Back to Journals » Cancer Management and Research » Volume 13
miRNA-382-5p Suppresses the Expression of Farnesoid X Receptor to Promote Progression of Liver Cancer
Authors Nie X , Liu H, Wei X, Li L, Lan L, Fan L, Ma H, Liu L, Zhou Y, Hou R, Chen WD
Received 9 June 2021
Accepted for publication 11 October 2021
Published 22 October 2021 Volume 2021:13 Pages 8025—8035
DOI https://doi.org/10.2147/CMAR.S324072
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Harikrishna Nakshatri
Xiaobo Nie,1 Huiyang Liu,1 Xiaoyun Wei,1 Lanqing Li,1 Linhua Lan,2 Lili Fan,1 Han Ma,1 Lei Liu,1 Yun Zhou,1 Ruifang Hou,1 Wei-Dong Chen1,3
1Key Laboratory of Receptors-Mediated Gene Regulation and Drug Discovery, School of Basic Medical Sciences, People’s Hospital of Hebi, Henan University, Kaifeng, People’s Republic of China; 2Key Laboratory of Diagnosis and Treatment of Severe Hepato-Pancreatic Diseases of Zhejiang Province, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, People’s Republic of China; 3Key Laboratory of Receptors-Mediated Gene Regulation and Drug Discovery, Key Laboratory of Molecular Pathology, School of Basic Medical Science, Inner Mongolia Medical University, Hohhot, People’s Republic of China
Correspondence: Wei-Dong Chen Email [email protected]
Background: The dysregulation of microRNAs (miRNAs) and hepatotoxicity due to the aberrant accumulation of bile acids (BAs) are notorious causes that predispose an individual to the development of hepatocellular carcinoma (HCC). Farnesoid X receptor (FXR), encoded by NR1H4 gene, has been identified as a crucial BA receptor to maintain the homeostasis of BA pool and its expression is decreased in HCC. miR-382-5p plays an important role in the pathogenesis of many human malignancies and was reported to promote the proliferation and differentiation of normal liver cells and liver regeneration. However, there is still some controversy about its role in HCC microenvironment. This study aims to explore the expression pattern of miR-382-5p in HCC and its role in regulating FXR during the development of HCC.
Methods: Tissues collected from 30 HCC patients were subjected to extraction of total RNA and quantitative real-time PCR (qRT-PCR) for the analyses of miR-382-5p expression and NR1H4 mRNA levels, and their expressions were verified by analyzing the online HCC-related GSE datasets. The role of miR-382-5p in regulating cellular proliferation and expression of FXR in different HCC cell lines was analyzed by qRT-PCR, Western Blot, real-time cellular analysis (RTCA) and luciferase reporter assays. The role of miR-382-5p in regulating downstream genes of FXR in HCC cells was also analyzed.
Results: miR-382-5p was upregulated in HCC tissues and inversely associated with the downregulation of NR1H4 mRNA levels. The luciferase reporter assay proved that miR-382-5p directly targeted the 3ʹ-untranslated region (3ʹ-UTR) of human NR1H4 mRNA. Overexpression of miR-382-5p led to a malignant proliferation of HCC cells by suppressing the expression of FXR. In contrast, blocking the endogenous miR-382-5p was sufficient to suppress the cellular proliferation rate of HCC through increasing FXR expression. Additionally, miR-382-5p inhibited the expression of some target genes of FXR, including SHP, FGF19 and SLC51A, and this inhibitory effect was FXR-dependent.
Conclusion: Therefore, miR-382-5p promotes the progression of HCC in vitro by suppressing FXR and could serve as a valuable therapeutic target for HCC treatment.
Keywords: hepatocellular carcinoma, miRNA-382-5p, FXR, proliferation
Introduction
HCC is the sixth most prevalent malignancy worldwide and represents the second leading cause of cancer-associated deaths. Over 850,000 new cases are diagnosed and 810,000 cancer deaths occur annually after 2015.1 Major risk factors of HCC include chronic viral hepatitis infection (hepatitis B or C), excessive alcohol or tobacco intake, non-alcoholic steatohepatitis and long-term aflatoxin exposure.2,3 The routine use of serum biomarkers testing and imaging scan are helpful for the early diagnosis of HCC, and measures such as limiting alcohol use, hepatitis B vaccination and body weight control could efficiently reduce the incidence of HCC,4 whereas there is no way of preventing its progression and the outlook for HCC is still very poor. Most HCC patients are often diagnosed at advanced stages or have metastasis without noticeable symptoms, and the survival rate is even down to 3% once the cancer metastasizes. Unfortunately, the pathogenic mechanism of HCC remains ambiguous, which is urgent to be elucidated.
