellite instability and noncoding genes, especially miRNA alteration pattern.
Ovarian cancer is the cause of greatest number of cancer-related deaths worldwide according to the latest statistical data, with high mortality rate due to the asymptomatic profile in early stages and nonsensitive diagnostic methods.7–9 Ovarian cancer is frequently diagnosed in final stages when massive ascites and peritoneal spreading has already occurred, a stage that usually is nonresponsive to classic but to aggressive treatment methods,10 and the survival rate remains low at 30%.8,11 In this regard, there is an urgent need not only for more sensitive and precise diagnostic methods but also for methods that are able to discriminate this type of malignant pathology in the early stages of development, or more preferably, before the installation of this asymptomatic disease, in the form of prevention biomarkers.9,12,13 The recently discovered miRNAs may lead to early diagnostic and therapeutic approaches, due to their gene expression regulation ability of the genes that play crucial roles in the installation and development of ovarian cancer, being actively involved in the etiology of this pathology.14
Ovarian epithelial tumors are classified as: type I tumors defined by a slow growth (comprising low-grade serous carcinoma, mucinous carcinoma, endometrioid carcinoma and clear-cell carcinomas) and type II tumors defined as a more aggressive form (including high-grade serous carcinoma, mixed mesodermal tumors and undifferentiated tumors).15 The endometriomas or ovarian endometriosis cysts are retrieved in ~17%–44% of patients diagnosed with endometriosis, which are considered as the common precursor lesions to ovarian cancer. EAOC is especially noticeable and retrieved as endometrioid and clear cell ovarian tumor.1
Another important concern in ovarian cancer biology consists in a low amount of information about the primary site of origin, due to the absence of glandular epithelia in the ovary which acts like a direct precedent for cancer.5 The most suitable candidate for the primary source of ovarian cancer nowadays is considered to be the ovarian surface epithelium, tissue formed of a monolayer of cells that covers the exterior of the ovaries.5,10 However, this covering tissue is not the only origin for this malignant disease. Increased evidences show that endometriosis can be related to benign-malignant transition to ovarian cancer, particularly in the case of endometrioid and clear cell adenocarcinoma of the ovary (also named in the literature as EAOC); however, these conditions present discrete features which are difficult to be identifed.16,17 The malignant transformation of endometriosis into ovarian cancer, particularly in endometrioid, clear cell and low-grade serous histotypes, is related to genetic and nongenetic factors or a combination of these factors.15
An increased number of studies incriminate miRNA in pathological status. miRNAs are presented today as key regulators involved in numerous processes in mammals and other multicellular organisms;18–20 they control biological pathways involved in proliferation, apoptosis, migration, cell cycle control, differentiation and processes that under aberrant functioning can lead to pathological status, including malignant transformation; hence, there is an increasing interest to use these short transcripts as biomarkers for prevention,21 detection and prognostic application and also as therapeutic targets.20,22
Formalin-fixed paraffin-embedded (FFPE) tissue is a key source of biological material for retrospective analysis, evaluation of molecular profiling and developing novel biomarkers for personalized health care.23,24 Taking into consideration the crucial role of miRNA transcripts in the regulation of numerous signaling pathways,19,20,25,26 there is a possibility to associate the expression of miRNAs in endometriosis and ovarian cancer samples with the risk for malignant transformation.21,27,28 Identification of a specific panel of miRNAs that are differentially expressed in these diseases, in particular among these two pathologies, can enable the discovery of precise biomarkers for diagnostic and prognostic21,25,28 purpose and in particular for assessing the risk involved in the transition from benign to malignant status.
Materials and methods
Sampling procedures
FFPE samples were collected from patients diagnosed at the Institute of Oncology “Prof Dr Ion Chiricuta” in Cluj-Napoca, Romania, with endometriosis and ovarian cancer between the years 2007 and 2014. Approval for the study was obtained from the ethical committee of the same institution (no 72/26). All the procedures for sample selection and processing were done in agreement with the Romanian and international legislation. All the clinical data obtained from the Pathology Department were used anonymously, being in agreement with ethical and legal requirements. This study is in agreement with the institutional policy that imposes a signed informed consent, mentioning that the data can be used for additional future research focused on molecular profiling.
The patient cohort was selected based on clinical data prior to PCR-array or quantitative real-time PCR (qRT-PCR) evaluation. The control patient cohort was selected from a group of patients who had undergone benign surgical resection for uterine pathologies. PCR-array patient cohorts included two cases of endometrial polyps, one uterine fibroid and one leiomyoma. qRT-PCR control group included surgically resected tissue from three cases of ovarian or uterine fibroid, five cases of leiomyoma, two cases of salpingitis and four cases of benign endometrial polyps. The endometriosis patient cohort included cases with left/right adnexal endometriosis and bilateral endometriosis but showing no signs of malignity. The endometroid tissue represents over 90%.
Then the histopathological diagnosis was confirmed. The selection between endometriosis lesions and tumor area was done based on hematoxylin and eosin staining and macrodissected regions from serial unstained FFPE sections. All the cases were pathologically reviewed and the regions were isolated separately based on the tissue type. For ovarian cancer, only the endometriod carcinoma cases (Table S1) with hyperplasia more than 80% were selected, using four slides with macrodissected tissue.
For extraction, we used an average of eight 10 μm FFPE sections from the tissue block for miRNA profiling and for qRT-PCR data validation. None of the selected patients included in this study had undergone preoperative chemotherapy or radiation therapy.
miRNA isolation from FFPE samples
Total RNA was extracted from FFPE tissue using miRCURY RNA Isolation Kit–FFPE (Exicon cat no 300115) based on the instructions furnished by the manufacturer’s protocol. Then the quantity of total RNA was evaluated using NanodropND-2000.
PCR array analysis
The cDNA synthesis was done using 100 ng of total RNA extracted from FFPE tissue, based on the recommended protocol, using the miScript HiSpec Buffer in a volume of 20 μL and the following program: 37°C for 60 minutes and then at 95°C for 5 minutes. The cDNA was then diluted and used for PCR array based on a SybrGreen protocol and Human Ovarian Cancer miScript miRNA PCR array plate (MIHS-110Z; Qiagen NV, Venlo, the Netherlands) following the protocol recommended by the manufacturer. These arrays contain probes for 84 miRNAs whose expression is known or expected to be altered in ovarian cancer, three wells for miRNA reverse transcription control assay and a positive PCR control. The qRT-PCR reaction was performed in Roche LightCycler480 instrument.
qRT-PCR measurements
For the validation of the most relevant altered miRNAs, we selected a subset of 17 normal samples, 33 endometriosis samples and 28 ovarian cancer samples. We used a two-step qRT-PCR. cDNA was generated using miScriptII RT Kit and then was diluted 1:5. The diluted cDNA sample was used as a template for performing qRT-PCR using QuantiTect (QuantiTect SYBR Green PCR kit, Qiagen) with specific miScript Primer (Qiagen). qRT-PCR assays were performed on ViiA 7 Real Time PCR System using the recommended amplification protocol (45 cycles of 30seconds at 95°C, 30seconds at 55°C and 30seconds at 72°C) and a melting curve step.
Data analysis
The analysis of miRNA relative expression was done using the data analysis portal furnished by Qiagen, USA (http://sabiosciences.com/pcr/arrayanalysis.php). For the normalization of data, we used the average Ct value of the cel-miR-39, SNORD68, SNORD95, SNORD96A and RNU6-6P. The quantification for determining the relative expression level of miRNAs was done in endometriosis (nine samples) versus normal tissue (five samples) and ovarian cancer tissue (eight samples).
The altered miRNA pattern in endometriosis versus control, in ovarian cancer versus normal tissue and in ovarian cancer versus endometriosis was analyzed using Ingenuity Pathways Analysis (www.ingenity.com) for biological significance identification.
The qRT-PCR data analysis was done using ΔΔCt method, as previously described by Berindan-Neagoe et al,29 statistical analysis was done using Graphpad Software (https://www.graphpad.com), the ROC (receiving-operating characteristic) curve was used for evaluating the AUC (area under the curve) offering information for the sensitivity of evaluated miRNA transcript.
Results
Evaluation of miRNA relative expression in endometriosis and ovarian cancer
This retrospective molecular profiling study was conducted on nine samples of endometriosis tissue, eight samples of ovarian cancer tissue and five samples of normal ovarian tissue using macrodissected specimens. The average age of endometriosis patients was 40.3±5.57 years, for ovarian cancer patients was 59.33±10.36 years and for control group patients was 60±4.48 years.