Farnesoid X receptor (FXR), encoded by NR1H4 gene and expressing highly in the liver and intestine, belongs to the nuclear receptor family and takes part in regulating BA homeostasis, glucose and lipid metabolism, liver regeneration, cholestasis and hepatic fibrosis.5–7 Chronic hepatic injury due to the excessive accumulation of BAs is considered a major culprit for the pathogenesis of HCC.8 Previously, it was reported that mice would develop liver tumors spontaneously in the absence of FXR,9 and whole-body loss of NR1H4 gene led to Myc and cyclin-dependent kinase 4 (Cdk4) inductions and BA elevation, eventually resulting in age-dependent hepatic tumorigenesis.10 Conversely, FXR can prevent BA-induced cytotoxicity and plays a protective role in hepatocarcinogenesis by transcriptionally regulating the expression of genes involved in BA metabolism, hepatic inflammation and tumorigenesis.11–13 However, FXR was found to decrease significantly in HCC tissues and closely related to clinicopathological characteristics.14 At present, the detailed mechanism by which FXR is inhibited during hepatocarcinogenesis remains undetermined.
MicroRNAs (miRNAs) are a family of short (about 18–25 nucleotides) noncoding RNAs that suppress the expression of target genes by preferentially targeting their 3ʹ-UTR, leading to the translational repression or mRNA degradation.15 miRNAs have been found to participate in the pathogenesis of HCC and exert an oncogenic or antineoplastic role through inhibiting the translation of target genes involved in cellular proliferation, differentiation, metastasis and apoptosis.16,17 However, miRNAs associated with the maintenance of BA homeostasis and modulating HCC arising therefrom have been rarely reported.18 Recently, a study showed that miR-192 was inversely correlated with FXR protein level in colonic adenocarcinoma tissues and functions as an oncogenic gene by directly targeting the 3ʹ-UTR of NR1H4 mRNA in human hepatoma cells and colon adenocarcinoma cells.19 Similarly, Zhang et al found that silencing of endogenous miR-421 suppressed the proliferative and invasive capacities of HCC cells through the disinhibition on the expression of FXR.20 miR-382-5p is another miRNA that has a putative binding site of human NR1H4 mRNA based on our preliminary analysis using the miRANDA database and was reported to promote the proliferation and differentiation of normal liver cells and liver regeneration through different mechanisms,21,22 but whether it has a similar effect and regulates the expression of FXR in HCC microenvironment is not known.
Here, we showed that miR-382-5p was aberrantly increased and contrary to the downregulation of FXR in HCC samples. Remarkably, FXR was identified as a direct target of miR-382-5p. Overexpression of miR-382-5p accelerated the proliferation of HCC cells by suppressing the expression of FXR and downstream target genes. Conversely, inhibiting the endogenous miR-382-5p had opposite effects. These data suggest that miR-382-5p serves as a potential molecular target for the development of novel therapies for HCC.
Patients and Methods
Collection of Hepatic Tissue Samples
Human hepatic samples were collected during surgery on patients with hepatic cancer, who underwent hepatectomy at the Department of General Surgery, Affiliated Huaihe Hospital of Henan University in 2018. Patients were selected based on the pathological diagnosis of hepatic cancer with no other malignancies or histories of preoperative anticancer treatments. All tissues were immediately snap-frozen and stored in liquid nitrogen until subsequent analysis. The study complied with the Declaration of Helsinki and was approved by the Biomedical Research Ethics Committee of Henan University, Kaifeng, and written informed consent was obtained from all patients.
Cell Culture and Transient Transfection with miR-382-5p
HCC cell lines (HepG2 and Huh-7) used in this study were purchased from School of Basic Medicine of Peking Union Medical College (Beijing, China). Cells were cultured in Dulbecco’s Modified Eagle’s Medium (Thermo, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo, USA) and 1% antibiotics (100 U/mL penicillin and 0.1 mg/mL streptomycin) (Gibco, USA) at 37 °C in a humidified atmosphere with 5% CO2.
To determine the effect of miR-382-5p on the mRNA and protein expression levels of NR1H4, HepG2 or Huh-7 cells were seeded into a 6-well plate at a density of 3×105 cells per well 18 hours prior to transfection. Cells were transfected with cholesterol-modified agonist (agomir, Ago) or inhibitor (antagomir, ANT) of hsa-miR-382-5p (RiboBio, China) or their corresponding negative controls at a final concentration of 100 nM (agomir), 150 nM or 200 nM (antagomir) using 5 μL of Lipofectamine 2000 (Invitrogen, USA) in serum- and antibiotic-free medium. The medium was replaced with fresh complete growth medium 6 h later and incubated for another 12 h. Cells were then treated with GW4064 (Sigma-Aldrich, USA) at the final concentration of 2 μM in serum-free medium for 24 h till the harvest.