First step in this study was to perform a comparative analysis of miRNA profile based on ΔΔCt method in endometriosis versus normal tissue, in ovarian cancer tissue versus normal tissue and then in endometriosis versus ovarian cancer tissue. Table 1 summarizes the differentially expressed miRNAs in endometriosis tissue versus normal tissue, ovarian cancer tissue versus normal tissue and ovarian cancer tissue versus endometriosis tissue. Of considerable interest were the miRNAs that had 1.5-fold expression difference as a cut-off level and a P-value of <0.05. Four miRNAs were identified to be overexpressed in endometriosis tissue, and in ovarian cancer tissue, 15 miRNAs were differentially expressed (nine overexpressed and six downregulated). A higher number of altered miRNAs (14 overexpressed and 23 downregulated) were observed when endometriosis versus ovarian cancer tissue was analyzed. Figure 1 presents Venn diagram for the main miRNAs expressed across the analyzed subgroups.
  | Table 1 Differentially expressed microRNAs considering as cut-off value a fold change ≤1.5 or ≥1.5 and P-value <0.05 |
  | Figure 1 Venn diagram presenting the altered expression levels of common and differentially expressed miRNAs in the analyzed subgroups. |
The present study had identified a particular molecular signature based on the main oncogenic miRNAs overexpressed in endometriosis and ovarian pathology (Figure S1), and also we were able to identify an miRNA panel that was able to discriminate between ovarian cancer and endometriosis.
The differential expression of miRNA proved to be statistically significant, when analyzed using interpretative program Ingenuity Pathway Analysis (IPA), leading to summarization of their biological significance and to defining the molecular interaction between altered miRNAs and their related target gene (Table 2). The results are displayed as networks (Figures 2 and 3) and classified in terms of disease and cellular and molecular functions of the altered miRNAs (Table 3).
  | Table 2 IPA analysis based on altered miRNA pattern in the analyzed subgroups, focusing on diseases |
  | Figure 2 Network generated for the altered miRNAs in the analysis of endometriosis versus normal tissue. The upregulated miRNAs are displayed in red. |
  | Figure 3 Network generated for the altered miRNAs in the analysis of ovarian cancer versus control. The upregulated miRNAs are displayed in red and the downregulated miRNAs in green. |
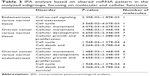  | Table 3 IPA analysis based on altered miRNA pattern in the analyzed subgroups, focusing on molecular and cellular functions |
qRT-PCR data validation
qRT-PCR data validation comprises 17 control samples with an average age of 48.53±9.14 years, 33 endometriosis patients of 41.68±8.78 years, and 33 ovarian cancer patients of 55.75±10.98 years.
miR-93 was proved to be overexpressed in endometriosis and ovarian cancer patients, the expression level being much higher in ovarian cancer compared to endometriosis. Fold change (FC) for endometriosis was 2.42±1.63 and for ovarian cancer was 26.19±30.59, the results being statistically significant in agreement with the PCR-array data (Figure 4). ROC curves for miR-92 reveal a value of AUC for endometriosis group of 0.69 and for ovarian cancer patients of 0.82.
miR-200c was found to be overexpressed in ovarian cancer samples when they were analyzed versus control or versus endometriosis samples. The expression level for miR-200c in normal tissue was 1.63±1.53, in endometriosis 1.27±2.91 and in ovarian cancer 12.29±14.61. AUC values for miR-200c was 0.69 for endometriosis and 0.84 for ovarian cancer, displaying a good sensitivity and specificity for ovarian cancer. Similar expression level was observed for the other two representatives of the same family, namely miR-141 and miR-429, which have an expression level in endometriosis of 8.035±25.09 and 16.17±47.13 and in ovarian cancer of 203.1±275.6 and 104.6±172.6, respectively. A reduced sensitivity was observed for miR-141, whereas highest AUC values were obtained for miR-492 in both the analyses.
An increased level of miR-155 expression was observed in endometriosis and ovarian cancer tissues, but this difference was not statistically significant. For the endometriosis versus control group, FC was 1.416±0.7981 (P-value =0.0507) and for ovarian cancer group, FC was 2.703±2.830 (P-value =0.0552). miR-205 expression level was 7.235±15.49 (AUC =0.69) in endometriosis and 390.2±466.3 (AUC=0.79) in ovarian cancer. miR-492 was overexpressed in both pathologies, with a fold change of 5.40±6.48 for endometriosis and 10.17±10.29 for ovarian cancer. The AUC values for miR-492 revealed a good sensitivity for both pathologies.
A good level of reproducibility was observed among the two patient cohorts used for PCR-array evaluation and qRT-PCR, with an exception for miR-155 which showed the same expression level but the P-value was not statistically significant, emphasizing the potential prognostic significance for the validated miRNA.