To further determine the effect of miR-382-5p on the expression of FXR target genes on the basis of FXR overexpression, HepG2 or Huh-7 cells cultured in 12-well plate (1.5 × 105 cells/well) were co-transfected with FXR overexpression plasmid (700 ng/well, NC-OE vector as control) (GenePharma, China) and miR-382-5p-Ago (100 nM, NC-Ago as control) or miR-382-5p-ANT (150 nM, NC-ANT as control) using 3 μL of Lipofectamine 2000. The medium was replaced with fresh complete growth medium 6 h later, and cells were cultured for another 12 h, followed by the treatment of 2 μM GW4064 in serum-free medium for 24 h till the harvest.
RNA Extraction and qRT-PCR for Mature miRNA
miRNAs were isolated from cells or tissue samples using a miRNA fast purification kit (BioTeke Corporation, China) according to the instruction. miRNAs were conjugated with Poly (A) tails and reverse transcribed to cDNA by using miRNA-First Strand cDNA Synthesis kit (BioTeke Corporation, China). The qRT-PCR analyses were performed by using miDETECT A Track™ miRNA qPCR Kit with SYBR green (RiboBio, China). In brief, mature miR-382-5p (25ng of cDNA/reaction) was amplified with the forward primer (5ʹ-CCGGAAGTTGTTCGTGGTGGATTC-3ʹ) and universal reverse qRT-PCR primer according to the following conditions: denaturation at 95 °C for 3 min, 40 cycles of denaturation at 95 °C for 30 seconds and annealing and extension at 62 °C for 40 seconds. Melting curve analysis (65–95 °C) was routinely performed to verify the specificity of primers at the end of the assay. The relative levels of mature miR-382-5p were normalized with human U6 small nuclear RNA (snRNA) by using the comparative ΔΔCt method, and relative fold change of miRNA was calculated by the equation 2−ΔΔCt. The forward primer for U6 snRNA in the qRT-PCR was also supplemented in the kit.
qRT-PCR Detection for mRNA
Total RNA was isolated from cells or tissue samples using the microRNA fast purification kit according to the instruction, then 2 μg of RNA was reverse transcribed to cDNA by using the Strand cDNA Synthesis kit (Thermo, USA). The qRT-PCR analyses were performed by using 50ng of cDNA per reaction with SYBR green primer sets designed to amplify target genes according to the above described conditions. The relative mRNA levels of target genes were normalized against human β-actin by using the comparative ΔΔCt method23. Sequences of primers used for real-time PCR are listed in Table 1.
 |
Table 1 The Sequences of Primers Used in qRT-PCR |
Protein Extraction and Immunoblot
Whole-cell extracts from cells were lysed in RIPA buffer consisting 1% NP-40, 0.5% deoxycholate, 0.1% SDS (Beyotime, China) in the presence of Protease Inhibitor Cocktail (Roche, Switzerland) and centrifuged at 15,000 × g for 15 min at 4 °C. The protein concentrations of cell lysates were determined by bicinchoninic acid (BCA) assay (Beyotime, China). Equal amounts of protein extract (30 μg/well) were resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred onto a 0.2 μm polyvinylidene fluoride (PVDF) membrane followed by block with 5% non-fat milk in 1×TBST for 1 h. The blot was probed with specific primary antibodies against FXR (1:1000) or β-actin (1:1000) (Omnimabs, USA) overnight at 4 °C and then incubated with corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (Proteintech, China) at room temperature for 1 h. The blot was then incubated with SuperSignal West Pico Chemiluminescent Substrates (Thermo, USA) and developed under Automatic Multifunction Chemiluminescent Detection System (Tanon, China). The signal was quantified by densitometry by using ImageJ Software v 1.8.0.
Plasmid Construction and Luciferase Reporter Assays
The DNA fragment corresponding to 3ʹ-UTR (1873–2284 nt) of human NR1H4 mRNA containing the potential miR-382-5p recognition site (FXR-pmiGLO) was amplified by PCR with specific primers and cloned into pmiGLO dual-luciferase vector (Promega, USA) at Sac I and Xbal I sites, the recombinant plasmid was named as FXR-pmiGLO after confirmed by sequencing.
HepG2 or Huh-7 cells were grown to 60% confluence in 12-well plate. Cells were transiently co-transfected with FXR-pmiGLO plasmid (400 ng/well) and NC-agomir, miR-382-5p-agomir, miR-382-5p-ANT or miR-149-5p-Ago at a final concentration of 100 nM using 3 μL of Lipofectamine 2000. The medium was replaced with fresh complete growth medium 6 h later, and cells were cultured for another 24 h, followed by the measure of activities of firefly luciferase and Renilla luciferase using the Dual-Luciferase® Reporter Assay System (Promega, USA) according to the manufacturer’s instruction. In brief, cells in each well were lysed with 250 μL of passive lysis buffer, then 20 μL of cell lysate was transferred into the luminometer tube containing 100 μL of LAR II to measure the firefly luciferase activity, 100 μL of Stop & Glo® Reagent was then delivered into the tube to measure the Renilla luciferase activity. The relative luciferase activity of each well was calculated by the ratio of firefly luciferase activity to Renilla luciferase activity. Normalized firefly luciferase activity for each group was compared to that of the NC-Ago control group. Three independent luciferase reporter assays were carried out in quadruplicate wells for each group.