Discussion
In this study, we have demonstrated that miRNAs are aberrantly expressed in endometriosis versus normal tissue and ovarian cancer versus normal tissue, and we were also able to identify a panel of miRNAs that are differentially expressed in ovarian cancer versus endometriosis tissue, the details of which can be observed in Figures 1 and S1. Thus, miRNAs can be used as prognostic and diagnostic markers, emphasizing the functional differences that are able to regulate the key processes. The global miRNA expression pattern might undoubtedly discriminate endometriosis from normal tissue, ovarian cancer from normal tissue and also ovarian cancer from endometriosis tissue. The analysis has detected important number of miRNAs with an altered expression pattern being involved in malignant transformation. For qRT-PCR validated miRNAs, good sensitivity and specificity can be observed from ROC curves, in particular for ovarian cancer pathology. These panels of miRNAs can be taken into consideration for possible use as prognostic or diagnostic markers, especially for endometriosis patients undergoing surgical procedures.
One of the identified transcripts that was found to be overexpressed in endometriosis and ovarian cancer is miR-325, a transcript whose overexpression was confirmed even when comparing the degree of expression between the two pathologies using endometriosis samples as control group. The increased expression of miR-325 in ovarian cancer against endometriosis suggests a possible application for its use as a prognostic marker in regard to a possible transition from endometriosis to malignant pathologies. miR-325 has the potential of a tumor suppressor which is reflected by the promotion of tumor progression, increased angiogenesis, invasion and metastasis.30 There is a lack of specific data regarding miR-325 expression in the context of ovarian cancer or endometriosis, but there are several studies reporting its role in other malignant pathologies.31,32 This way, the degree of expression of miR-325 can be utilized as a clinical parameter for detecting ovarian cancer invasion,31 whose expression is inhibited in these tumors. miR-203, an inflammatory transcript,33 was observed to be downregulated in the endometriosis group.
Regarding the expression of miR-200 family, our study has highlighted a significant overexpression value for miR-200a, miR-200b, miR-200c, miR-429, miR-141 and members of the ovarian tumor samples, these members topping the list for positive aberrant expression. Of the five members, miR-200a, miR-200b and miR-200c are currently proposed for use as biomarkers in ovarian cancer.34–38 The ROC data being in agreement with our study34 encourage their application in clinical practice. Furthermore, the expression level of miR-200a and miR-200c has been identified to be associated with the degree of progress for ovarian pathology, whereas the upregulation of miR-200a was found to be correlated with the histology and tumor stage; patients with metastasis in lymph nodes presented a significant elevation of mirR-200c.38 Also, miR-200b has been characterized as a possible therapeutic target related with drug resistance.35 With regard to the expression of miR-141, an increased resistance for cisplatin was found to be proportional to the grade of positive aberrancy of this transcript.39 Members of the miR-200 family (miR-141 and miR-200a) have also been identified in the endometriosis samples but with an expression opposite to that found in ovarian cancer.40 These two transcripts are currently proposed as potential noninvasive biomarkers, their expression from tissue samples being correlated with the values from blood samples.41 Given the role of miR-200 family members in numerous biologic and pathologic processes, especially in epithelial–mesenchymal transition (EMT), we can assume that these transcripts may be involved in the transition from benign endometriosis to malignant pathologies such as ovarian cancer.
Another overexpressed transcript found with an altered expression level in endometriosis samples is miR-520e. This transcript has been studied in hepatocarcinoma42 and breast cancer,43 where its role as a tumor suppressor has been proven.44 This feature places the transcript in a list of potential therapeutic elements. Given the similarity of endometriosis and malignant pathologies in terms of high levels of cell proliferation and also the presence of miR-520 in samples from endometriosis patients, there is a possibility of using this transcript as a therapeutic target for inhibiting cellular expansion.
miR-492 has been identified as overexpressed in both pathologies in the present study, with a more noticeable level in ovarian cancer; however, information on its exact role in progression of endometriosis and ovarian cancer is absent in literatures. miR-492 promotes the progression of hepatic cancer by targeting the PTEN gene and increasing the level of AKT activation in cancer cells, thus having an oncogenic role.45 Patients with an overexpression of miR-492 and an inhibition of the PTEN gene show a lower rate of survival,46 thus sustaining metastatic features.47 Considering the different expression levels of miR-492 in endometriosis and ovarian cancers, with a higher level in the latter, follow-up studies could confirm the quality of miR-492 as a prognostic biomarker in terms of transition from benign to malignant; given its role in abnormal cellular proliferation, this transcript could be placed on the list of potential therapeutic targets for inhibiting aberrant cell growth.
miR-182 was proved to be overexpressed in ovarian cancer, sustaining the present findings. This transcript is presented as a prognostic marker for ovarian cancer,48 as it plays a role in malignant transformation by sustaining cell proliferation, invasion and migration processes, as was emphasized by Liu et al.49,50
The let-7 family is composed of 13 distinct members found on nine different chromosomes.51 Of these 13 members, our study has identified three of them (let-7a, let-7b and let-7c) as being overexpressed in ovarian cancer samples.52 Because of their inhibiting activity on oncogenes associated with ovarian cancer, such as KRAS,52 HRAS,53,54 c-MYC54 and HMGA-2,55 downregulation of the let-7 family members correlates with advanced tumor stages and a lower survival rate.56 Together with the miR-200 family, the let-7 group represents an important potential prognostic and diagnostic biomarker for ovarian cancer as well as a therapeutic element involved in the regulation of key oncogenes.54 An interesting study on miR-30a reported this to be related to decidualization and to endometriosis tissue transformation,57 sustaining our finding related to malignant transformation and therapeutic target for ovarian serous adenocarcinoma.58 miR-30a was found to have an oncogenic role and to modulate BCL2A1, IER3 and cyclin D2 expression via FOXL2,59 the expression of which is related to drug resistance mechanisms.60
miR-145 was found to be downregulated in ovarian cancer and thus has a prognostic role.61 A functional study on ovarian cancer cell lines reveals that treatment with agomiR-145 caused inhibition of TRIM2 and leads to the inhibition of Bim, a proapoptotic Bcl-2 family member.62 miR-145 is related to ovarian cancer chemoresistance by regulation of cell cycle-related proteins: Cdk6 and Sp1.61 This transcript was not found to be statistically significant in endometriosis group but was downregulated in ovarian cancer versus control or ovarian cancer versus endometriosis analysis. Therefore, miR-145 can be involved in malignant transformation, due to the fact that miR-145 was related to an increased cell proliferation, invasiveness and stem cells features in endometrioid primary cell cultures and stabilized cell lines.63
miR-205 is a highly controversial miRNA that was retrieved statistically significantly only in ovarian cancer versus endometriosis group. There are no data available in literature with regard to miR-205 implication in endometriosis, but in ovarian cancer, it is presented to be connected to the regulation of cell proliferation, EMT and invasion mechanism and metastasis processes.64 miR-205 directly targets VEGF-A65 or promotes cell motility via ZEB1,66 which is related with the suppression of SMAD4 and PTEN, an event that favors ovarian cancer progression and67 therefore sustaining malignant transition as showed in our data. miR-27a regulates cancer cell growth, EMT and migration in ovarian cancer.68 miR-27a is an important transcript responsible for drug resistance by targeting MDR1/P-gp and HIPK2 in ovarian cancer cells.69
The EMT-related miRNA has an important role in modulation and favoring proangiogenic mechanism; therefore, this miRNA can be considered as both a prognostic and a therapeutic target in the selected pathologies. These EMT-related miRNAs are prone for activation by angiogenic mechanims70,71 and, in particular, can influence the implantation of endometrial cells on ectopic sites.40
Conclusion
Our data have demonstrated that differentially expressed miRNAs might contribute significantly to regulating the functions of pathological processes sustaining cell proliferation. Endometriosis still remains an enigmatic disease, but now it is clear that miRNAs play an important role in this pathology and can be related to malignant transition. miRNAs that are related to inflammation (miR-325) and those regulating the EMT (miR-200a, miR-200b, miR-200c, miR-141, miR-429, miR-30a, miR-145 and miR-205) can contribute to malignant transformation.
The association between endometriosis and ovarian carcinoma remains debated but is sustained by the common miRNAs. At the same time, we have identified a particular miRNA signature that is capable of discriminating between endometriosis and ovarian cancer, this finding being the ground for additional functional studies needed to clarify the prospective role of miRNAs in endometriosis and its related malignancies.
This panel of identified miRNAs can enable the discovery of dysregulated miRNAs that can act as precise biomarkers for diagnostic and prognostic applications and in particular for assessing the risk involved in the transition from benign to malignant status.
Acknowledgment
This study was sustained by the Iuliu Hatieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania, for PhD research project (grant no 7690/13/15.04.2016).