Cell Proliferation Assay
Cell proliferation was determined by using Real Time Cellular Analysis (RTCA, ACEA Biosciences, USA). In brief, HepG2 and Huh-7 cells were transfected with miR-382-5p-Ago (100 nM), miR-382-5p-ANT (150 nM for HepG2 cells, 200 nM for Huh-7 cells) or corresponding control and cultured for another 12 h, followed by seeding into E-plate 16 (1 × 104 cells/well). The cellular proliferation index was recorded every 15 minutes to 48 hours or 72 hours by using RTCA placed in a cell incubator. The cellular growth index was generated from the average values in triplicate wells for each group.
Statistical Analysis
Statistical analysis and figures were carried out using the GraphPad Prism 8.0. Unpaired two-sided Student’s t-test was performed to evaluate statistical significance of the difference between two groups, and unpaired two-sided Student’s t-test with Welch’s correction was used for the data with abnormal distribution. Particularly, paired two-sided Student’s t-test was performed to compare miR-382-5p levels and NR1H4 mRNA levels in hepatic cancer tissues and paired normal tissues. Error bar for the experiments represents the standard deviation of the mean value (mean value ± S.D.) from at least three independent experiments. P values lower than 0.05 were considered as statistically significant. One asterisk, two asterisks, and three asterisks represent P<0.05, P<0.01, and P<0.001, respectively.
Results
MiR-382-5p Was Increased and Opposite to NR1H4 mRNA Level in HCC Tissues
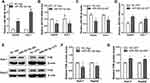
To determine the role of miR-382-5p in HCC microenvironment, we examined the miR-382-5p expression level and NR1H4 mRNA level by qRT-PCR analyses in 30 pairs of HCC and paired non-neoplastic hepatic samples, the latter of which are from normal hepatic tissues adjacent to cancer. The expression level of miR-382-5p was upregulated in HCC tissues compared with corresponding normal ones (Figure 1A), whereas the NR1H4 mRNA level was obviously downregulated in HCC tissues (Figure 1B). To explore the relationship between HCC progression and miR-382-5p or NR1H4 mRNA levels, HCC tissues were divided into two groups according to the clinical pathological stage. As shown in Figure 1C and D, the expression level of miR-382-5p was gradually elevated from normal to early HCC (stage I/II) and then to advance HCC (stage III/IV) tissues. Conversely, the NR1H4 mRNA level showed an opposite changing trend. However, due to the limited number of fresh samples, we only found a negative correlation without statistical significance (R=−0.37, P=0.123) between the levels of miR-382-5p expression and NR1H4 mRNA in all pairs of HCC tissues (data not shown). Through the GEO datasets analysis, upregulation of miR-382-5p was also found in HCC tissues in GSE12717 cohort (n=6) (Figure 2A), and NR1H4 mRNA level was found to be decreased in HCC tissues in GSE57957 cohort (Figure 2B), and gradually decreased from normal livers to early HCC (eHCC) and then to advanced HCC (aHCC) in GSE54238 cohort (Figure 2C). Taken together, these data suggested that downregulation of NR1H4 transcript level might be associated with the upregulation of miR-382-5p level in HCC tissues.
MiR-382-5p Promoted the Proliferation of HCC Cells
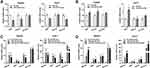
To investigate whether miR-382-5p could affect the proliferation of HCC cells, HepG2 or Huh-7 cells were transiently transfected with miR-382-5p-Ago or NC-Ago. The results from the RTCA assay showed that overexpression of miR-382-5p could accelerate the proliferation of HCC cell lines compared with NC-Ago (Figure 3A). We further examined the effect of silencing miR-382-5p on cellular proliferation rate. As expected, blocking the endogenous mature miR-382-5p with its antisense antagomir inhibited the proliferation rates of both HCC cell lines (Figure 3B).
MiR-382-5p Downregulated the Transcription and Translation of NR1H4 Gene in HCC Cells
To understand the effect of miR-382-5p on the transcription and translation of NR1H4 gene, HepG2 or Huh-7 cells were transfected with 100 nM miR-382-5p-Ago or NC-Ago for 36 h, separately. The overexpression of miR-382-5p was verified through miRNA qRT-PCR analysis (Figure 4A). As shown in Figure 4B, the endogenous NR1H4 mRNA levels were suppressed obviously in both HCC cell lines. To assess whether a competitive combination of endogenous miR-382-5p with its antagomir could reverse the inhibitory effect on NR1H4 mRNA, HepG2 or Huh-7 cells were then transfected with 150 nM miR-382-5p-ANT or NC-ANT for 36 h. As expected, NR1H4 mRNA levels in both cell lines were upregulated significantly when the endogenous miR-382-5p was suppressed by its antagomir (Figure 4C and D). The protein levels of FXR had similar changes when overexpressing or silencing miR-382-5p in both HCC cell lines (Figure 4E and G). These data clearly showed that miR-382-5p inhibited the transcription and translation of NR1H4 gene in HCC cells.
FXR Was Confirmed as a Direct Target of MiR-382-5p in HCC Cells
The bioinformatic tool miRANDA predicates a putative recognition element for the seed sequence of hsa-miR-382-5p in the 3ʹ-UTR of NR1H4 mRNA, and the potential binding site is located on +1969 to +1990 in the 3ʹ-UTR of NR1H4 mRNA (Figure 5A). To elucidate whether miR-382-5p could interfere the translation process of FXR protein by targeting the 3ʹ-UTR of NR1H4 mRNA, we successfully constructed the recombinant FXR-pmiGLO plasmid and co-transfected it with miR-382-5p-Ago, the luciferase assays showed that overexpression of miR-382-5p decreased the luciferase activity of FXR-pmiGLO plasmid. On the contrary, its activity was not affected when co-transfected with control miRNA, miR-382-5p-ANT or another miRNA with no binding site of 3ʹ-UTR of NR1H4 mRNA (Figure 5B). These results demonstrated FXR was a direct target of miR-382-5p in HCC cells.
MiR-382-5p Affected the Expression of FXR-Target Genes in HCC Cells
In the process of FXR-dependent BA metabolism, small heterodimer partner (SHP), fibroblast growth factor 19 (FGF19) and organic solute transporters alpha (OSTα, encoded by SLC51A gene) are typical target genes of FXR and their expression are upregulated when FXR is activated. The response of these target genes to miR-382-5p agomir or antagomir in HepG2 and Huh-7 cells was then tested using qRT-PCR. The results showed the mRNA level of SHP was inhibited in both cells lines when overexpressing miR-382-5p (Figure 6A), and an opposite changing trend was found when blocking the endogenous mature miR-382-5p (Figure 6B). However, possibly due to the difference in cytogenetic background, not all target genes had response to miR-382-5p agomir or antagomir in these two cell lines, we found a similar changing trend for FGF19 mRNA level in HepG2 cells and for SLC51A mRNA level in Huh-7 cells. To further explore the effect of miR-382-5p on the expression of these genes on the basis of FXR overexpression, HCC cell lines were co-transfected with FXR overexpressing plasmid and miR-382-5p-Ago or miR-382-5p-ANT. The results showed that overexpression of FXR in HCC cells resulted in an obvious increase of NR1H4, SHP, FGF19 and SLC51A mRNA levels, and this induction effect could be blocked when exogenous FXR was inhibited by the treatment of miR-382-5p agomir (Figure 6C). Conversely, the increase in the mRNA levels of these genes was further enhanced when endogenous FXR was disinhibited by the treatment of miR-382-5p antagomir (Figure 6D). Collectively, these results indicate that miR-382-5p can inhibit the expression and function of FXR in HCC cells.
Discussion
Abundant evidences have reported that the aberrant expression of miRNAs is closely related to the generation and progression of human malignancies by playing an oncogenic or tumor-suppressing role. Thus far, the role of miR-382-5p in the progression of human malignancies has been documented, whereas its expression pattern and function showed some discrepancies in human cancers. In particular, miR-382-5p was detected to be decreased in cancer tissues of leukemia, osteosarcoma, colorectal, esophageal, gastric, lung and pancreatic cancers,24–30 and overexpression of miR-382-5p could effectively inhibit the proliferative and invasive capacities of cancer cells and the tumor growth in nude mice. Furthermore, overexpression of miR-382 increases the sensitivity of HCC cells to γδ T lymphocytes, one subgroup of T lymphocytes that display potent anti-cancer activity.31 On the contrary, miR-382-5p was proved to be an onco-miRNA in infantile hemangioma, glioma, pancreatic and breast cancers,32–35 and acts as an angiogenic miRNA by repressing PTEN.36 Additionally, circ_0008285 could inhibit the proliferation and migration of colorectal cancer cells by sponging miR-382-5p to elevate the expression of PTEN.35
miR-382-5p was reported to promote liver regeneration in mice and cell proliferation in mouse and human normal liver cell lines via targeting PTEN-AKT axis.21 Similarly, miR-382-5p promoted the differentiation of rat liver progenitor cells into hepatocytes by targeting Ezh2,22 demonstrating a cellular proliferation-promoting role in liver. However, the expression pattern and function of miR-382-5p in HCC microenvironment is still obscure. A recent study found the expression of miR-382-5p was downregulated in HCC cell lines compared to immortalized normal liver L02 cells, and overexpression of miR-382 inhibited the metastasis of HCC by targeting GOLM1.37 However, another group oppositely reported that miR-382-5p was overexpressed in hepatitis B virus core protein (HBc) positive HCC cells and tissues, and HBc enhanced HCC metastasis through activating miR-382-5p/DLC-1 axis.38 The discrepancy in these data might be attributed to different controls. The immortalized non-tumorigenic cell lines often lose the cell cycle checkpoint pathways,39 therefore, they might be unsuitable as the controls when comparing the expression of proliferation-related genes with cancer tissues. In this study, matched normal liver tissue was used as the control of HCC tissue, and the results showed that miR-382-5p was upregulated in HCC tissues. After transfection with agomir or antagomir of miR-382-5p, cholesterol modified miR-382-5p mimic or inhibitor, which sustains longer time in cells, the proliferate rate of HCC cells was inhibited or accelerated separately, suggesting that miR-382-5p plays a carcinogenic role during HCC progression.