Disclosure
The authors report no conflicts of interest in this work.
References
Aznaurova YB, Zhumataev MB, Roberts TK, Aliper AM, Zhavoronkov AA. Molecular aspects of development and regulation of endometriosis. Reprod Biol Endocrinol. 2014;12:50. | ||
Liu FS. Molecular carcinogenesis of endometrial cancer. Taiwan J Obstet Gynecol. 2007;46(1):26–32. | ||
Guo SW. Endometriosis and ovarian cancer: potential benefits and harms of screening and risk-reducing surgery. Fertil Steril. 2015;104(4):813–830. | ||
Laudanski P, Charkiewicz R, Tolwinska A, Szamatowicz J, Charkiewicz A, Niklinski J. Profiling of selected microRNAs in proliferative eutopic endometrium of women with ovarian endometriosis. Biomed Res Int. 2015;2015:760698. | ||
Mandai M, Yamaguchi K, Matsumura N, Baba T, Konishi I. Ovarian cancer in endometriosis: molecular biology, pathology, and clinical management. Int J Clin Oncol. 2009;14(5):383–391. | ||
Pavone ME, Lyttle BM. Endometriosis and ovarian cancer: links, risks, and challenges faced. Int J Womens Health. 2015;7:663–672. | ||
Buis CC, van Leeuwen FE, Mooij TM, Burger CW; OMEGA Project Group. Increased risk for ovarian cancer and borderline ovarian tumours in subfertile women with endometriosis. Hum Reprod. 2013;28(12):3358–3369. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Manchanda R, Halaska MJ, Piek JM, et al. The need for more workshops in laparoscopic surgery and surgical anatomy for European gynaecological oncology trainees: a survey by the European Network of Young Gynaecological Oncologists. Int J Gynecol Cancer. 2013;23(6):1127–1132. | ||
Vlad C, Kubelac P, Alexandru I, Achimas-Cadariu P. The role of primary debulking in advanced ovarian cancer patients. J BUON. 2016;21(5):1320. | ||
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. | ||
Buys SS, Partridge E, Black A, et al. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA. 2011;305(22):2295–2303. | ||
Irimie A, Achimas-Cadariu P, Burz C, Puscas E. Multiple primary malignancies – epidemiological analysis at a single tertiary institution. J Gastrointestin Liver Dis. 2010;19(1):69–73. | ||
Kinose Y, Sawada K, Nakamura K, Kimura T. The role of microRNAs in ovarian cancer. Biomed Res Int. 2014;2014:249393. | ||
Pavone ME, Lyttle BM. Endometriosis and ovarian cancer: links, risks, and challenges faced. Int J Womens Health. 2015;7:663–672. | ||
Kondi-Pafiti A, Papakonstantinou E, Iavazzo C, Grigoriadis C, Salakos N, Gregoriou O. Clinicopathological characteristics of ovarian carcinomas associated with endometriosis. Arch Gynecol Obstet. 2012;285(2):479–483. | ||
Suryawanshi S, Vlad AM, Lin H-M, et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res. 2013;19(5):1213–1224. | ||
Mahdian-Shakib A, Dorostkar R, Tat M, Hashemzadeh MS, Saidi N. Differential role of microRNAs in prognosis, diagnosis, and therapy of ovarian cancer. Biomed Pharmacother. 2016;84:592–600. | ||
Braicu C, Calin GA, Berindan-Neagoe I. MicroRNAs and cancer therapy – from bystanders to major players. Current Med Chem. 2013;20(29):3561–3573. | ||
Muresan M, Zaharie F, Bojan A, et al. MicroRNAs in liver malignancies. Basic science applied in surgery. J BUON. 2015;20(2):361–375. | ||
Braicu C, Cojocneanu-Petric R, Chira S, et al. Clinical and pathological implications of miRNA in bladder cancer. Int J Nanomedicine. 2015;10:791–800. | ||
Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610. | ||
Chen X, Lu P, Wang DD, et al. The role of miRNAs in drug resistance and prognosis of breast cancer formalin-fixed paraffin-embedded tissues. Gene. 2016;595(2):221–226. | ||
Irimie AI, Braicu C, Cojocneanu-Petric R, Berindan-Neagoe I, Campian RS. Novel technologies for oral squamous carcinoma biomarkers in diagnostics and prognostics. Acta Odontol Scand. 2015;73(3):161–168. | ||
Berindan-Neagoe I, Calin GA. Molecular pathways: microRNAs, cancer cells, and microenvironment. Clin Cancer Res. 2014;20(24):6247–6253. | ||
Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64(5):311–336. | ||
Braicu C, Catana C, Calin GA, Berindan-Neagoe I. NCRNA combined therapy as future treatment option for cancer. Curr Pharm Des. 2014;20(42):6565–6574. | ||
Braicu C, Tomuleasa C, Monroig P, Cucuianu A, Berindan-Neagoe I, Calin GA. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ. 2015;22(1):34–45. | ||
Berindan-Neagoe I, Chiorean R, Braicu C, et al. Quantitative mRNA expression of genes involved in angiogenesis, coagulation and inflammation in multiforme glioblastoma tumoral tissue versus peritumoral brain tissue: lack of correlation with clinical data. Eur Cytokine Netw. 2012;23(2):45–55. | ||
Li H, Huang W, Luo R. The microRNA-325 inhibits hepatocellular carcinoma progression by targeting high mobility group box 1. Diagn Pathol. 2015;10:117. | ||
Yao S, Zhao T, Jin H. Expression of MicroRNA-325-3p and its potential functions by targeting HMGB1 in non-small cell lung cancer. Biomed Pharmacother. 2015;70:72–79. | ||
Banno K, Yanokura M, Iida M, et al. Application of microRNA in diagnosis and treatment of ovarian cancer. Biomed Res Int. 2014;2014:232817. | ||
Wang Y, Dong Q, Gu Y, Groome LJ. Up-regulation of miR-203 expression induces endothelial inflammatory response: potential role in preeclampsia. Am J Reprod Immunol. 2016;76(6):482–490. | ||
Lee H, Choi HJ, Kang CS, Lee HJ, Lee WS, Park CS. Expression of miRNAs and PTEN in endometrial specimens ranging from histologically normal to hyperplasia and endometrial adenocarcinoma. Mod Pathol. 2012;25(11):1508–1515. | ||
Cao Q, Lu K, Dai S, Hu Y, Fan W. Clinicopathological and prognostic implications of the miR-200 family in patients with epithelial ovarian cancer. Int J Clin Exp Pathol. 2014;7(5):2392–2401. | ||
Hu X, Macdonald DM, Huettner PC, et al. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114(3):457–464. | ||
Chen Y, Zhang L, Hao Q. Candidate microRNA biomarkers in human epithelial ovarian cancer: systematic review profiling studies and experimental validation. Cancer Cell Int. 2013;13(1):86. | ||
Zuberi M, Mir R, Das J, et al. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin Transl Oncol. 2015;17(10):779–787. | ||
van Jaarsveld MT, Helleman J, Boersma AW, et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene. 2013;32(36):4284–4293. | ||
Yang RQ, Teng H, Xu XH, et al. Microarray analysis of microRNA deregulation and angiogenesis-related proteins in endometriosis. Genet Mol Res. 2016;15(2). | ||
Rekker K, Saare M, Roost AM, et al. Circulating miR-200-family micro-RNAs have altered plasma levels in patients with endometriosis and vary with blood collection time. Fertil Steril. 2015;104(4):938–946.e932. | ||
Li B-A. A novel tumor suppressor miRNA miR-520e contributes to suppression of hepatoma. Acta Pharmacol Sin. 2012;33(1):3–4. | ||
Yi M, Li M, Long X, et al. miR-520e regulates cell proliferation, apoptosis and migration in breast cancer. Oncol Lett. 2016;12(5):3543–3548. | ||
Zhang S, Shan C, Kong G, Du Y, Ye L, Zhang X. MicroRNA-520e suppresses growth of hepatoma cells by targeting the NF-[kappa]B-inducing kinase (NIK). Oncogene. 2012;31(31):3607–3620. | ||
Shen F, Cai WS, Feng Z, et al. MiR-492 contributes to cell proliferation and cell cycle of human breast cancer cells by suppressing SOX7 expression. Tumour Biol. 2015;36(3):1913–1921. | ||
Jiang Y, Han Y, Sun C, et al. Rab23 is overexpressed in human bladder cancer and promotes cancer cell proliferation and invasion. Tumour Biol. 2016;37(6):8131–8138. | ||
von Frowein J, Pagel P, Kappler R, von Schweinitz D, Roscher A, Schmid I. MicroRNA-492 is processed from the keratin 19 gene and up-regulated in metastatic hepatoblastoma. Hepatology. 2011;53(3):833–842. | ||