Previous studies revealed hepatocarcinogenesis was inversely associated with the downregulation of FXR, which may be due to the upregulation of some oncogenic miRNAs, such as miR-192 and miR-421.19,40 In this study, we found miR-382-5p was also upregulated and opposite to the expression of FXR in HCC tissues. However, a larger cohort of HCC samples should be collected to determine their negative association. We further proved that overexpression of miR-382-5p decreased the NR1H4 mRNA and protein levels in HCC cells on the basis of FXR agonist treatment. Conversely, blocking the endogenous miR-382-5p with its complementary sequences had the opposite effects. According to the miRANDA database, there is a putative recognition element for the seed sequence of miR-382-5p in the 3ʹ-UTR of NR1H4 mRNA, and the luciferase assays proved a decreased activity of NR1H4-3ʹ-UTR luciferase caused by miR-382-5p, suggesting that miR-382-5p directly suppresses the translation of FXR by binding to the 3ʹ-UTR of NR1H4 mRNA.
FXR provides a negative feedback to the BA synthesis pathway by inducing some downstream target genes, such as SHP, FGF19 and SLC51A. Previously, the level of SHP was found to be decreased significantly in human HCC samples, and overexpression of SHP could partially protect FXR knockout mice against liver tumor development.41,42 Human FGF19 or murine homolog FGF15 has anti-fibrotic and anti-cholestatic activity in the liver but takes part in hepatocarcinogenesis through the FGF15/19-FGFR4 pathway.43 As a primary BA transporter involved in the excretion of BA from the enterocytes, SLC51A was reported to be upregulated and play a potential protective role during cholestasis.44 Therefore, it can be speculated that silencing miR-382-5p may inhibit the progression of HCC by restoring the expression of FXR and downstream target genes. Interestingly, our data are in accordance with this prediction, and conversely, overexpressing miR-382-5p accompanied by FXR agonist can accelerate the proliferation of HCC cells by suppressing the expression of FXR and some target genes to different extent, including SHP, FGF19 and SLC51A. In addition, our data showed that on the basis of FXR overexpression, the inductive effect of exogenous FXR on the expression of these target genes could be blocked obviously by miR-382-5p. On the contrary, silencing the endogenous FXR by miR-382-5p antagomir could further enhance the inductive effect of FXR on the expression of these genes, which strongly suggest the inhibitory effect of miR-382-5p on the expression and function of FXR in HCC cells.
Conclusion
In conclusion, the current study demonstrates that miR-382-5p is upregulated in HCC tissues in accordance with the downregulation of FXR. miR-382-5p promotes the proliferation of HCC cells by inhibiting the expression and function of FXR. Our results highlight miR-382-5p as oncogenic miRNA and a promising therapeutic target to treat HCC.
Acknowledgments
We thank Drs Yan-Dong Wang for helpful advices and discussions.
Funding
There is no funding to report.
Disclosure
The authors have declared that no conflict of interest exists.
References
1. McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of hepatocellular carcinoma. Hepatology. 2021;73(Suppl 1):4–13. doi:10.1002/hep.31288
2. Huang Y, Ge W, Zhou J, et al. The role of tumor associated macrophages in hepatocellular carcinoma. J Cancer. 2021;12(5):1284–1294. doi:10.7150/jca.51346
3. Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi:10.1038/s41575-019-0186-y
4. Erstad DJ, Razavi AA, Li S, et al. Prevention strategies for hepatocellular carcinoma. In: Hoshida Y, editor. Hepatocellular Carcinoma: Translational Precision Medicine Approaches. Cham (CH);2019:255–289.
5. Jung K, Kim M, So J, et al. Farnesoid X receptor activation impairs liver progenitor cell-mediated liver regeneration via the PTEN-PI3K-AKT-mTOR axis in zebrafish. Hepatology. 2020;74(1):397–410.
6. Keitel V, Droge CHaussinger D. Targeting FXR in cholestasis. Handb Exp Pharmacol. 2019;256:299–324.
7. Panzitt K, Wagner M. FXR in liver physiology: multiple faces to regulate liver metabolism. Biochim Biophys Acta Mol Basis Dis. 2021;1867(7):166133. doi:10.1016/j.bbadis.2021.166133
8. Kundu S, Kumar SBajaj S, Bajaj A. Cross-talk between bile acids and gastrointestinal tract for progression and development of cancer and its therapeutic implications. IUBMB Life. 2015;67(7):514–523. doi:10.1002/iub.1399
9. Yang F, Huang X, Yi T, et al. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67(3):863–867. doi:10.1158/0008-5472.CAN-06-1078
10. Takahashi S, Tanaka N, Fukami T, et al. Role of Farnesoid X receptor and bile acids in hepatic tumor development. Hepatol Commun. 2018;2(12):1567–1582. doi:10.1002/hep4.1263
11. Wang YD, Yang F, Chen WD, et al. Farnesoid X receptor protects liver cells from apoptosis induced by serum deprivation in vitro and fasting in vivo. Mol Endocrinol. 2008;22(7):1622–1632. doi:10.1210/me.2007-0527
12. Wang YD, Chen WD, Li C, et al. Farnesoid X receptor antagonizes JNK signaling pathway in liver carcinogenesis by activating SOD3. Mol Endocrinol. 2015;29(2):322–331. doi:10.1210/me.2014-1225
13. Huang XF, Zhao WY, Huang WD. FXR and liver carcinogenesis. Acta Pharmacol Sin. 2015;36(1):37–43. doi:10.1038/aps.2014.117
14. Su H, Ma C, Liu J, et al. Downregulation of nuclear receptor FXR is associated with multiple malignant clinicopathological characteristics in human hepatocellular carcinoma. Am J Physiol Gastrointest Liver Physiol. 2012;303(11):G1245–G1253. doi:10.1152/ajpgi.00439.2011
15. Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi:10.1016/S0092-8674(01)00616-X
16. Nie X, Liu Y, Chen WD, et al. Interplay of miRNAs and canonical Wnt signaling pathway in hepatocellular carcinoma. Front Pharmacol. 2018;9:657. doi:10.3389/fphar.2018.00657
17. Lu X, Yang C, Hu Y, et al. Upregulation of miR-1254 promotes hepatocellular carcinoma cell proliferation, migration, and invasion via inactivation of the Hippo-YAP signaling pathway by decreasing PAX5. J Cancer. 2021;12(3):771–789.
18. Marin JJ, Bujanda L, Banales JM. MicroRNAs and cholestatic liver diseases. Curr Opin Gastroenterol. 2014;30(3):303–309. doi:10.1097/MOG.0000000000000051
19. Krattinger R, Bostrom A, Schioth HB, et al. microRNA-192 suppresses the expression of the farnesoid X receptor. Am J Physiol Gastrointest Liver Physiol. 2016;310(11):G1044–51. doi:10.1152/ajpgi.00297.2015
20. Zhang Y, Gong W, Dai S, et al. Downregulation of human farnesoid X receptor by miR-421 promotes proliferation and migration of hepatocellular carcinoma cells. Mol Cancer Res. 2012;10(4):516–522. doi:10.1158/1541-7786.MCR-11-0473
21. Bei Y, Song Y, Wang F, et al. miR-382 targeting PTEN-Akt axis promotes liver regeneration. Oncotarget. 2016;7(2):1584–1597. doi:10.18632/oncotarget.6444
22. Zheng Y, Zhou J, Li X, et al. Mir-382 promotes differentiation of rat liver progenitor cell WB-F344 by targeting Ezh2. Cell Physiol Biochem. 2018;48(6):2389–2398. doi:10.1159/000492654
23. Nie X, Xia F, Liu Y, et al. Downregulation of Wnt3 suppresses colorectal cancer development through inhibiting cell proliferation and migration. Front Pharmacol. 2019;10:1110. doi:10.3389/fphar.2019.01110
24. Liu D, Zhong L, Yuan Z, et al. miR-382-5p modulates the ATRA-induced differentiation of acute promyelocytic leukemia by targeting tumor suppressor PTEN. Cell Signal. 2018;54:1–9. doi:10.1016/j.cellsig.2018.11.012
25. Xu M, Jin H, Xu CX, et al. miR-382 inhibits osteosarcoma metastasis and relapse by targeting Y box-binding protein 1. Mol Ther. 2015;23(1):89–98. doi:10.1038/mt.2014.197
26. Feng J, Qi B, Guo L, et al. miR-382 functions as a tumor suppressor against esophageal squamous cell carcinoma. World J Gastroenterol. 2017;23(23):4243–4251. doi:10.3748/wjg.v23.i23.4243
27. Wang Y, Bu P, Li F, et al. [Effects of miR-382 on cell migration, invasion and proliferation of gastric cancer cell lines MGC-803]. Zhonghua Yi Xue Za Zhi. 2017;97(8):612–615. Chinese.