Marzec-Kotarska B, Cybulski M, Kotarski JC, et al. Molecular bases of aberrant miR-182 expression in ovarian cancer. Genes Chromosomes Cancer. 2016;55(11):877–889. | ||
Liu Z, Liu J, Segura MF, et al. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J Pathol. 2012;228(2):204–215. | ||
Meng X, Joosse SA, Muller V, et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer. 2015;113(9):1358–1366. | ||
Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. | ||
Zaman MS, Maher DM, Khan S, Jaggi M, Chauhan SC. Current status and implications of microRNAs in ovarian cancer diagnosis and therapy. J Ovarian Res. 2012;5(1):44. | ||
Oh JJ, Grosshans DR, Wong SG, Slamon DJ. Identification of differentially expressed genes associated with HER-2/neu overexpression in human breast cancer cells. Nucleic Acids Res. 1999;27(20):4008–4017. | ||
Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. | ||
Slack FJ, Weidhaas JB. MicroRNA in cancer prognosis. N Engl J Med. 2008;359(25):2720–2722. | ||
Yang N, Kaur S, Volinia S, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68(24):10307–10314. | ||
Aoyagi Y, Nasu K, Kai K, et al. Decidualization differentially regulates microRNA expression in eutopic and ectopic endometrial stromal cells. Reprod Sci. 2017;24(3):445–455. | ||
Zhou J, Gong G, Tan H, et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. 2015;33(6):2915–2923. | ||
Wang T, Li F, Tang S. MiR-30a upregulates BCL2A1, IER3 and cyclin D2 expression by targeting FOXL2. Oncol Lett. 2015;9(2):967–971. | ||
Liu J, Wu X, Liu H, et al. Expression of microRNA-30a-5p in drug-resistant and drug-sensitive ovarian cancer cell lines. Oncol Lett. 2016;12(3):2065–2070. | ||
Zhu X, Li Y, Xie C, et al. miR-145 sensitizes ovarian cancer cells to paclitaxel by targeting Sp1 and Cdk6. Int J Cancer. 2014;135(6):1286–1296. | ||
Chen X, Dong C, Law PT, et al. MicroRNA-145 targets TRIM2 and exerts tumor-suppressing functions in epithelial ovarian cancer. Gynecol Oncol. 2015;139(3):513–519. | ||
Adammek M, Greve B, Kassens N, et al. MicroRNA miR-145 inhibits proliferation, invasiveness, and stem cell phenotype of an in vitro endometriosis model by targeting multiple cytoskeletal elements and pluripotency factors. Fertil Steril. 2013;99(5):1346–1355.e1345. | ||
Zhou J, Liu H, Chen Y, Wen J, Li L, Wu X. [Expression and significance of VEGF, miR-205 and target protein Ezrin and Lamin A/C in ovarian cancer]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2014;39(2):142–150. Chinese. | ||
Li J, Li L, Li Z, et al. The role of miR-205 in the VEGF-mediated promotion of human ovarian cancer cell invasion. Gynecol Oncol. 2015;137(1):125–133. | ||
Niu K, Shen W, Zhang Y, Zhao Y, Lu Y. MiR-205 promotes motility of ovarian cancer cells via targeting ZEB1. Gene. 2015;574(2):330–336. | ||
Li J, Hu K, Gong G, et al. Upregulation of MiR-205 transcriptionally suppresses SMAD4 and PTEN and contributes to human ovarian cancer progression. Sci Rep. 2017;7:41330. | ||
Xu L, Xiang J, Shen J, et al. Oncogenic MicroRNA-27a is a target for genistein in ovarian cancer cells. Anticancer Agents Med Chem. 2013;13(7):1126–1132. | ||
Li Z, Hu S, Wang J, et al. MiR-27a modulates MDR1/P-glycoprotein expression by targeting HIPK2 in human ovarian cancer cells. Gynecol Oncol. 2010;119(1):125–130. | ||
Onisim A, Achimas-Cadariu A, Vlad C, Kubelac P, Achimas-Cadariu P. Current insights into the association of Nestin with tumor angiogenesis. J BUON. 2015;20(3):699–706. | ||
Gherman C, Braicu OL, Zanoaga O, et al. Caffeic acid phenethyl ester activates pro-apoptotic and epithelial-mesenchymal transition-related genes in ovarian cancer cells A2780 and A2780cis. Mol Cell Biochem. 2016;413(1–2):189–198. |
Supplementary materials
  | Table S1 Patient characteristics for PCR-array and qRT-PCR patient cohort |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