28. Chen T, Ren H, Thakur A, et al. miR-382 inhibits tumor progression by targeting SETD8 in non-small cell lung cancer. Biomed Pharmacother. 2017;86:248–253. doi:10.1016/j.biopha.2016.12.007
29. Zhang H, Zhu C, He Z, et al. LncRNA PSMB8-AS1 contributes to pancreatic cancer progression via modulating miR-382-3p/STAT1/PD-L1 axis. J Exp Clin Cancer Res. 2020;39(1):179. doi:10.1186/s13046-020-01687-8
30. Xie L, Pan Z. Circular RNA circ_0000467 regulates colorectal cancer development via miR-382-5p/EN2 axis. Bioengineered. 2021;12(1):886–897. doi:10.1080/21655979.2021.1889130
31. Chen Z, Zheng Z, Feng L, et al. Overexpression of miR-382 sensitizes hepatocellular carcinoma cells to gammadelta T cells by inhibiting the expression of c-FLIP. Mol Ther Oncolytics. 2020;18:467–475. doi:10.1016/j.omto.2020.07.012
32. Li D, Li P, Guo Z, et al. Downregulation of miR-382 by propranolol inhibits the progression of infantile hemangioma via the PTEN-mediated AKT/mTOR pathway. Int J Mol Med. 2017;39(3):757–763. doi:10.3892/ijmm.2017.2863
33. Ma Z. Downregulation of SETD8 by miR-382 is involved in glioma progression. Pathol Res Pract. 2018;214(3):356–360. doi:10.1016/j.prp.2018.01.004
34. Ho JY, Hsu RJ, Liu JM, et al. MicroRNA-382-5p aggravates breast cancer progression by regulating the RERG/Ras/ERK signaling axis. Oncotarget. 2017;8(14):22443–22459. doi:10.18632/oncotarget.12338
35. Wang J, Luo J, Liu G, et al. Circular RNA hsa_circ_0008285 inhibits colorectal cancer cell proliferation and migration via the miR-382-5p/PTEN axis. Biochem Biophys Res Commun. 2020;527(2):503–510. doi:10.1016/j.bbrc.2020.03.165
36. Seok JK, Lee SH, Kim MJ, et al. MicroRNA-382 induced by HIF-1alpha is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 2014;42(12):8062–8072. doi:10.1093/nar/gku515
37. Zhang S, Ge W, Zou G, et al. MiR-382 targets GOLM1 to inhibit metastasis of hepatocellular carcinoma and its down-regulation predicts a poor survival. Am J Cancer Res. 2018;8(1):120–131.
38. Du J, Bai F, Zhao P, et al. Hepatitis B core protein promotes liver cancer metastasis through miR-382-5p/DLC-1 axis. Biochim Biophys Acta Mol Cell Res. 2018;1865(1):1–11. doi:10.1016/j.bbamcr.2017.09.020
39. Maqsood MI, Matin MM, Bahrami AR, et al. Immortality of cell lines: challenges and advantages of establishment. Cell Biol Int. 2013;37(10):1038–1045. doi:10.1002/cbin.10137
40. Zhang Y, Gong W, Dai S, et al. Downregulation of human farnesoid X receptor by miR-421 promotes proliferation and migration of hepatocellular carcinoma cells. Mol Cancer Res. 2012;10(4):516–522. doi:10.1158/1541-7786.MCR-11-0473
41. He N, Park K, Zhang Y, et al. Epigenetic inhibition of nuclear receptor small heterodimer partner is associated with and regulates hepatocellular carcinoma growth. Gastroenterology. 2008;134(3):793–802. doi:10.1053/j.gastro.2008.01.006
42. Li G, Kong B, Zhu Y, et al. Small heterodimer partner overexpression partially protects against liver tumor development in farnesoid X receptor knockout mice. Toxicol Appl Pharmacol. 2013;272(2):299–305. doi:10.1016/j.taap.2013.06.016
43. Alvarez-Sola G, Uriarte I, Latasa MU, et al. Fibroblast growth factor 15/19 in Hepatocarcinogenesis. Dig Dis. 2017;35(3):158–165. doi:10.1159/000450905
44. Soroka CJ, Ballatori N, Boyer JL. Organic solute transporter, OSTalpha-OSTbeta: its role in bile acid transport and cholestasis. Semin Liver Dis. 2010;30(2):178–185. doi:10.1055/s-0030-1253226
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.